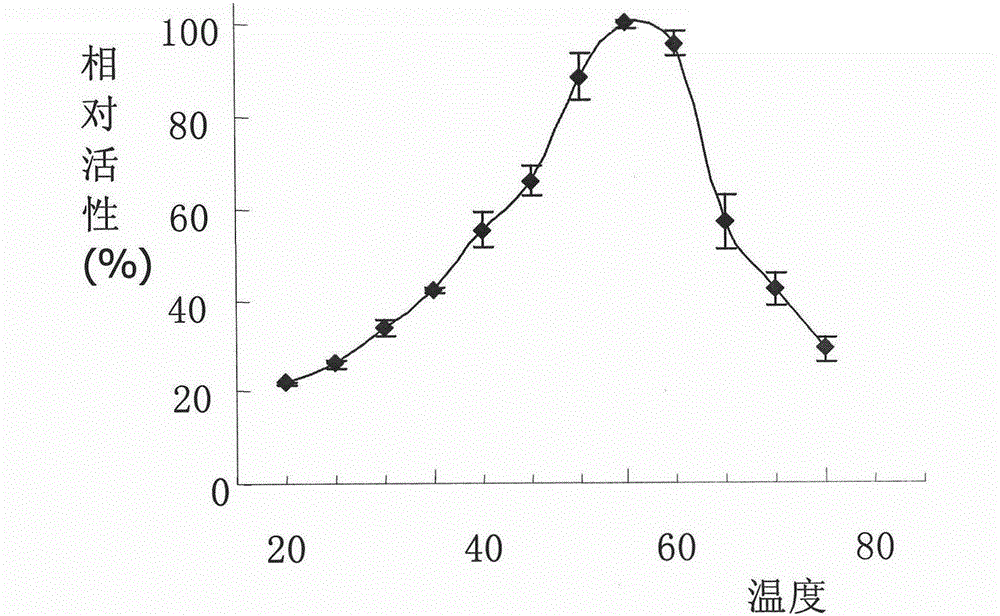
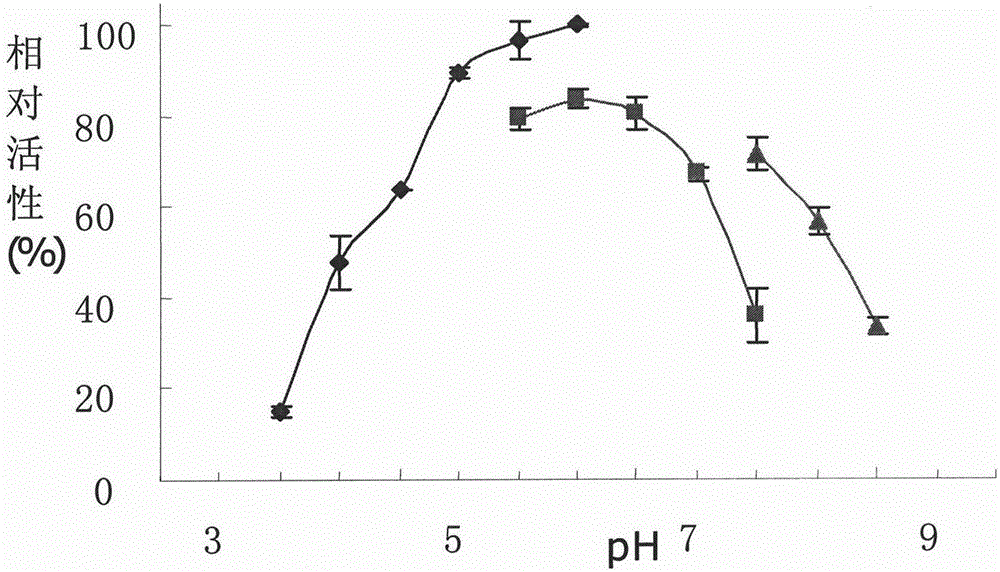
High-efficiency endoglucanase rucelb, its coding gene, preparation method and application
An endoglucanase and encoding gene technology, applied to the field of endoglucanase and its preparation, can solve problems such as low enzyme activity and achieve the effect of good thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 The construction of Chinese yak rumen microbial metagenomic DNA library
[0037] The rumen contents of 2 Chinese Qinghai yaks were collected from a slaughterhouse in Xining City, filtered through three layers of gauze, the filtrate was centrifuged to collect rumen microbial cells, and frozen at -80°C until use. Take 100-200ul bacteria samples, wash 2-3 times with 1ml PBS, add 650uL DNA extraction buffer (Tris-HCl, 100mMpH8.0; Na 2 EDTA100mMpH8.0; Na 3 PO 4 Buffer100mMpH8.0; NaCl1.5M; CTAB1%; pH8.0), after mixing, place in -80°C, then place in a 65°C water bath to melt, repeat three times; add 3-4μL lysozyme (100mg / L ) shake horizontally (37°C, 225rpm) in a shaker for about 30min; add 2-3μL proteinase K (20mg / mL) and continue to shake for about 30min; add 50-70uLSDS (20%), mix well, and incubate at 65°C for 1 -2h, invert the centrifuge tube up and down every 10-20min to mix; centrifuge at 12,000rpm for 10min at room temperature, collect the supernatant, add ...
Embodiment 2
[0040] Cloning and sequence analysis of RuCelB gene derived from embodiment 2 rumen microorganisms
[0041] The endoglucanase gene on 6C6 was cloned into the pGEM11z vector by subcloning method: the cosmid plasmid of the screened positive clone 6C6 was partially digested with Sau3AI into a 2-5kb fragment, ligated into a fragment digested with BamHI and In the dephosphorylated pGEM11z vector, transform DH5α, and perform functional screening on the subcloned library by the method described in Example 1. The obtained subclones are sequenced with T7 and SP6 universal primers, and the endoglucan is determined by homologous comparison The coding region sequence of the enzyme gene, its gene nucleotide sequence is shown in SEQIDNO1, and named as RuCelB.
[0042] The gene cds encodes 336 amino acids, the ORF sequence is shown in SEQ ID NO2, and the theoretical molecular weight is 38.4kD. Using SMART to analyze the structural domain, it was shown that the 24 amino acids from the N-term...
Embodiment 3
[0043] Recombinant expression of embodiment 3 RuCelB gene in escherichia coli
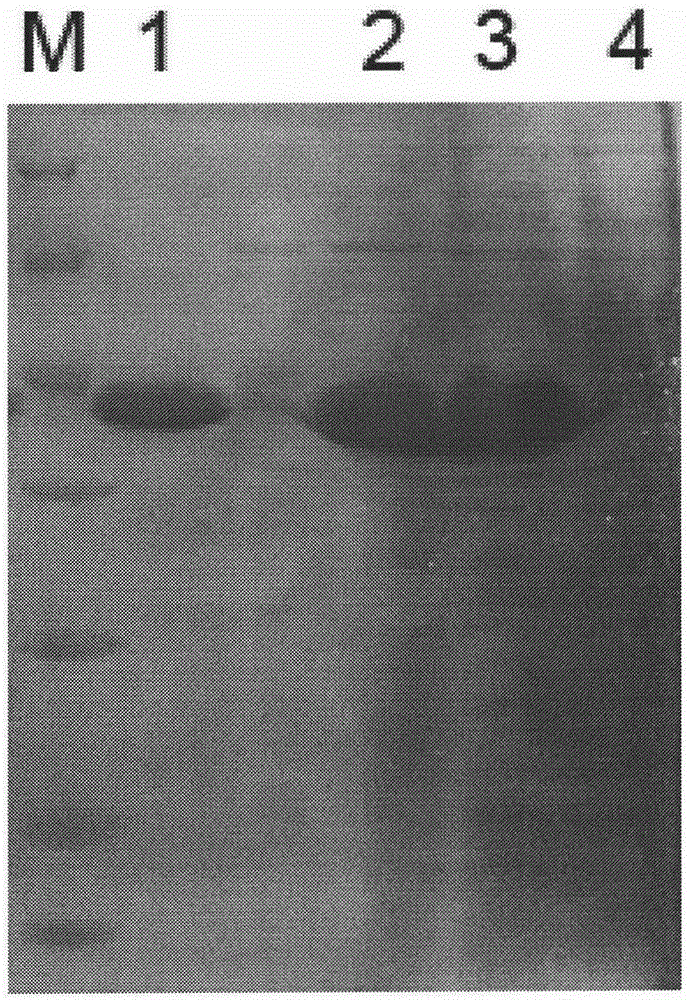
[0044] In order to clone the RuCelB gene sequence into the Escherichia coli expression vector pET-21a for recombinant expression, a pair of primers were designed: the forward primer added 6 His codons, skipped the signal peptide, and amplified from the 25th position of the protein, The sequence is RuCelBF:ATA GAATTC CATCATCACCATCATCACGGCAACGGCTGGGTC, reverse primer is RuCelBR:TGT AAGCTT ACCCGCCTGTCCCTG, the underlined restriction enzyme sequence. The RuCelB gene fragment was amplified by PCR reaction, and after gel recovery, it was digested with EcoRI and HindIII. After the fragment was recovered, it was ligated with the pET-21a vector that was also digested with EcoRI and HindIII, and the ligated product was transformed into E. coli Top10 strain. The obtained transformants were identified by cooking PCR using the above primers, and the enzyme activity and sequencing of the transformants contai...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com