Modified cellulase gene and expression vector and application thereof
A cellulase and cellulose technology, applied in the field of genetic engineering, can solve the problems of low utilization rate, low enzyme activity, and increased cost, and achieve the effects of reducing production cost, improving nutritional value, and improving palatability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] In this example, Aspergillus niger and Trichoderma konii were used as strains preserved in the laboratory. Escherichia coli DH5α and cloning plasmid pMD19-T were purchased from TAKARA Company. PCR amplification primers were designed. The complete gene sequences of Aspergillus niger and Trichoderma konei were obtained from NCBI, and primers were designed downstream of the signal peptide sequence in the coding region of the CBHI gene, and an XbaⅠ restriction site was added to the forward primer. A PstI restriction site was added to the reverse primer, and protective bases GC and CCG were added to the 5' end of the reverse primer respectively (these primers were synthesized by Shanghai Sangon Bioengineering Co., Ltd.).
[0038] Trichoderma konshii primers:
[0039] Forward primer K1 is shown in SEQ NO.5: 5'-GC T CTA GA A TGT ATC GGA AGT TGG CCG TC Xba Ⅰ
[0040] The reverse primer K2 is shown in SEQ NO.6: 5'-CCG CTC GAG TTA CAG GCA CTG AGA GTA GTA Pst Ⅰ
[0041] Asp...
Embodiment 2
[0044] Example 2 PCR Amplification of Cellulase Gene
[0045] Trichoderma konshii and Aspergillus niger were inoculated in PDA medium respectively, cultured on a shaker at 30°C at 120rpm for 48 hours, the bacteria were filtered with sterile filter paper and dried, and the RNA extraction kit was used to extract Trichoderma konii and Aspergillus niger of total RNA.
[0046] Among them, the PDA medium is prepared by weighing MgSO 4 0.15g, soluble starch 3g, glucose 10g, KH 2 PO 4 1g, 1g of yeast extract powder, 2.5g of tryptone, dissolved in 400mL of distilled water, stirred thoroughly to dissolve, and distilled to 500mL with distilled water. 121°C, 1.05×10 5 Pa high-pressure steam sterilization for 20min, set aside.
[0047] To establish the cDNA library of the cellulase gene, treat the total RNA with DNase I, react at 37°C for 20-30min, and then inactivate it in a PCR instrument at 80°C for 10min; configure the following system in the microtube according to Table 1.
[...
Embodiment 3
[0060] Example 3 Ligation transformation of cellulase gene
[0061] For the connection of the cellulase gene to the pMD19-vector, configure the connection system of the cellulase gene PCR recovery product according to Table 4, and connect overnight at 4°C.
[0062] Table 4 PCR product and pMD19-T vector ligation reaction system (μL)
[0063]
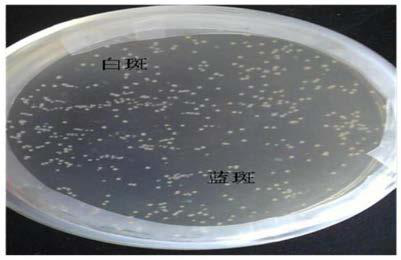
[0064] Place well-grown E. coli plates in a refrigerator at 4°C, and develop color for several hours. Such as figure 2 As shown, the Escherichia coli without the recombinant plasmid has β-galactosidase activity, the center of the colony is light blue, and the periphery is dark blue; while the Escherichia coli containing the recombinant plasmid has no β-galactosidase activity, the colony Milky white for initial screening.
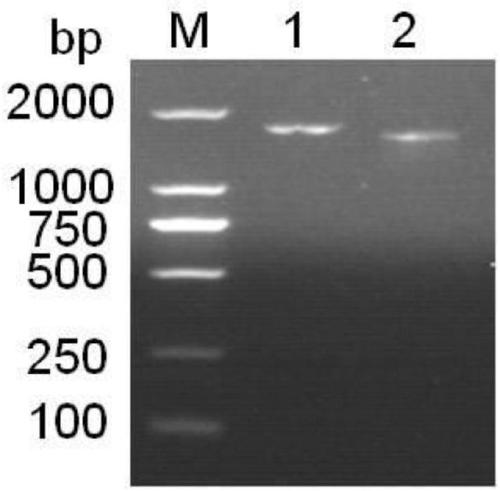
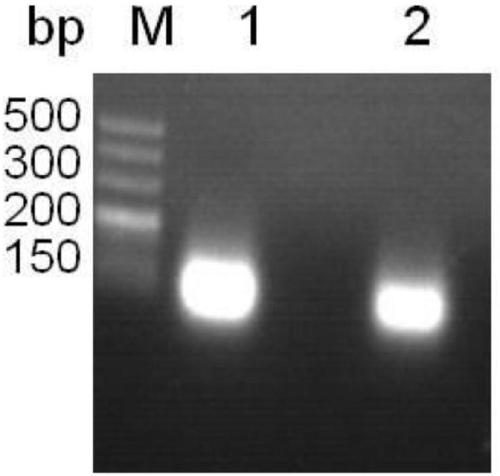
[0065] For gene preparation, perform PCR amplification on the cellulase gene that has been sequenced, and use an agarose purification and recovery kit to recover the PCR product to obtain the cellulase gene.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com