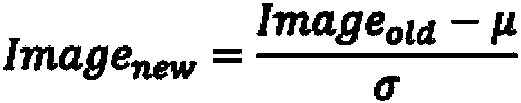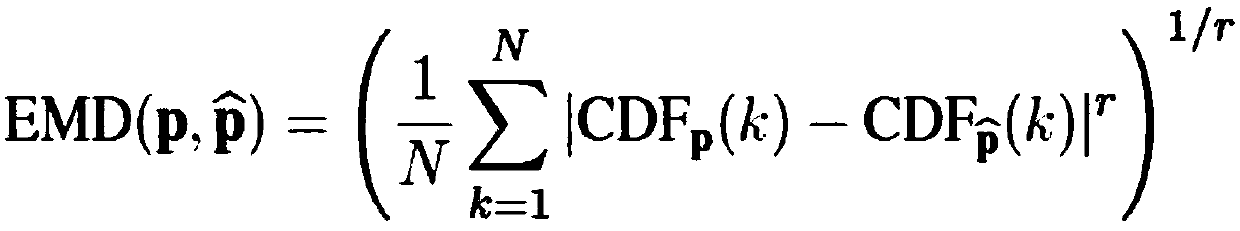Patents
Literature
52 results about "Chromosome maps" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
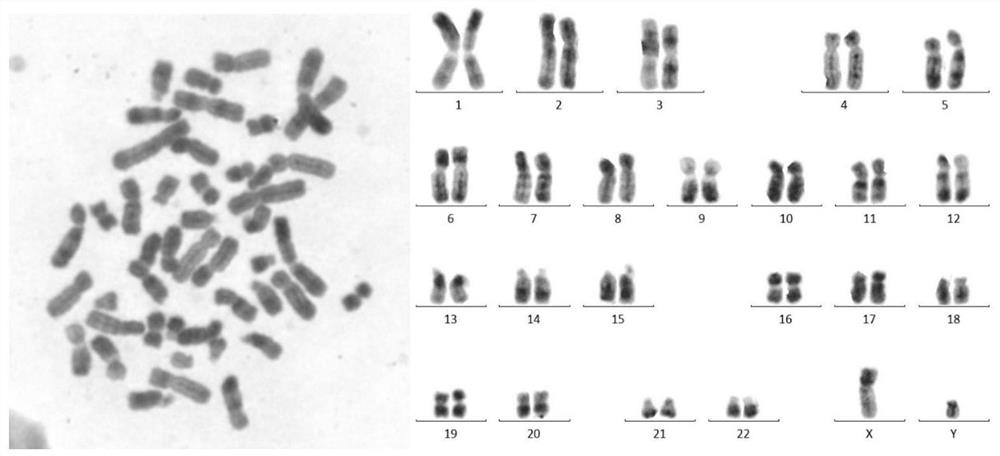
Computer-assisted karyotyping
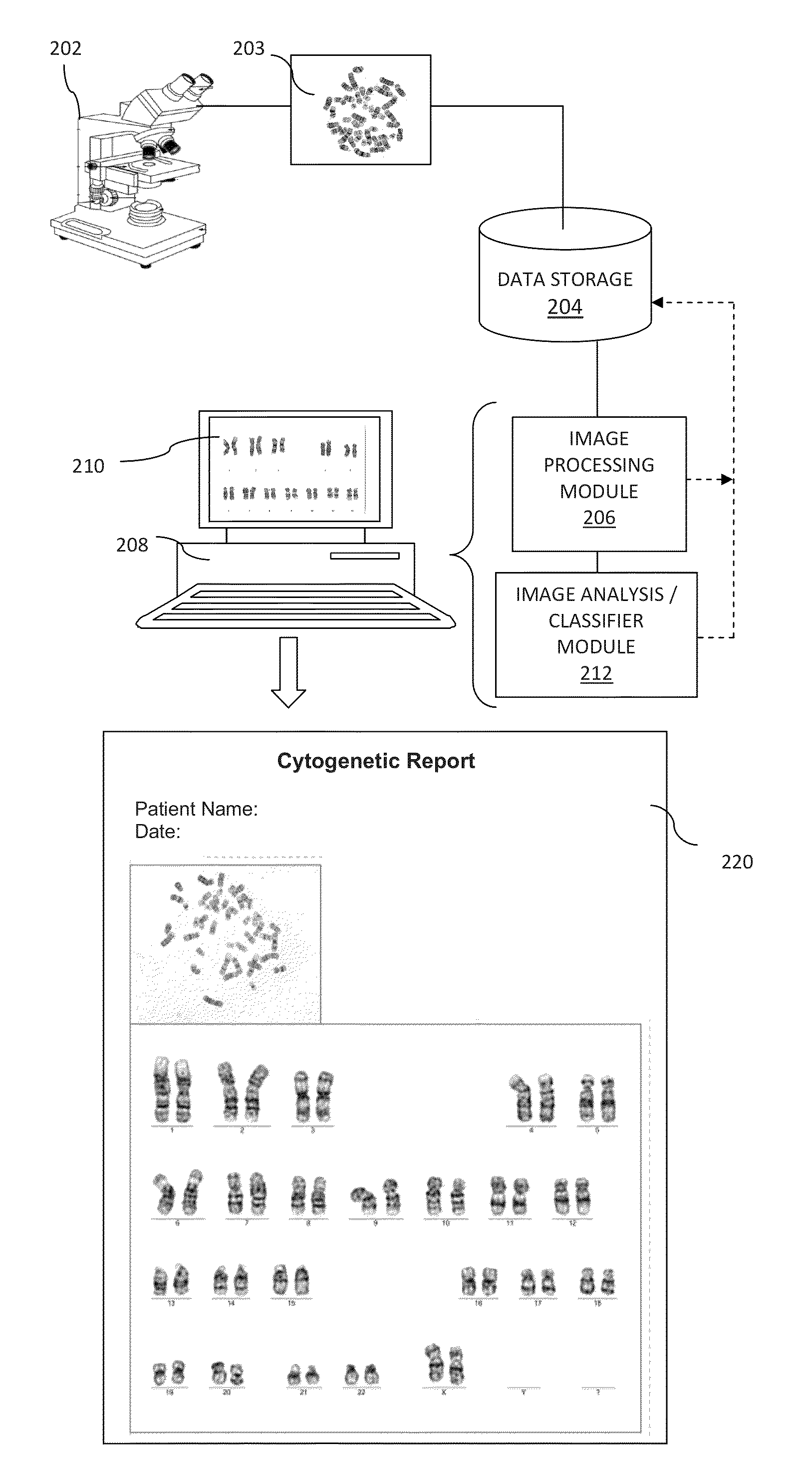
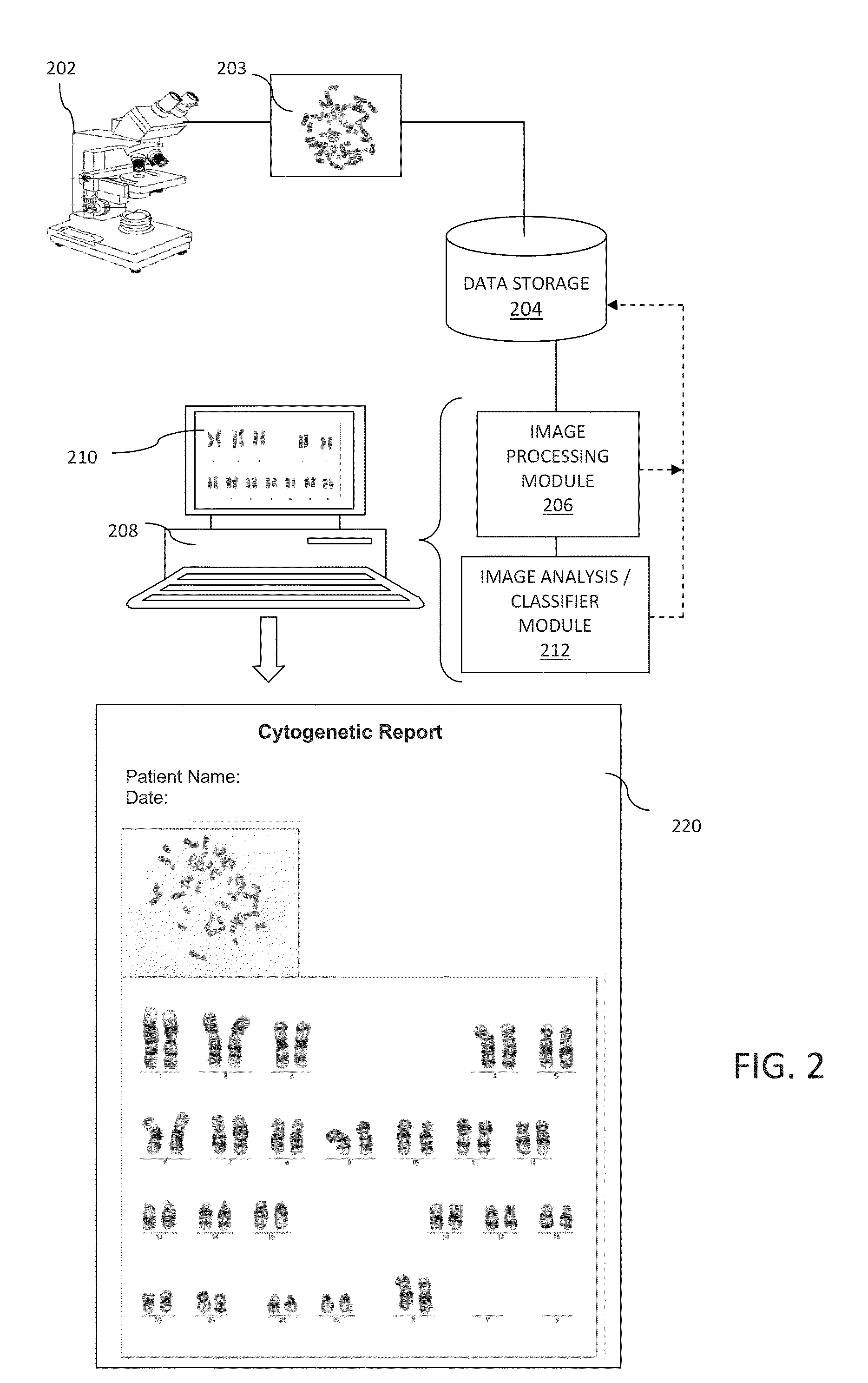
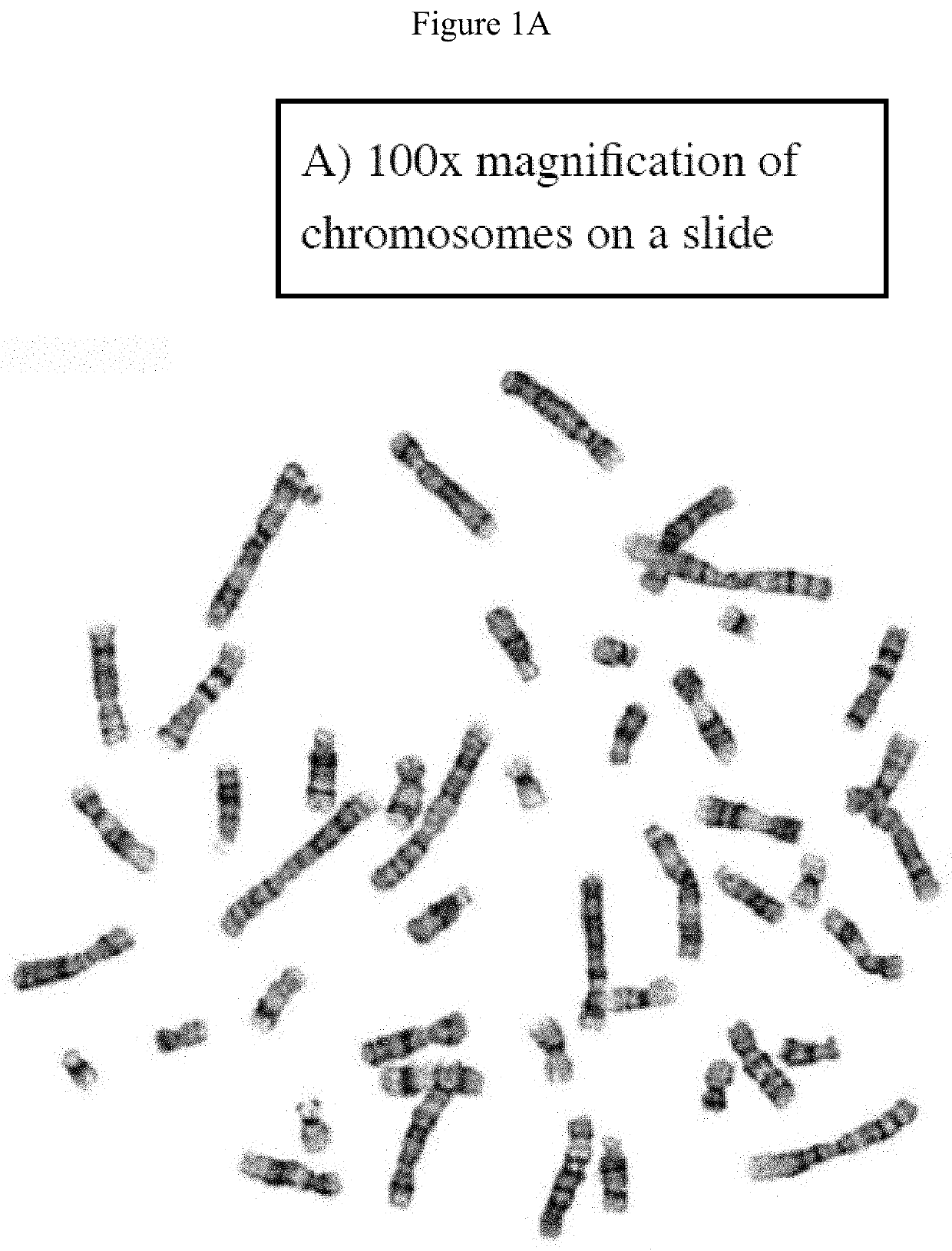
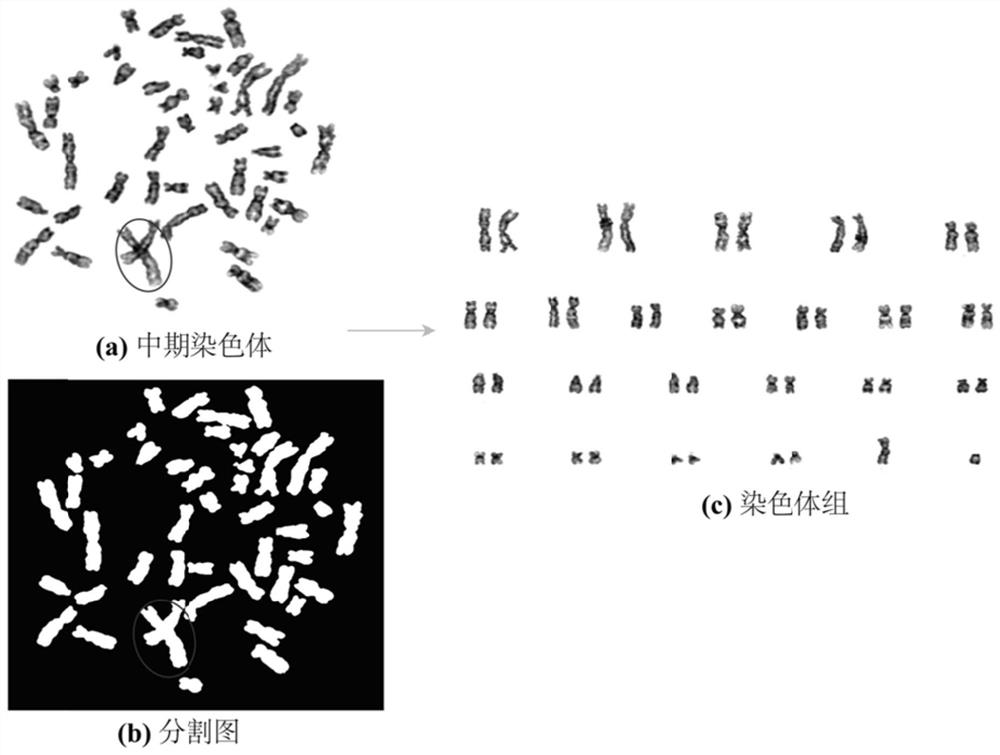
A system and method for computer-assisted karyotyping includes a processor which receives a digitized image of metaphase chromosomes for processing in an image processing module and a classifier module. The image processing module may include a segmenting function for extracting individual chromosome images, a bend correcting function for straightening images of chromosomes that are bent or curved and a feature selection function for distinguishing between chromosome bands. The classifier module, which may be one or more trained kernel-based learning machines, receives the processed image and generates a classification of the image as normal or abnormal.
Owner:HEALTH DISCOVERY CORP
An automatic chromosome counting method based on depth learning
ActiveCN109523520ASimple data sourceLow costImage enhancementImage analysisAlgorithmCorrection method
Owner:中科伊和智能医疗科技(北京)有限公司
Automatic chromosome karyotype analysis and anomaly detection method
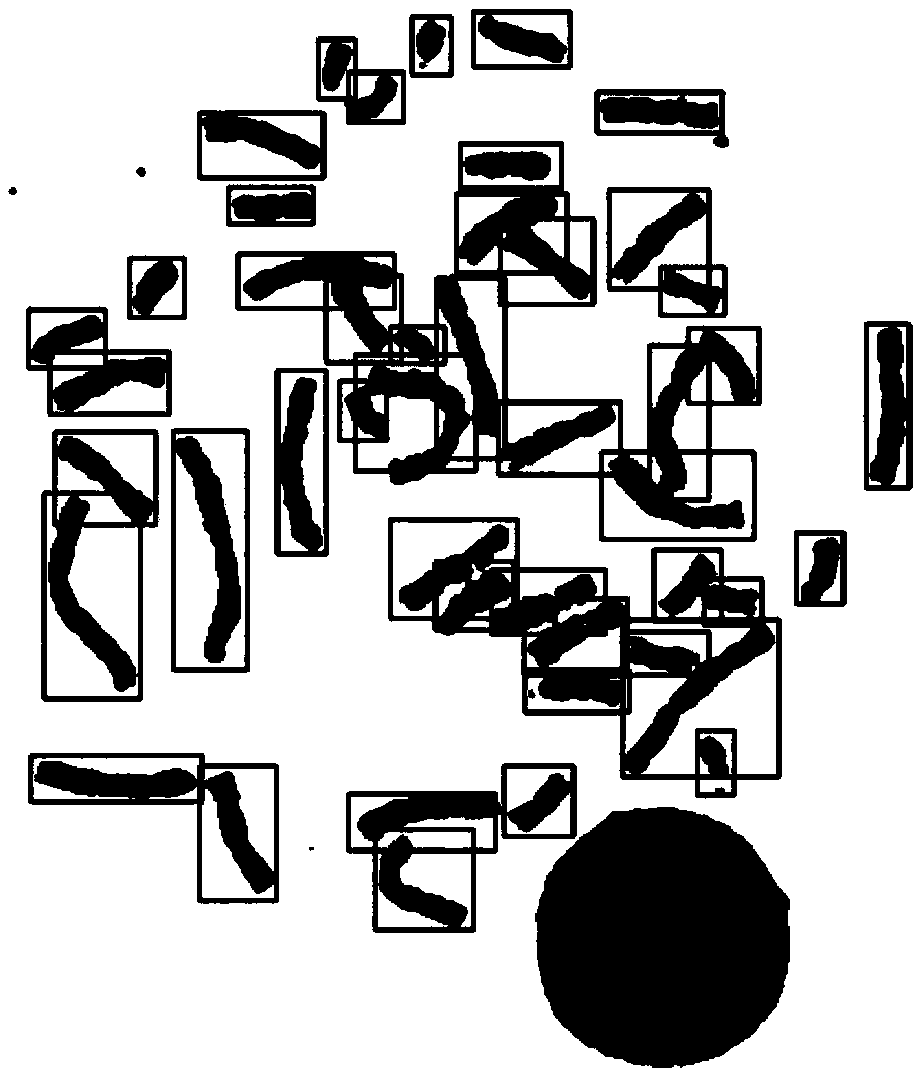
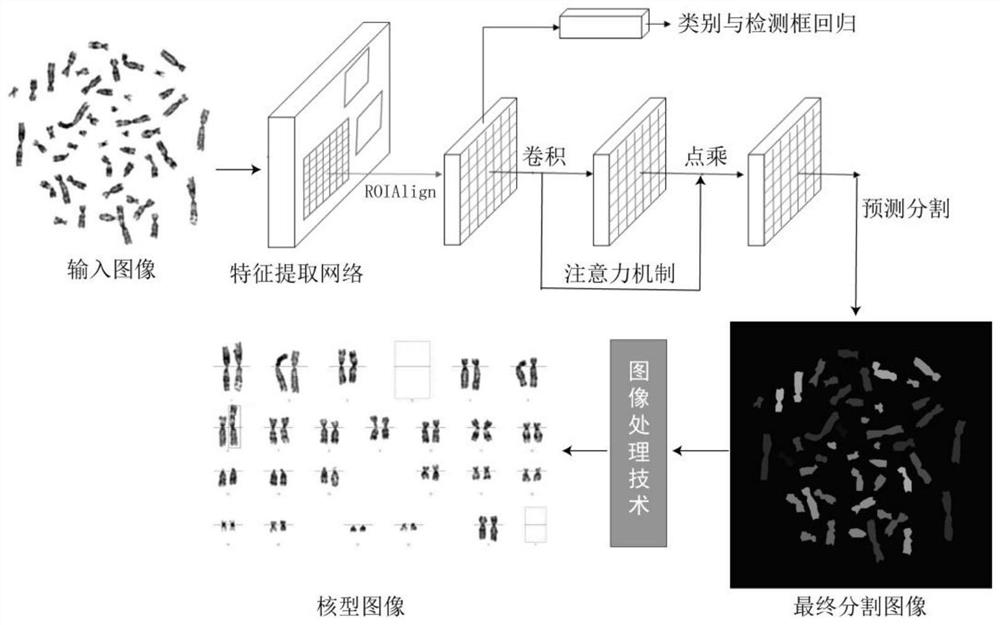
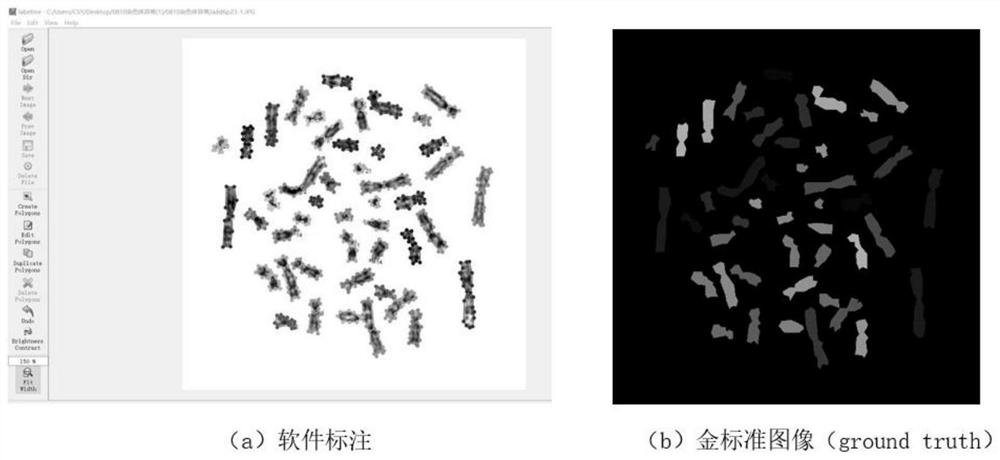
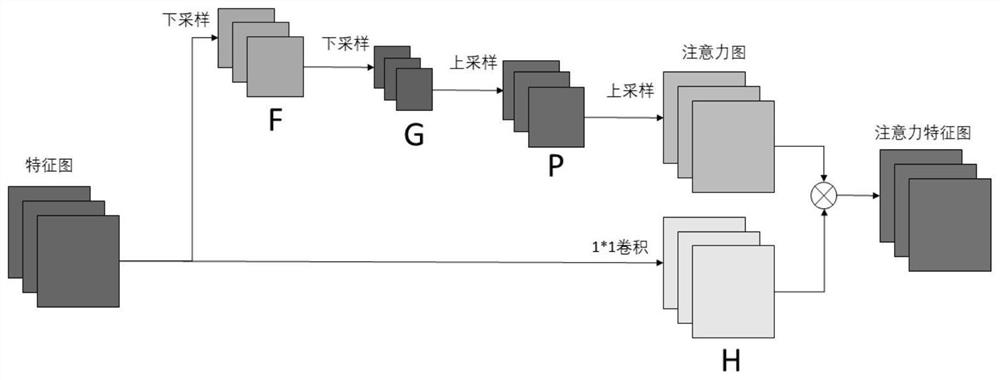
The invention provides an automatic chromosome karyotype analysis and anomaly detection method. According to the method, an attention mechanism and a convolutional neural network are combined, the target is positioned in two stages, and the area of the target is roughly positioned in the first stage; in the second stage, an attention mechanism is added to the target area in the first stage, deepersemantic features are extracted to predict a mask of the target, meanwhile, the position of the target is roughly positioned by means of category prediction and a detection box regression task, and segmentation is conducted; finally, the trained model is used for segmenting and detecting the chromosome image, chromosome segmentation and anomaly detection can be accurately achieved, and thereforechromosome karyotype analysis automation is achieved. The method provided by the invention can overcome the defects that a traditional chromosome karyotype analysis method mainly depends on genetics to carry out artificial interaction analysis, time is consumed, overlapped chromosomes are not easy to separate, abnormal chromosomes cannot be automatically detected and the like.
Owner:WUHAN UNIV
Method for detecting and extracting chromosome contour based on directional searching
InactiveCN101414358AMeet the needs of accurate detection of object contoursImprove accuracyCharacter and pattern recognitionRadiologyOperand
The invention discloses a chromosome contour detecting and extracting method based on directed search, comprising: giving binaryzation treatment to collected images of chromosome; making a point on the contour of the chromosome as a starting point in the image after the binaryzation treatment; searching for and extracting border information continuously along the edge of the chromosome contour in a clockwise direction till reaching the detecting and extracting starting point again, so as to detect the contour of the chromosome wholly. The detecting and extracting method disclosed by the invention satisfies the requirements that contours of objects should be accurately detected in medical treatment fields such as chromosome analysis, cell research, pathological analysis, etc. The method of the invention improves automation level of segregating and incising overlapped and adherent chromosomes according to the contour, perfects the shortcomings of traditional computing methods that the contour fractures, the contour is incontinuous and unevenness details of the contour are ignored, and improves the accuracy of detecting results, furthermore operand is greatly reduced.
Owner:GUANGDONG VTRON TECH CO LTD
A double chromosome image cutting method based on a Compact SegUnet self-learning model
ActiveCN109934828APrecise positioningEfficient and high-quality segmentationImage analysisCharacter and pattern recognitionPattern recognitionData set
The invention discloses a double chromosome cutting method based on a Compact SegUnet self-learning model. The method comprises steps of by learning a double-chromosome overlapping data set generatedbased on real chromosome picture simulation, extracting high-dimensional features of different areas of the image by the model, predicting the probability that each pixel of the image belongs to the overlapping area and each chromosome according to the difference between the chromosome overlapping area and the non-overlapping area and the difference between different chromosomes, and finally selecting the classification with the maximum probability to complete the segmentation on the overlapping chromosome pixel level. Compared with a traditional manual observation distinguishing method, the method has the advantages that the efficiency is greatly improved, and the working time and cost are saved; Compared with an existing geometric segmentation method, the problem that partial overlappingis not easy to segment is solved, and practicability is high; Compared with an existing deep learning model, the segmentation accuracy is improved.
Owner:XIAN JIAOTONG LIVERPOOL UNIV +1
Chromosome cutting data processing method and system and storage medium
PendingCN111223084AAccurate segmentationImprove accuracyImage enhancementImage analysisImaging processingGenetics
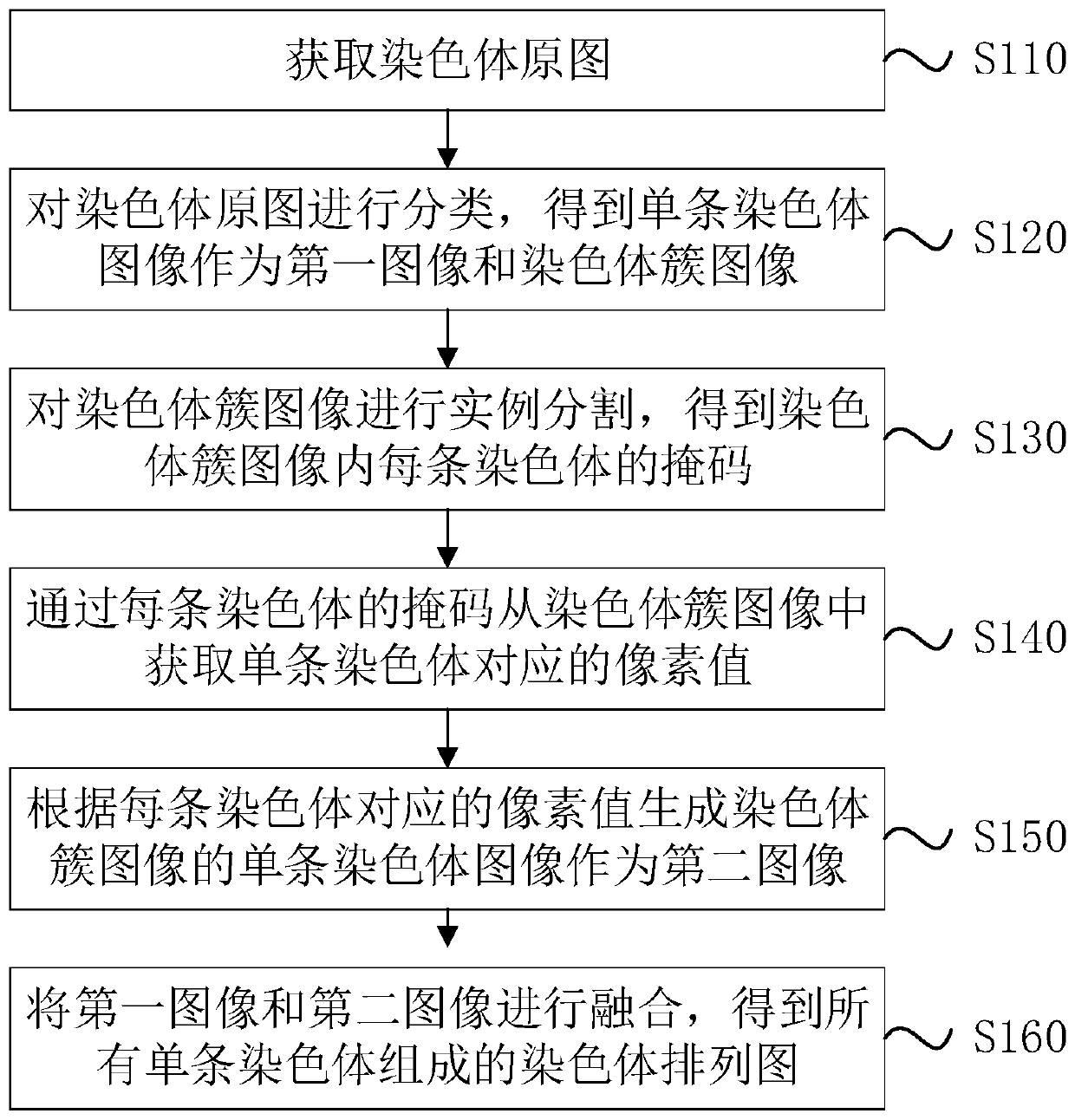
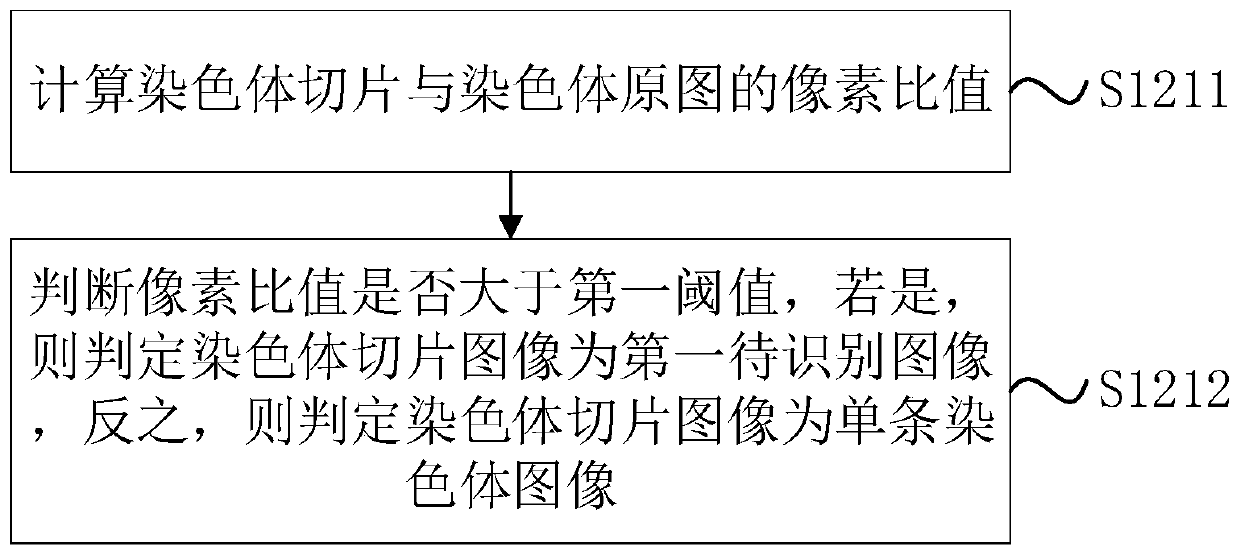
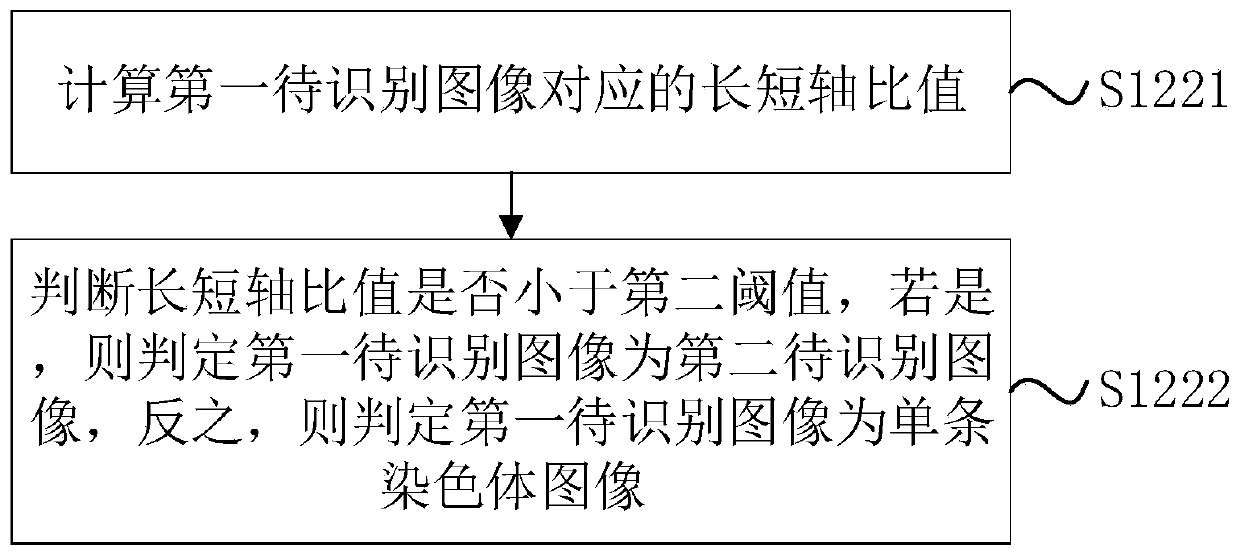
The invention discloses a chromosome cleavage data processing method and system and a storage medium. The method comprises the following steps: acquiring a chromosome original image; classifying the chromosome original images to obtain a single chromosome image as a first image and a chromosome cluster image; carrying out instance segmentation on the chromosome cluster image to obtain a mask of each chromosome in the chromosome cluster image; obtaining a pixel value corresponding to a single chromosome from the chromosome cluster image through the mask of each chromosome; generating a single chromosome image of the chromosome cluster image as a second image according to the pixel value corresponding to each chromosome; and fusing the first image and the second image to obtain a chromosomearrangement diagram composed of all single chromosomes. According to the method, chromosome images with chromosome intersection or overlapping can be accurately segmented, and the accuracy of chromosome classification and karyotype analysis by doctors is improved. The method can be applied to the field of image processing.
Owner:SOUTH CHINA NORMAL UNIVERSITY +1
Automatic chromosome segmentation and classification method based on deep learning
ActiveCN113658150AFully automatedRealize intelligenceImage enhancementImage analysisSupport vector machineClassification methods
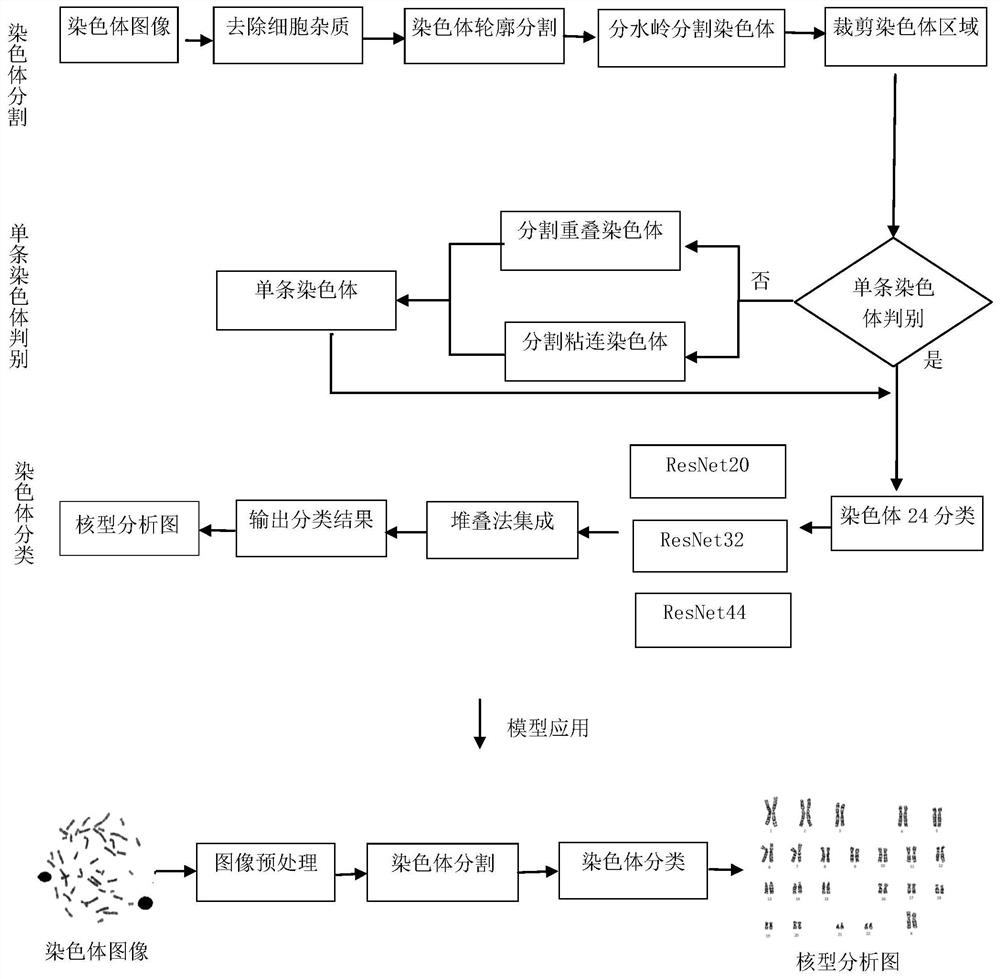
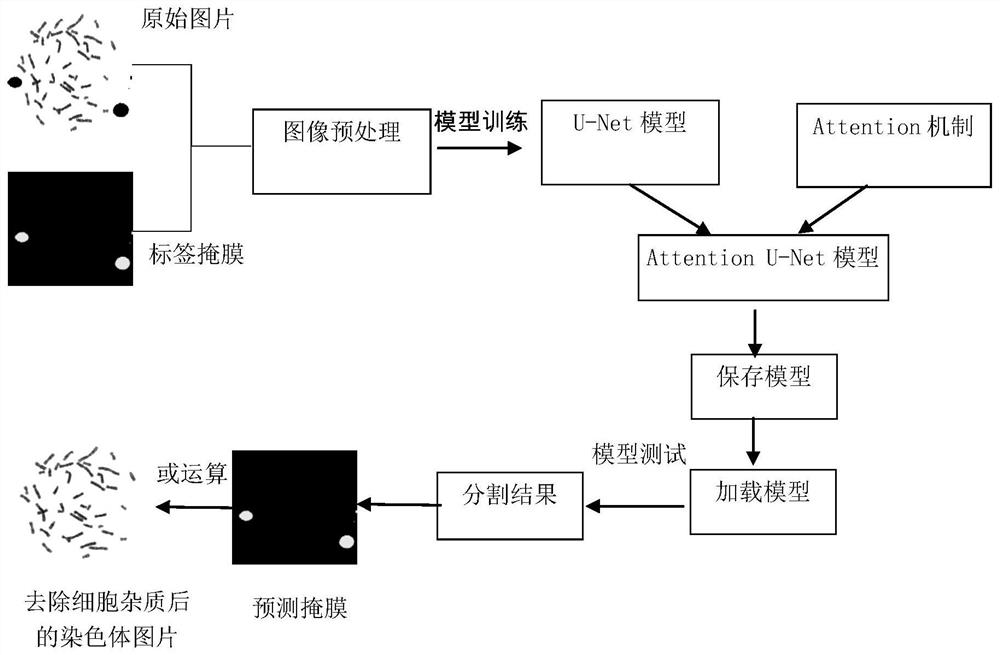
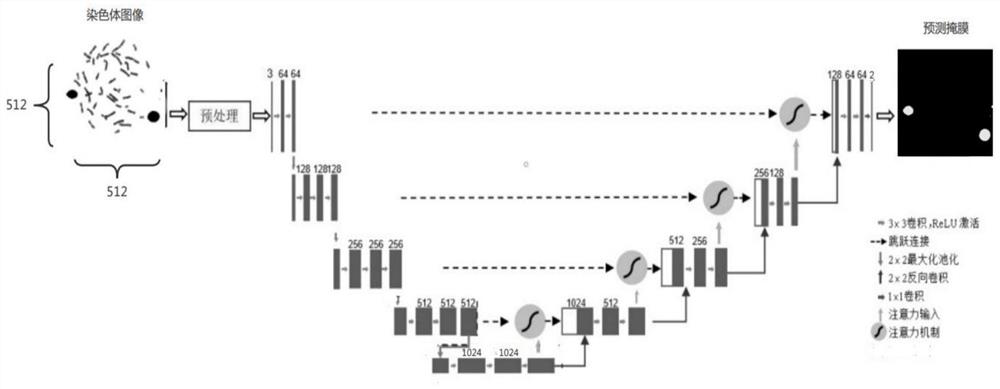
The invention discloses an automatic chromosome segmentation and classification method based on deep learning. The method comprises the following steps: obtaining a chromosome image and filtering cell impurities by using an Attention U-Net model; segmenting chromosomes and cutting out each chromosome region image; extracting features from the obtained chromosome region, training a support vector machine, a random forest and a logistic regression classifier, and carrying out model integration by adopting a voting method so as to identify overlapped adhesion chromosomes or single chromosomes; respectively designing independent segmentation modules for the overlapped adhesion chromosomes, segmenting the overlapped chromosomes by utilizing a method of separating and then splicing, and segmenting the adhesion chromosomes by utilizing a convex defect point detection method; and respectively inputting the chromosome training data with marked types into 24 classification models ResNet20, ResNet32 and ResNet44 for training, then carrying out model integration by using a stacking method, and outputting a final chromosome classification result and a chromosome karyotype analysis chart so as to carry out chromosome anomaly identification.
Owner:XI AN JIAOTONG UNIV +1
Chromosome division phase image quality evaluation method based on deep learning
PendingCN111325711AEvaluate image qualityImprove operational efficiencyImage enhancementImage analysisImaging processingAlgorithm
The invention discloses a chromosome division phase image quality evaluation method based on deep learning, and belongs to the technical field of chromosome image processing. Through the deep learningmodel, the quality of the chromosome split-phase images can be scored, the image quality of the chromosome images can be accurately evaluated, the robustness for evaluating different types of chromosome split-phase images is high, and the operation efficiency of doctors is improved. According to the invention, the chromosome division phase image quality can be evaluated accurately and efficiently; the method can effectively improve the evaluation and selection efficiency of the chromosome division phase images, shortens the screening time of the chromosome division phase images, can effectively reduce the workload of doctors, is not interfered by the outside, is simple and reasonable in process, can be popularized and applied to the outside in a large scale, and is simple in deployment.
Owner:HANGZHOU DIAGENS BIOTECH CO LTD
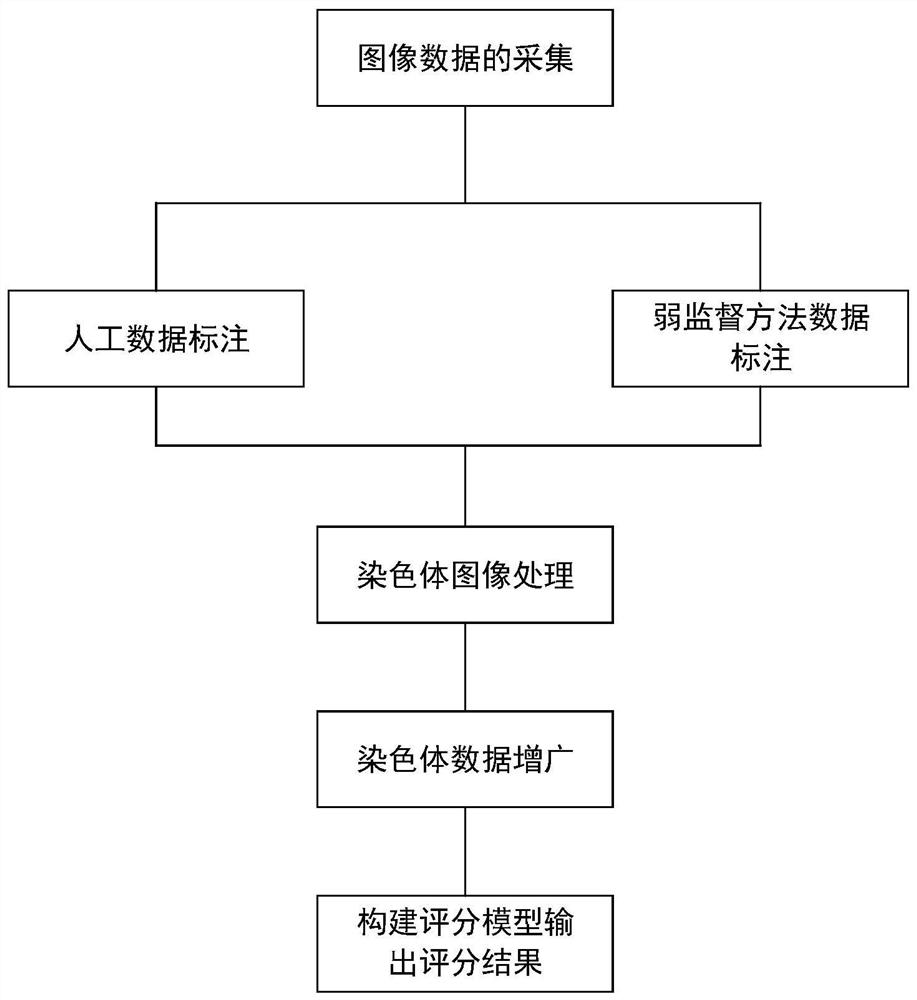
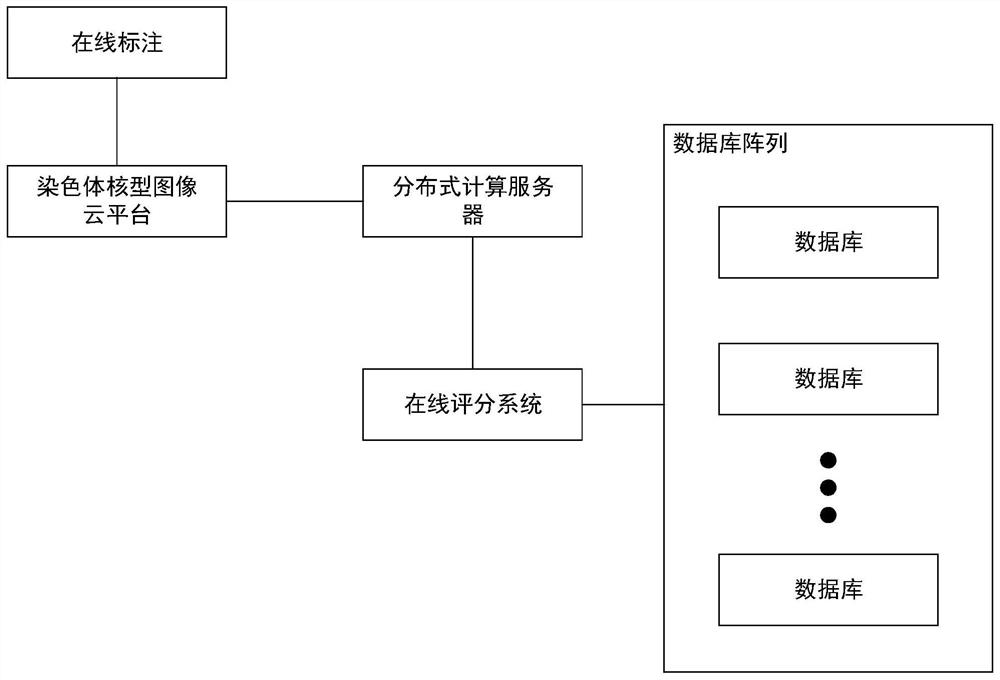
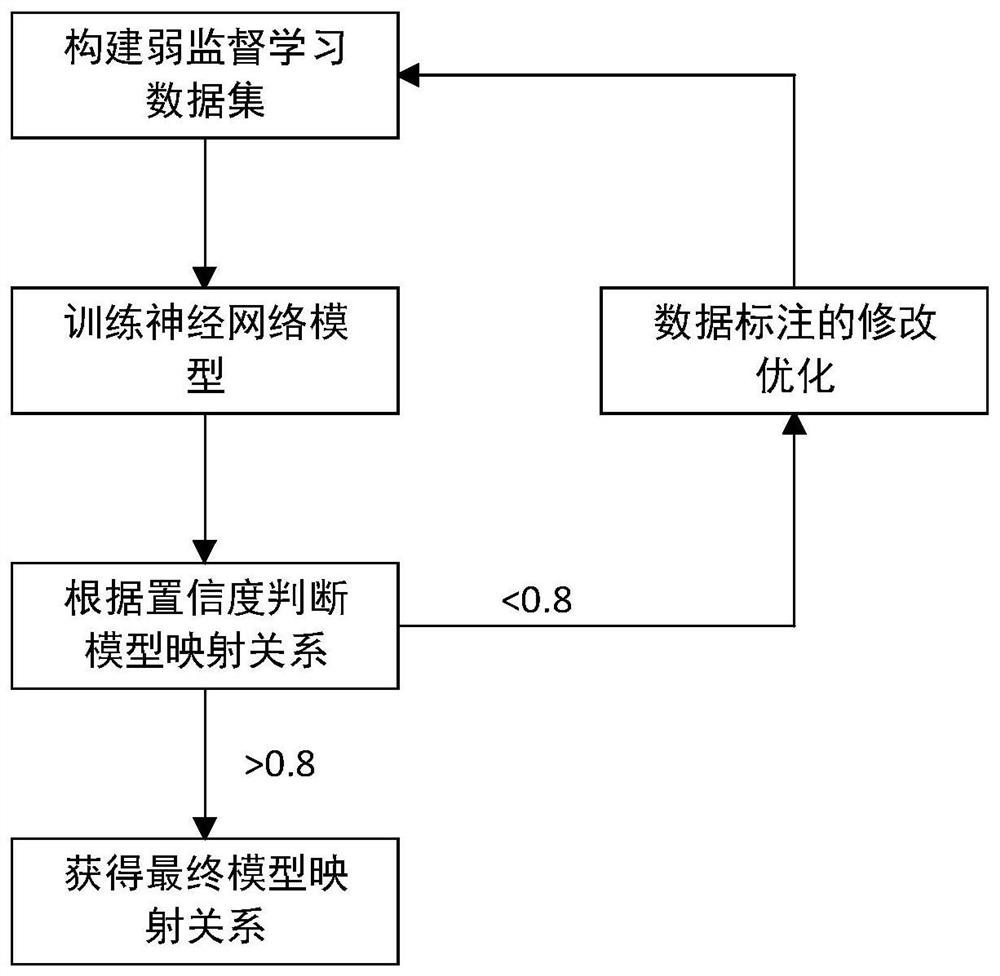
Chromosome karyotype image intelligent evaluation method and system
InactiveCN111652167ALow costImprove labeling efficiencyNeural architecturesRecognition of DNA microarray patternImaging processingImaging analysis
The invention provides a chromosome karyotype image intelligent evaluation method and system. The method relates to the technical field of image processing, and is characterized in that automatic labeling of chromosome image data is carried out through a weak supervised learning model; annotation costs is reduced, labeling efficiency is improved, extension modes such as random projection transformation are adopted for chromosome image data; an optimized residual network model is adopted to build a scoring network model, detail differences of chromosome images are better reserved, the accuracyof the scoring network model is improved, meanwhile, a scoring system is established by adopting a micro-service distributed frame through a cloud platform technology, cross-cloud image analysis is achieved, and the efficiency of the system is improved.
Owner:SICHUAN UNIV +1
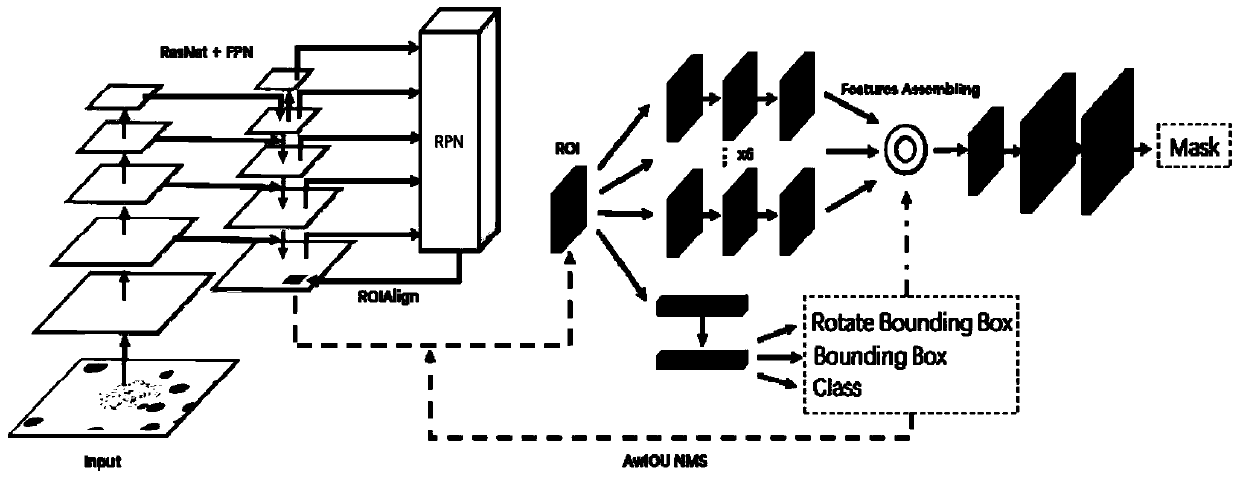
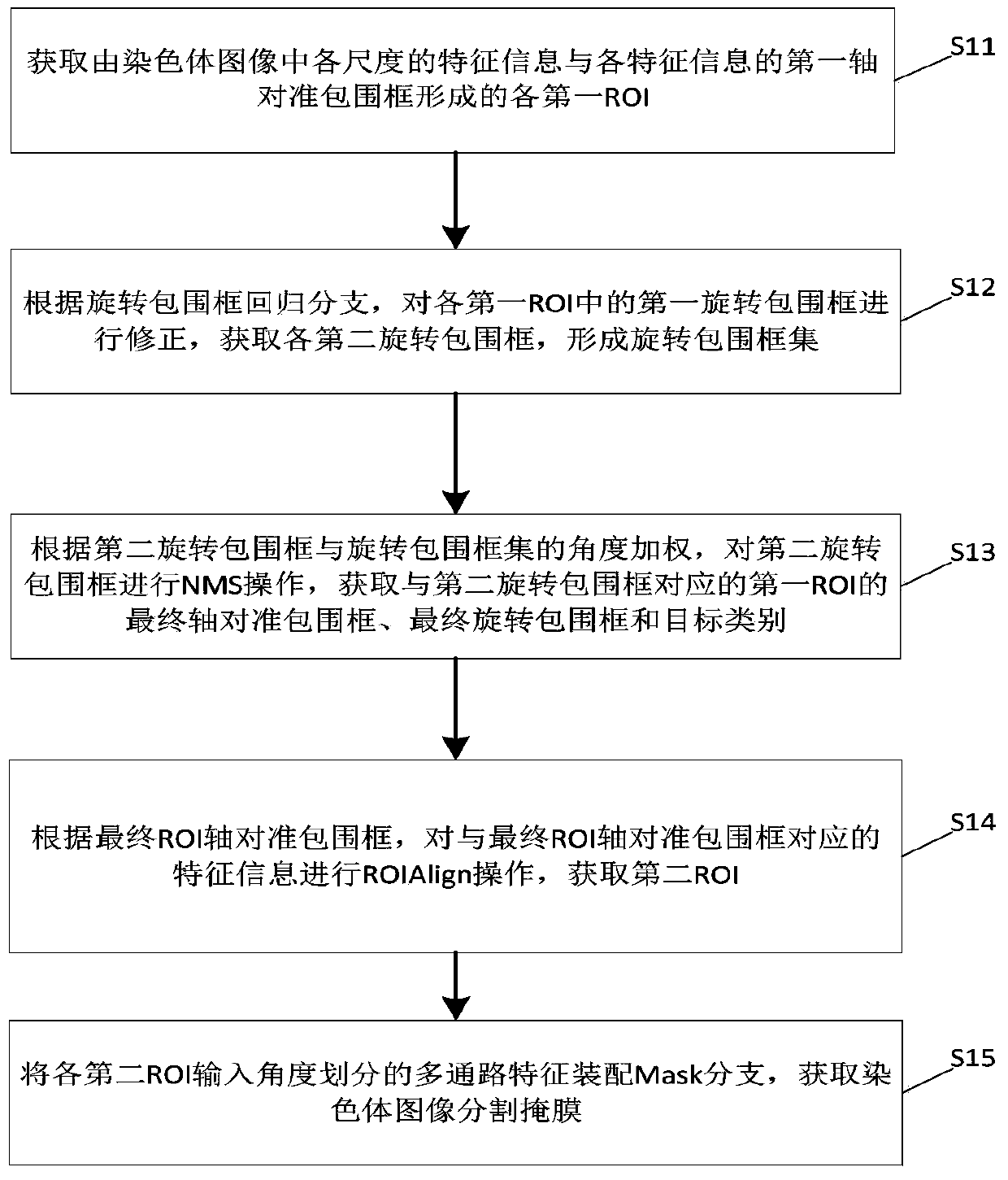
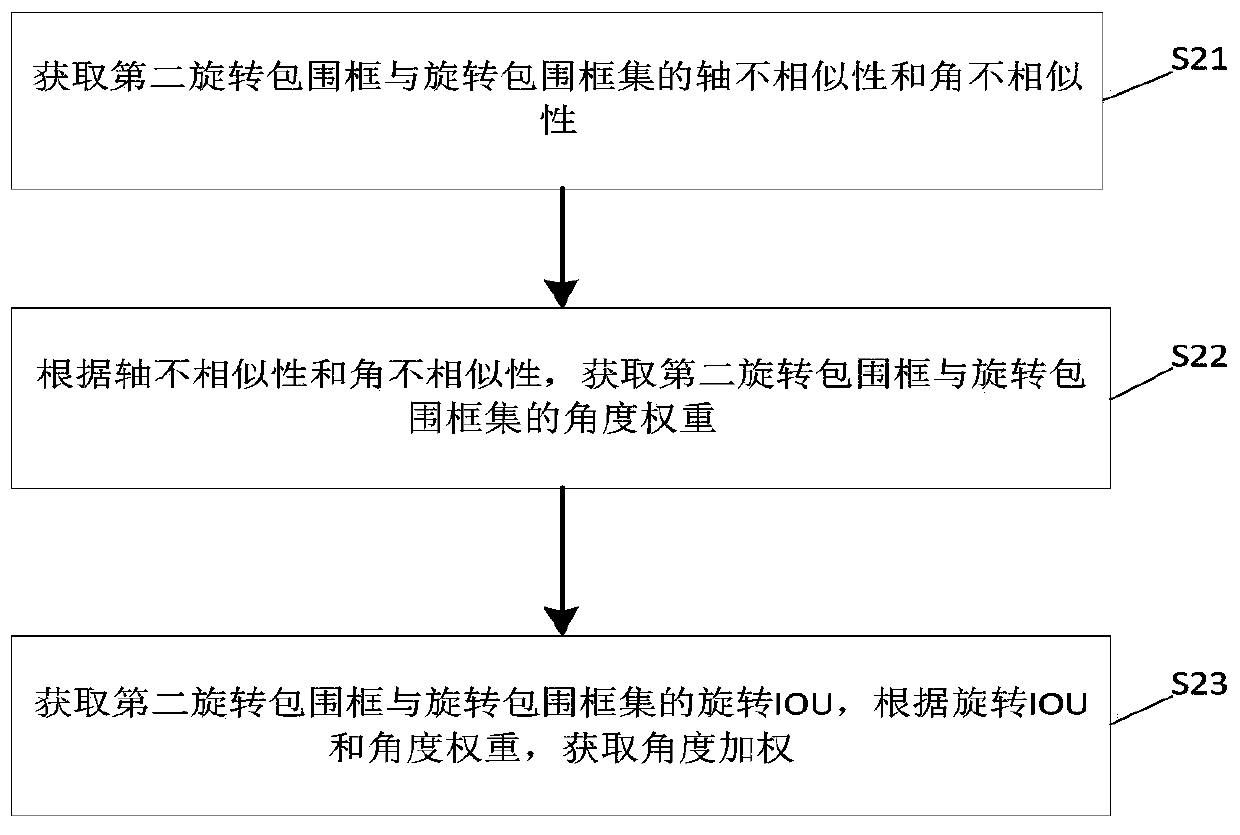
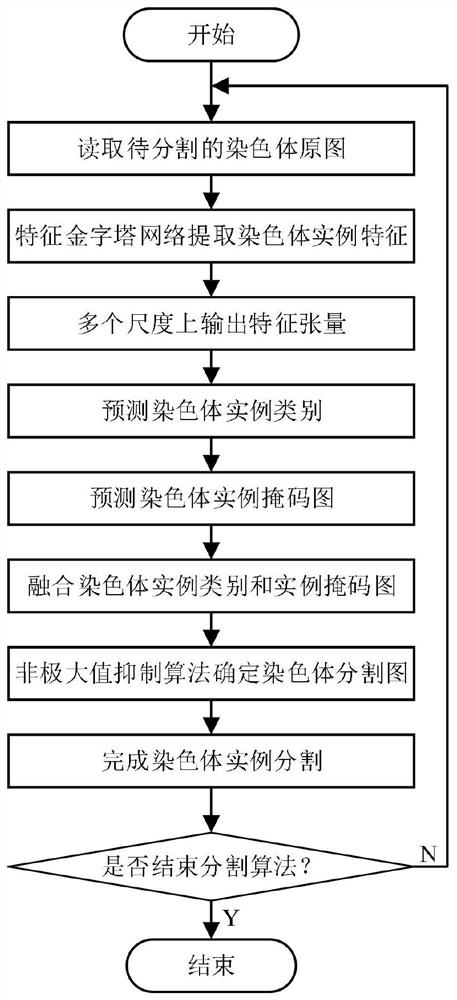
Chromosome image instance segmentation method and device based on improved Mask RCNN
ActiveCN110895810ASolve problems such as mutual interferenceImprove segmentationImage enhancementImage analysisPattern recognitionEngineering
The invention discloses a chromosome image instance segmentation method and device based on an improved Mask RCNN. The method comprises the steps of obtaining each first ROI formed by feature information of each scale in a chromosome image and each corresponding first axis alignment bounding box; correcting the first rotary bounding box in each first ROI to obtain each second rotary bounding box to form a rotary bounding box set; performing NMS operation on the second rotary bounding box according to the angle weighted IOU of the second rotary bounding box and the rotary bounding box set to obtain a final axis alignment bounding box of the first ROI; and according to the final ROI axis alignment bounding box and the corresponding feature information, obtaining a second ROI, inputting the second ROI into a Mask branch, and obtaining a chromosome image segmentation mask. Compared with the prior art, the obtained first ROI is subjected to the NMS operation combined with the angle weightedIOU, so that the problems of excessive inhibition of cross overlapping chromosomes, mutual interference of overlapping monomers during mask regression and the like are solved, and the segmentation effect of the chromosome image is improved.
Owner:GUANGZHOU ELECTRONICS TECH
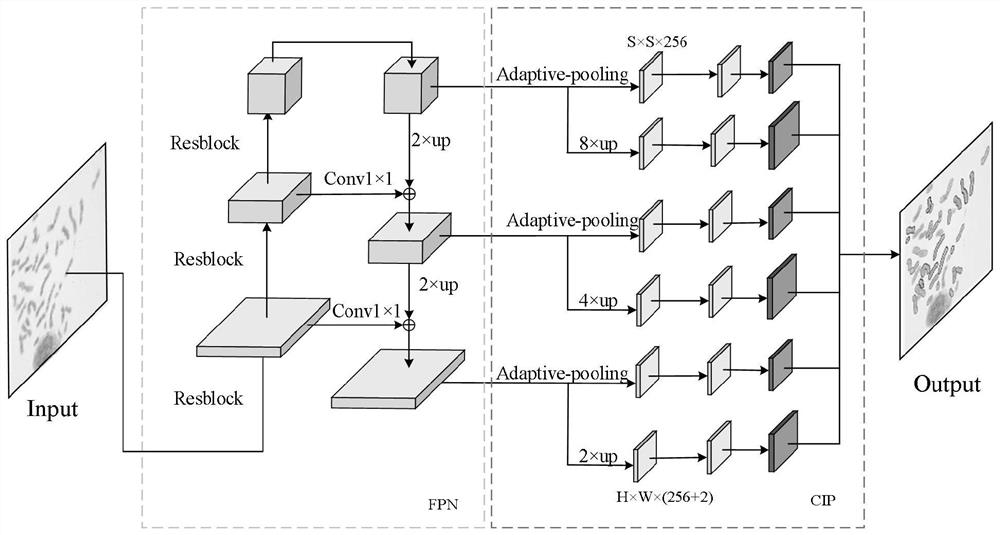
Cross chromosome image instance segmentation method based on chromosome trisection feature point positioning
ActiveCN112365482AImprove Segmentation AccuracyFast splitImage enhancementImage analysisPattern recognitionChiasma
The invention discloses a cross chromosome image instance segmentation method based on chromosome trisection feature point positioning, which comprises the following steps of: dividing an image containing chromosomes into S * S grid units, and allocating chromosome instances to a plurality of grid units according to the positions of chromosome instance trisection feature points, wherein each gridunit is responsible for predicting the instance category of one instance. Each output channel is responsible for predicting an instance mask map of a chromosome object to which the grid cell belongs.Compared with an instance positioning segmentation network which only adopts an instance center point to complete instance segmentation, the method provided by the invention adopts chromosome trisection feature points to complete chromosome instance segmentation, has higher segmentation accuracy for crossed and overlapped chromosomes, solves the problems that the segmentation effect of a chromosome image is still not ideal and the chromosomes with small size are easily undetected. Therefore, the robustness of the chromosome instance segmentation algorithm is greatly improved, and the segmentation speed of chromosome instances is increased.
Owner:SHANGHAI BEION MEDICAL TECH CO LTD
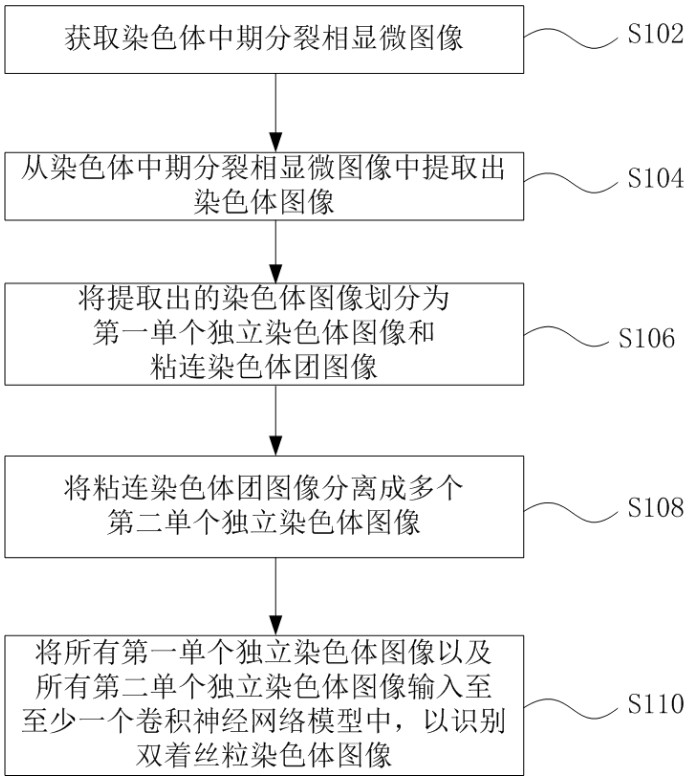
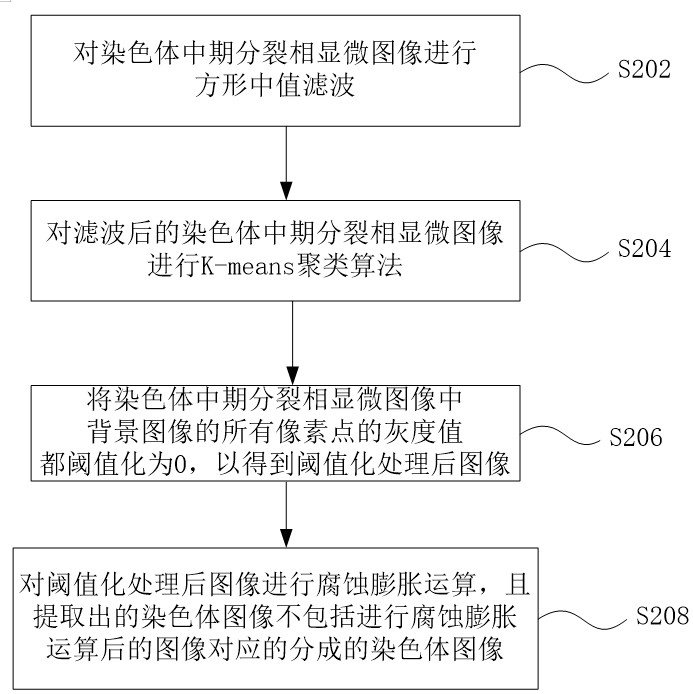
Method and system for automatically identifying microscopic image of metaphase splitting phase of chromosome
ActiveCN113537182AFully automatedRecognition speed is fastImage enhancementImage analysisMicroscopic imageChromosome metaphase
The invention discloses an automatic identification method and system for a chromosome metaphase splitting phase microscopic image based on a convolutional neural network. The automatic identification method comprises the following steps: acquiring the chromosome metaphasesplitting phase microscopic image; extracting a chromosome image from the chromosome metaphase splitting phase microscopic image; dividing the extracted chromosome image into a first single independent chromosome image and an adhesion chromosome group image; separating the adhesion chromosome group image into a plurality of second single independent chromosome images; and inputting all the first single independent chromosome images and all the second single independent chromosome images into at least one convolutional neural network model to identify a double centromere chromosome image. The double-centromere chromosome image is identified through the convolutional neural network model, so that automation in the identification process of the double-centromere chromosome is realized.
Owner:BEIJING HUIRONGHE TECH
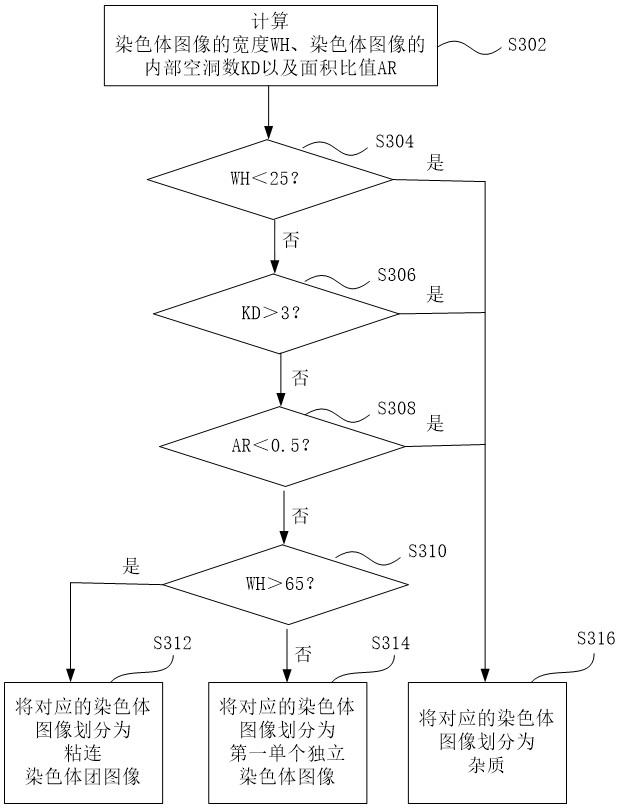
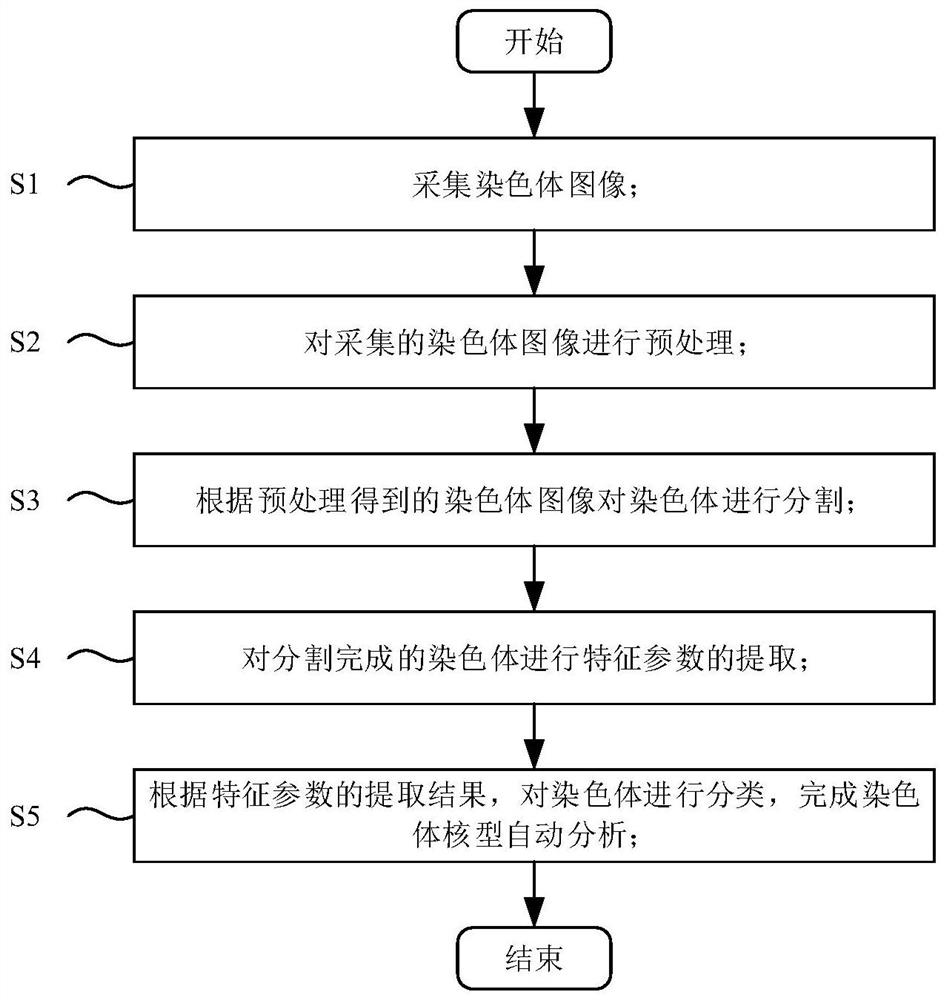
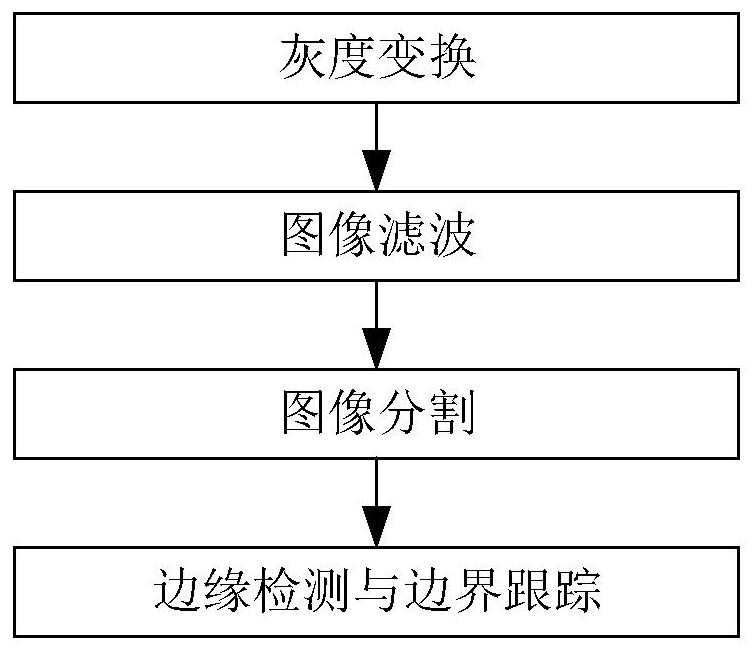
A blood tumor chromosome karyotype automatic analysis method and system
PendingCN113435285ARealize intelligenceImprove diagnostic accuracyImage enhancementImage analysisImaging processingKaryotype
The invention provides a blood tumor chromosome karyotype automatic analysis method. The method comprises the following steps: S1, acquiring a chromosome image; s2, preprocessing the acquired chromosome image; s3, segmenting the chromosome according to the chromosome image obtained by preprocessing; s4, extracting characteristic parameters of the segmented chromosomes; and S5, classifying the chromosomes according to an extraction result of the characteristic parameters, and completing the automatic analysis of the karyotype of the chromosome. The invention further provides a blood tumor chromosome karyotype automatic analysis system, intelligence of chromosome karyotype analysis is achieved; the cytogenetics experiment technology is combined with advanced technologies such as image processing and image analysis, the diagnosis accuracy is greatly improved, the detection time is shortened, the detection efficiency is greatly improved, and the technical defects of low intelligent degree and low detection efficiency in the prior art are effectively overcome.
Owner:THE THIRD AFFILIATED HOSPITAL OF SUN YAT SEN UNIV +1
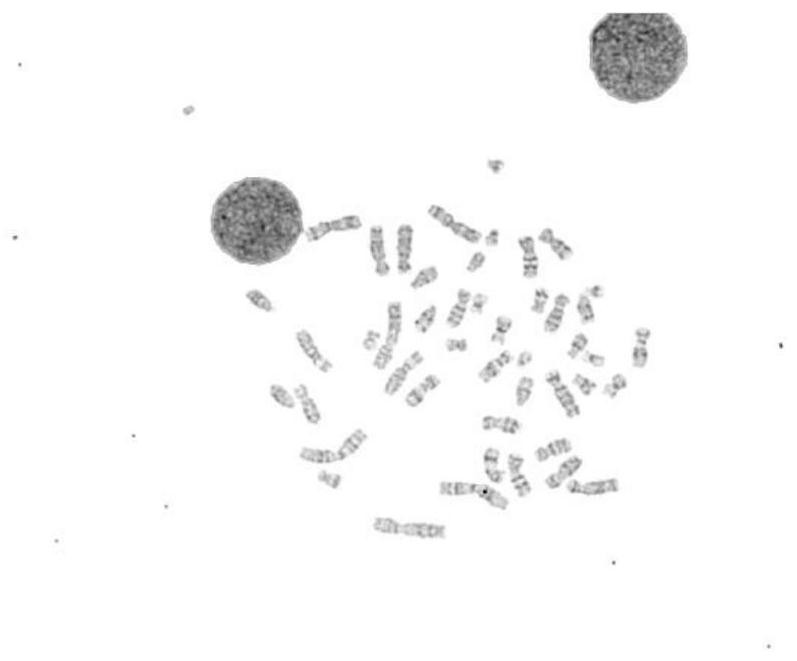
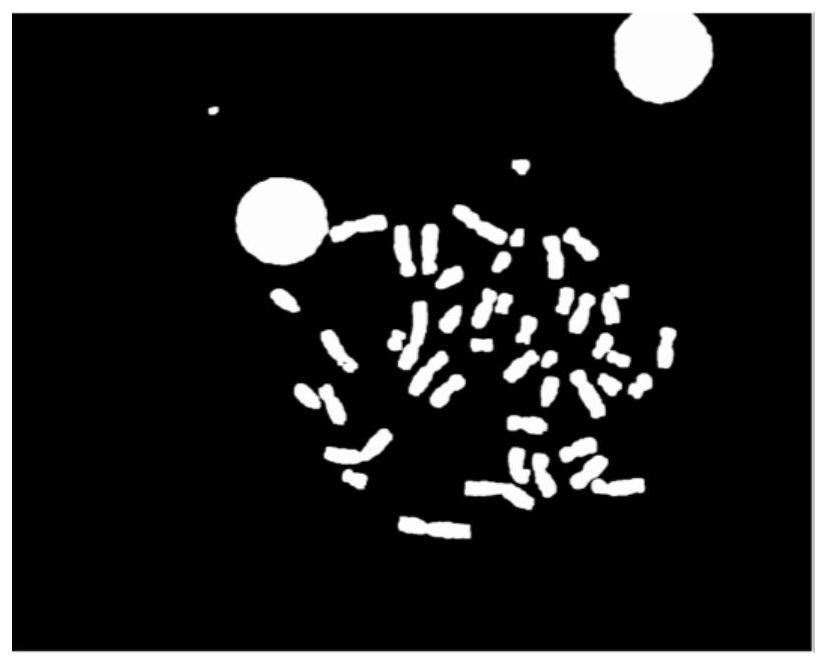
Chromosome scatter image automatic segmentation method
PendingCN113643306ARealize multi-level fine operationSegmentation process is stableImage enhancementImage analysisAutomatic segmentationKaryotype
The invention provides an automatic segmentation method for a chromosome scatter image. The automatic segmentation method comprises the following steps: S1, acquiring a connected domain binary image of the chromosome scatter image; S2, segmenting the connected domain binary image to obtain a connected domain image of the chromosome scatter image; S3, carrying out binary classification on the connected domain image, and separating an adherent chromosome image and a single chromosome image; S4, inputting the separated cohesive chromosome image into a deep learning segmentation network pre-training model for secondary segmentation to obtain a chromosome karyotype image. Compared with an existing chromosome scatter image segmentation method, the method has the advantages that the segmentation of most chromosomes is realized by using a traditional algorithm, and then the classification result, namely adherent chromosomes, is further segmented again by using image dichotomy, so the multi-level fine operation of chromosome segmentation is realized, and the segmentation process is more stable; meanwhile, the segmentation method only needs to label adhered chromosomes and does not need to label a large amount of data, so that the manual labeling cost is saved.
Owner:CHANGCHUN INST OF OPTICS FINE MECHANICS & PHYSICS CHINESE ACAD OF SCI +1
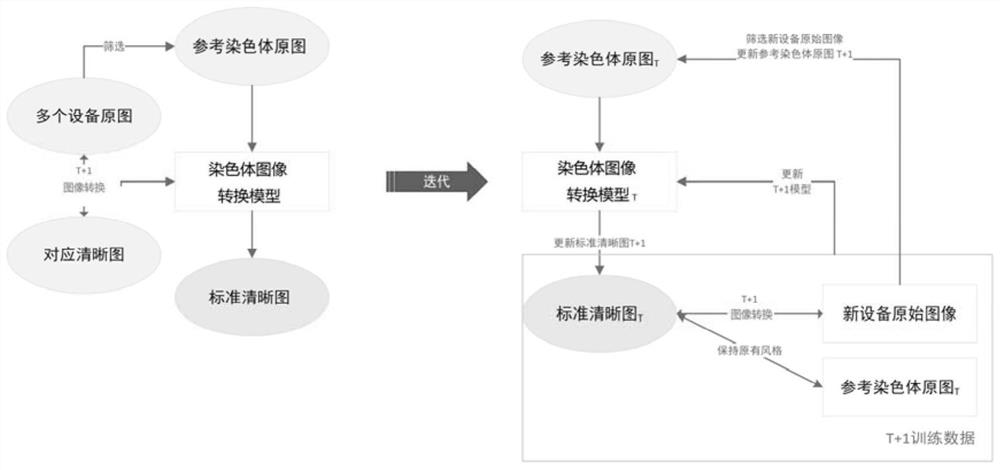
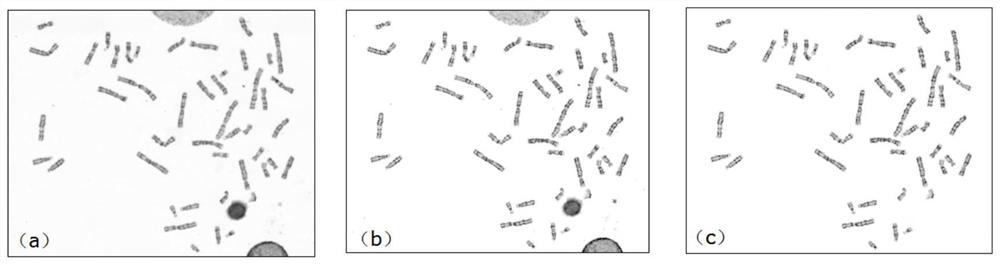
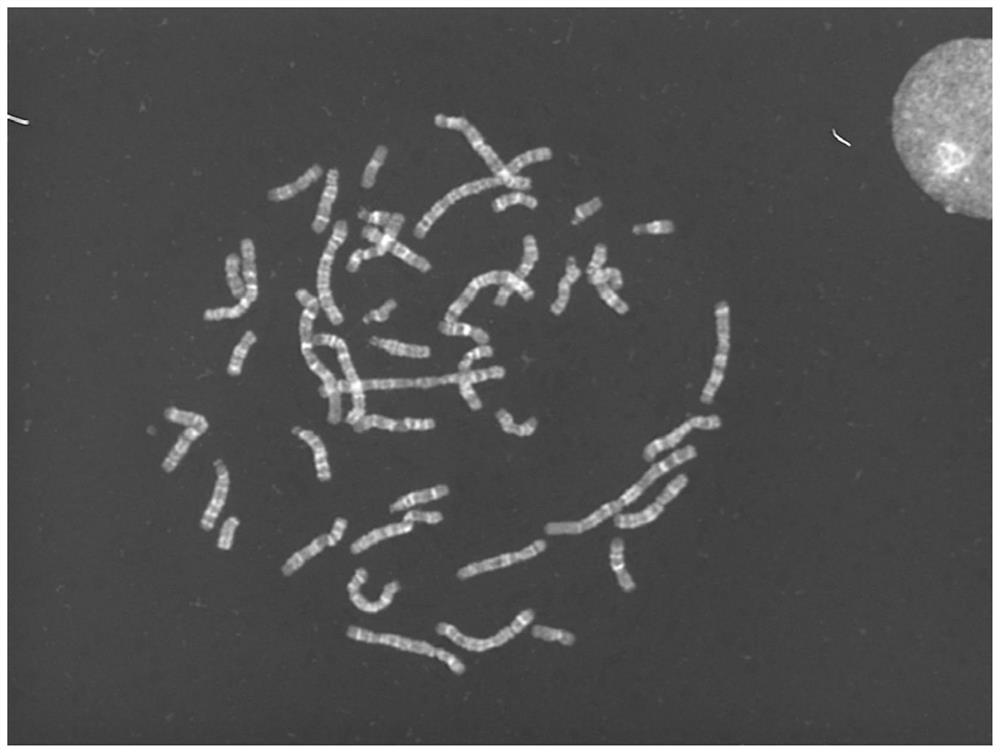
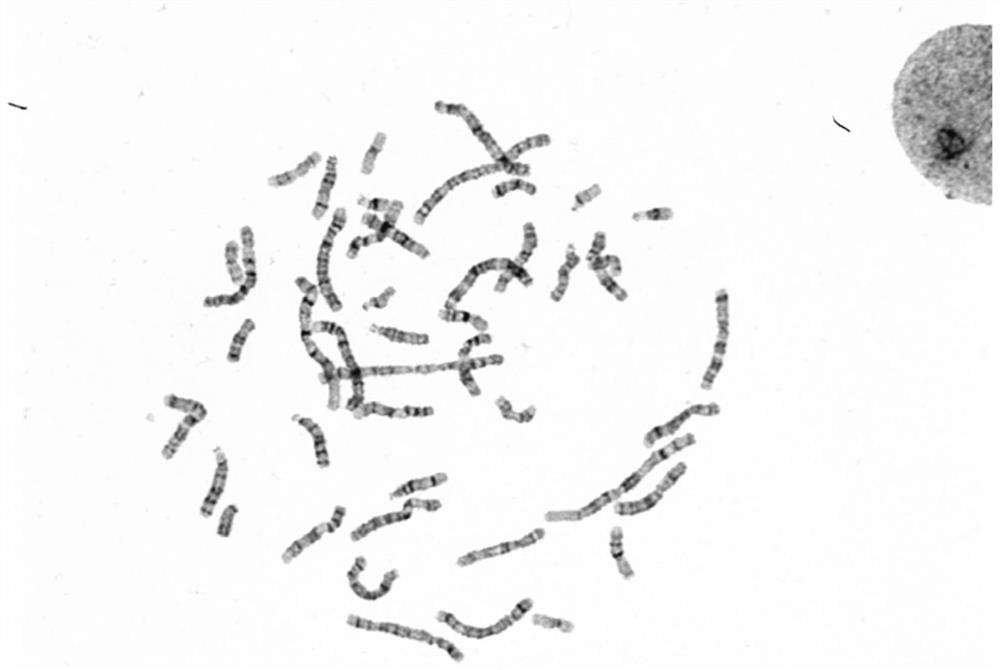
Chromosome image processing method and system
ActiveCN114240883AImprove generalizationImprove general performanceImage enhancementImage analysisImaging processingGenetics
The invention relates to the technical field of chromosome analysis, and provides a chromosome image processing method and system, and the method comprises the following steps: obtaining an original chromosome image and a clear chromosome image with stripes, and obtaining an original image set and a clear chromosome image set; preprocessing the image set, and dividing the image set into a training set and a test set; constructing a chromosome image conversion model based on a cyclic generative adversarial network; respectively inputting the training set and the test set into a chromosome image conversion model for training and testing; acquiring a historical chromosome original image of a target device or system, adding the historical chromosome original image into an original image set, preprocessing the updated image set, and dividing a training set and a test set; and training and testing the chromosome image conversion model through the updated training set and test set, carrying the trained chromosome image conversion model in a target device or system, and converting an original chromosome image obtained by the target device or system in real time into a chromosome image with clear stripes.
Owner:易构智能科技(广州)有限公司
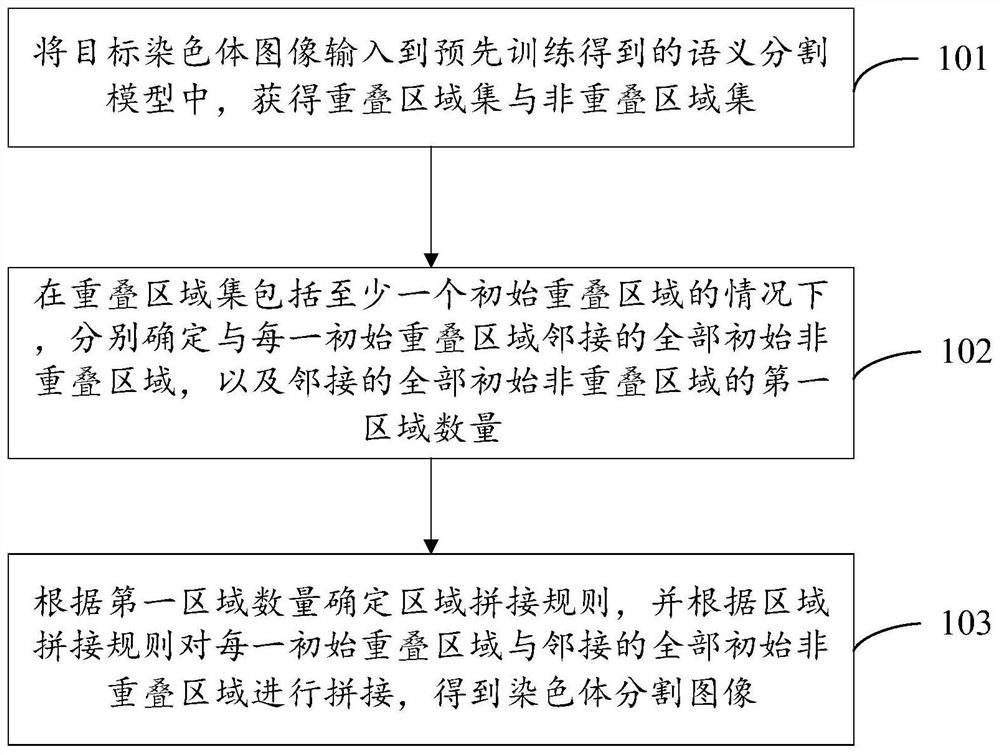
Chromosome segmentation method and device
ActiveCN112037180AImprove Segmentation AccuracyImprove segmentation efficiencyImage enhancementImage analysisPattern recognitionArtificial intelligence
The embodiment of the invention provides a chromosome segmentation method and device, and the method comprises the steps: inputting a target chromosome image into a pre-trained semantic segmentation model, obtaining an overlapping region set and a non-overlapping region set, and enabling the non-overlapping region set to comprise at least one initial non-overlapping region; under the condition that the overlapping region set comprises at least one initial overlapping region, determining all initial non-overlapping regions adjacent to each initial overlapping region and the number of first regions of all the adjacent initial non-overlapping regions; and determining a region splicing rule according to the number of the first regions, and splicing each initial overlapping region with all adjacent initial non-overlapping regions according to the region splicing rule to obtain a chromosome segmentation image. According to the embodiment of the invention, the chromosome segmentation accuracyand segmentation efficiency can be improved.
Owner:HUNAN ZIXING INTELLIGENT MEDICAL TECH CO LTD
Chromosomal enhancement and automatic detection of chromosomal abnormalities using chromosomal ideograms
PendingUS20210295518A1Accurate and sensitive identificationEasy to analyzeImage enhancementImage analysisGeneticsChromosomal Abnormality
A method for detecting chromosomal abnormalities comprising conversion of a chromosomal image into an ideogram which is compared to a control or standard ideogram of a chromosome; or converting an ideogram of a chromosome into a chromosomal image.
Owner:NAT GUARD HEALTH AFFAIRS +2
Small-molecule fluorescent probe and application thereof in chromosome analysis and cytogenetics
ActiveCN111303145AOrganic chemistryMicrobiological testing/measurementHomologous chromosomeMolecular fluorescence
The invention discloses a small-molecule fluorescent probe and an application thereof in chromosome analysis and cytogenetics, and the small-molecule fluorescent probe can be used as a universal toolfor chromosome image segmentation and is used for identifying and distinguishing overlapped, aggregated or contacted chromosomes. The small-molecule fluorescent probe is used as a reference marker forpositioning centromere positions, is used for classifying homologous chromosomes, distinguishing different forms of chromosomes and estimating chromosome arm lengths, is used in cooperation with karyotype analysis for chromosome deletion and polyploid, is used in cooperation with fluorescence in situ hybridization (FISH), and is used for positioning designated genes and judging chromosome abnormalities. The invention provides a rapid and reliable method for auxiliary chromosome analysis, and has great potential in conventional clinical practice.
Owner:陈斯杰 +2
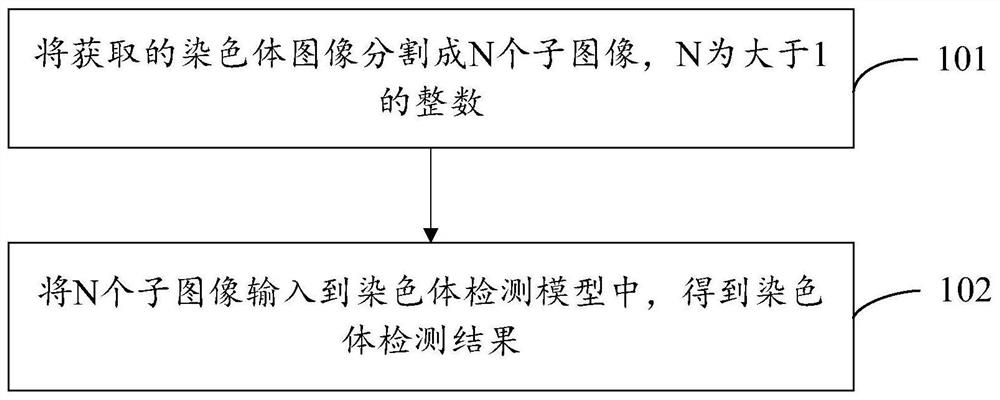
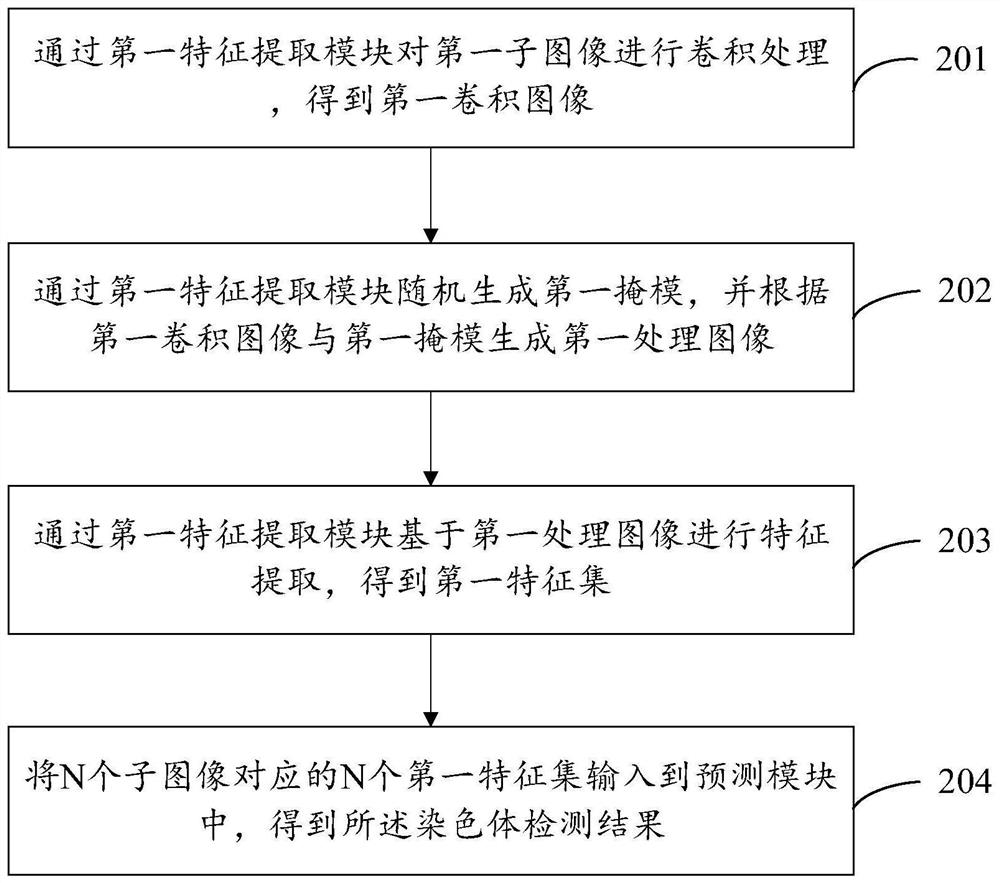
Chromosome detection method and device and electronic equipment
PendingCN112037173AImprove robustnessImprove accuracyImage enhancementImage analysisFeature extractionAlgorithm
The embodiment of the invention provides a chromosome detection method and device and electronic equipment, and the method comprises the steps: segmenting an obtained chromosome image into N sub-images, N being an integer greater than 1; inputting the N sub-images into a chromosome detection model to obtain a chromosome detection result, the chromosome detection model comprising a prediction module and N feature extraction modules, the N feature extraction modules being in one-to-one correspondence with the N sub-images, and the input ends of the N feature extraction modules being used for receiving the N sub-images, wherein the output ends of the N feature extraction modules are connected to the input end of the prediction module, and the output end of the prediction module is used for outputting the chromosome detection result. According to the embodiment of the invention, the problem of over-fitting of the chromosome detection model can be alleviated, and the robustness of the chromosome detection model and the accuracy of chromosome detection are improved.
Owner:HUNAN ZIXING INTELLIGENT MEDICAL TECH CO LTD
Overlapping chromosome segmentation method based on deformable U-shaped network
ActiveCN111414788AHave modeling abilityGood segmentation effectImage enhancementImage analysisTest sampleNetwork structure
The invention discloses an overlapping chromosome segmentation method based on a deformable U-shaped convolutional neural network, so as to further improve the accuracy of the existing segmentation technology. An experiment is performed around a human chromosome image of the same region in the metaphase of mitotic metaphase. The method comprises the following steps: 1) combining DAPI and Cy3 images into a grayscale image, and extracting 46 individual chromosomes one by one; 2) respectively carrying out image enhancement processing on the 46 chromosome images and superposing the 46 chromosome images in pairs to construct an overlapped chromosome image library; 3) dividing the overlapped chromosome image library into a training sample set, a verification sample set and a test sample set; and4) constructing a deformable U-shaped network, completing network training through the training sample set, verifying the sample set to evaluate and test the network, and finally performing overlapping chromosome segmentation on the test sample set. According to the method, various characteristics including chromosome morphological changes such as angles and curl states are extracted by utilizinga deformable U-shaped network structure, so that the accuracy of overlapping chromosome segmentation is improved.
Owner:CHINA UNIV OF MINING & TECH
Automatic overlapping chromosome segmentation method based on adversarial learning multi-scale features
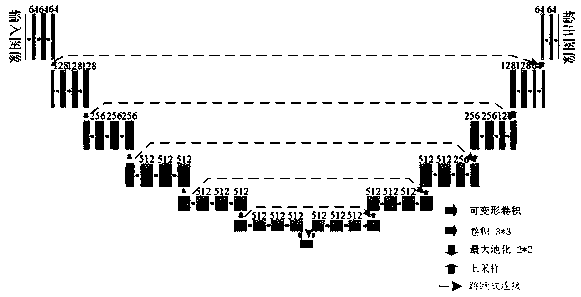
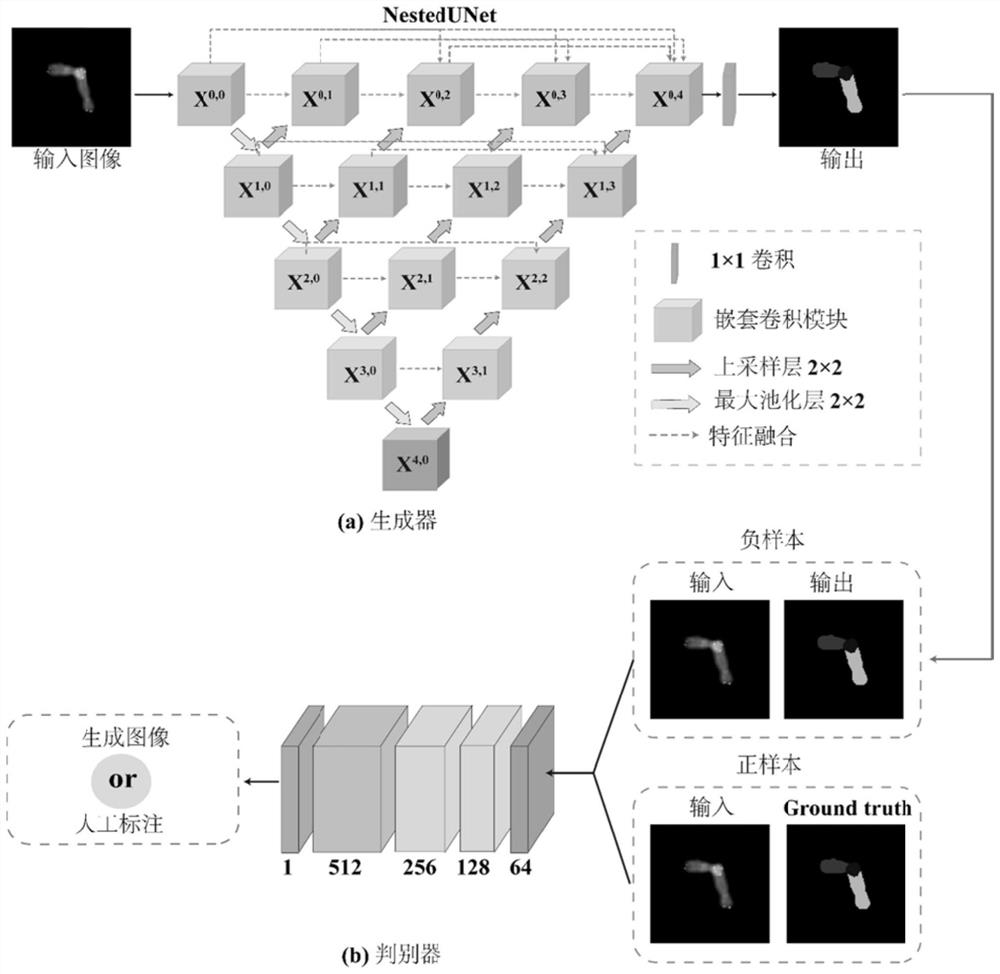
ActiveCN112215847AEasy accessImprove Segmentation AccuracyImage enhancementImage analysisFeature learningGenerative adversarial network
The invention discloses an automatic overlapping chromosome segmentation method based on adversarial learning multi-scale features. Challenges of human chromosome analysis are overlapping of automaticchromosome segmentation, which hinders medical diagnosis and biomedical research. Therefore, the invention provides an adversarial multi-scale feature learning framework, adopts a nested U-shaped network as a generator, and aims to explore 'optimal' representation of chromosome images by using multi-scale features. Conditional generative adversarial network (cGAN) adversarial learning are used topromote output distribution to be closer to a gold standard image; a least square GAN target is adopted to improve the training stability of the framework; and the continuity optimization is carriedout by using the Lovasz-Softmax loss, so that better performance is obtained. Experimental results show that the method is superior to other traditional algorithms in subjective visual effect and objective evaluation standard.
Owner:WUHAN UNIV
Chromosome cluster and chromosome instance identification method and system and storage medium
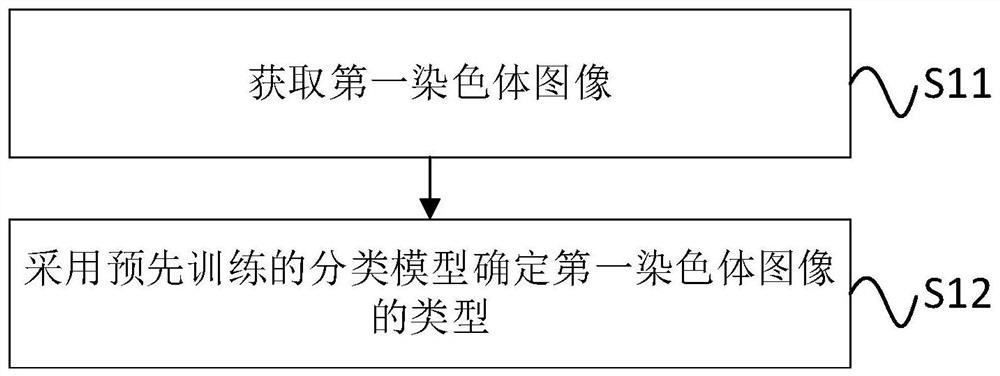
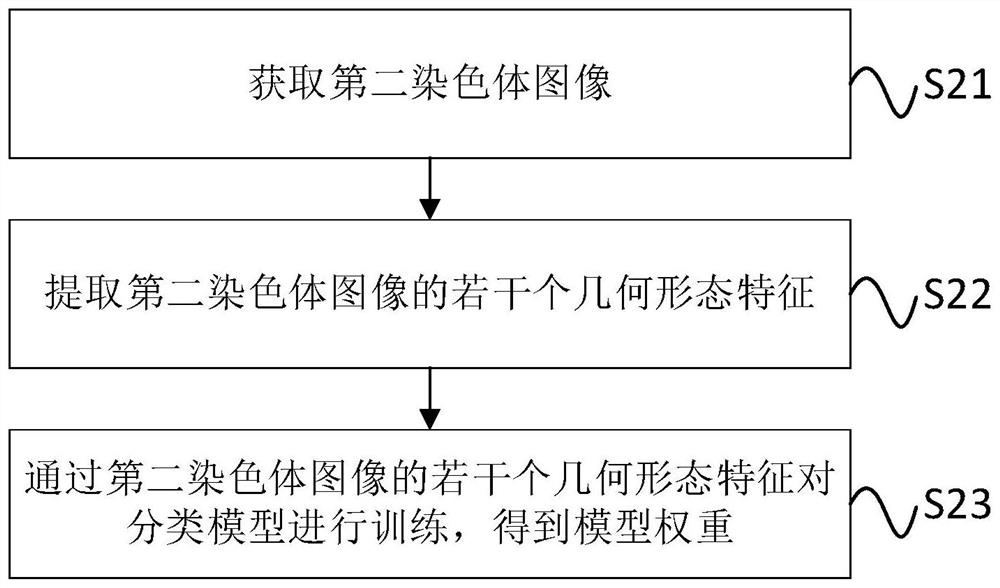
ActiveCN112487941AQuick classificationReduce workloadRecognition of DNA microarray patternMolecular structuresPattern recognitionImage extraction
The invention discloses a chromosome cluster and chromosome instance recognition method and system and a storage medium. The method comprises the following steps: obtaining a first chromosome image which is a to-be-recognized chromosome image; determining the type of the first chromosome image by adopting a pre-trained classification model, wherein the training step of the classification model comprises the following steps: acquiring a second chromosome image; extracting a plurality of geometrical morphology features of the second chromosome image; and training the classification model througha plurality of geometrical morphology features of the second chromosome image to obtain a model weight. According to the method, the classification model is trained by extracting the plurality of geometrical morphology features of the second chromosome image to obtain the model weight, and then the type of the chromosome image to be identified is determined through the pre-trained classificationmodel, so that the workload of workers is reduced, and the accuracy of the identification result of the chromosome cluster and the chromosome instance is improved. The method can be applied to the technical field of chromosome processing.
Owner:SOUTH CHINA NORMAL UNIVERSITY
R-band chromosome karyotype image identification method and system based on grouping information assistance
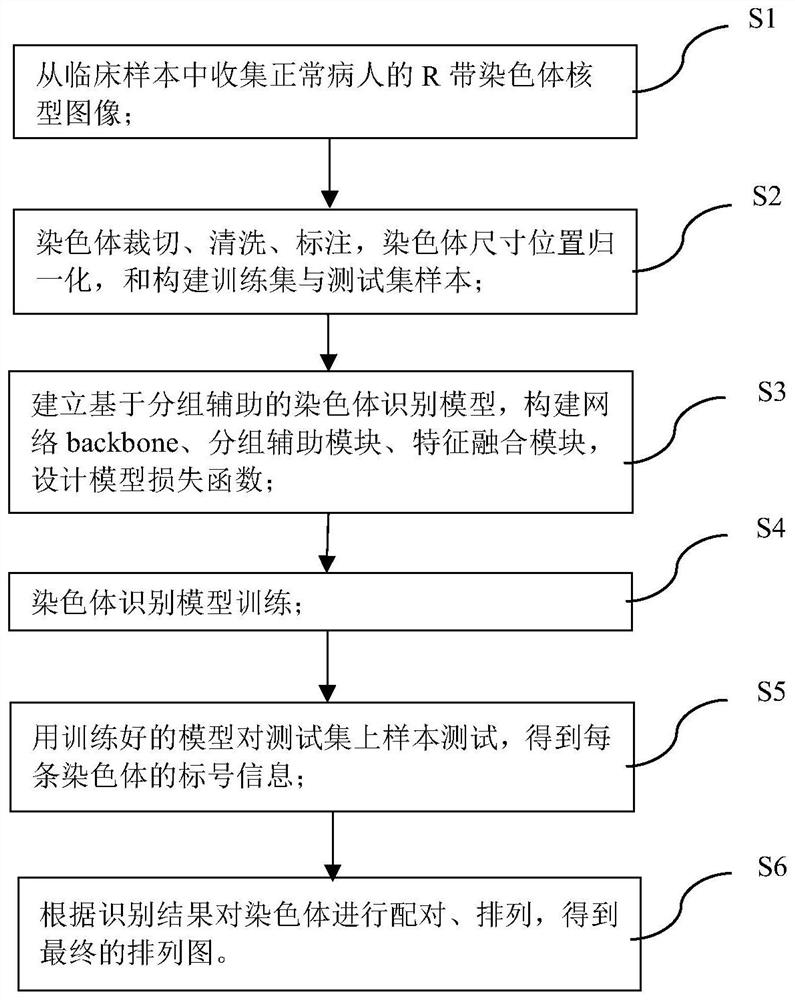
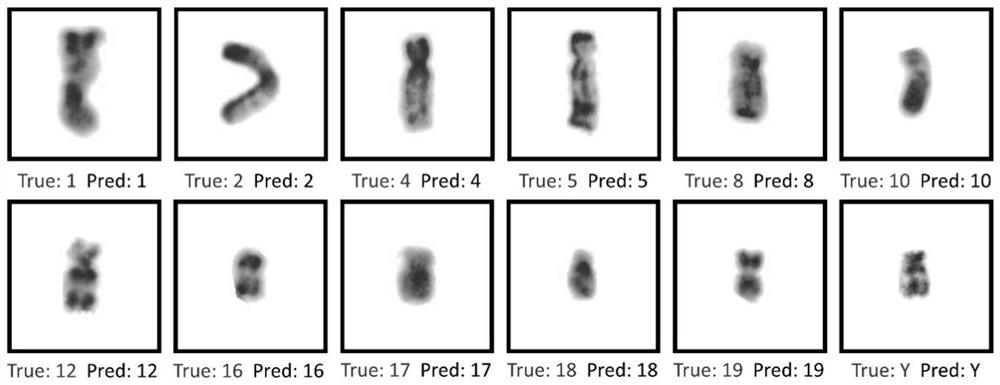
PendingCN114550170AAutomatic classification of accuracySort by accuracyAcquiring/recognising microscopic objectsNeural architecturesPattern recognitionMonosomal karyotype
The invention provides an R-band chromosome karyotype image identification method based on grouping information assistance. The method comprises the following steps: obtaining a chromosome image; performing normalization processing on the chromosome image; a dyeing identification model is constructed, the chromosome identification model takes chromosome length grouping and centroid position grouping as auxiliary tasks, and chromosome identification is taken as a main task; training the chromosome recognition model; and using the trained chromosome identification model to predict the category. According to the method, the analysis efficiency of the karyotype is effectively improved, automatic classification and sorting of the chromosomes are accurately completed, the process is simple, and the method has a relatively great application prospect.
Owner:SHANGHAI JIAO TONG UNIV
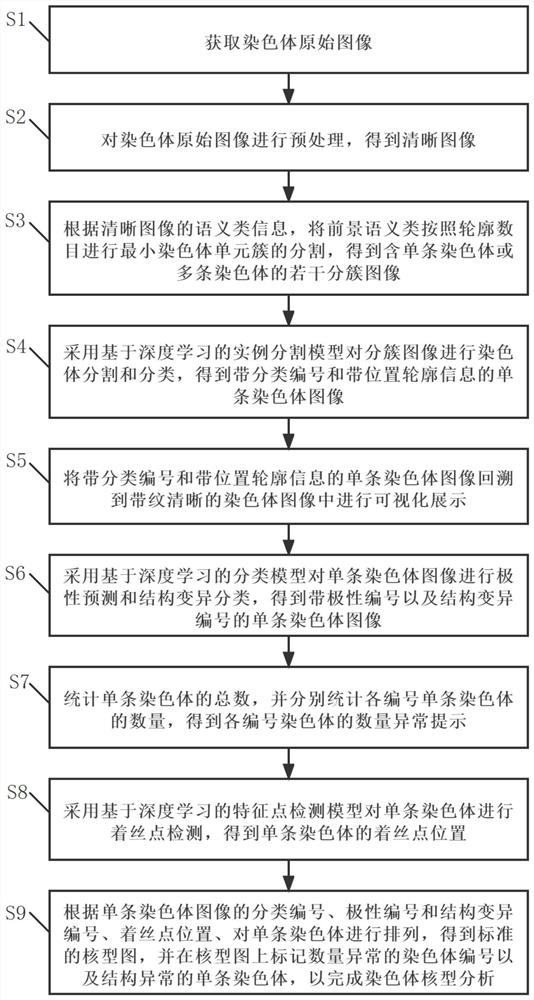
Deep learning-based karyotype analysis method and system
ActiveCN114219786AImprove work efficiencyEasy to operateImage enhancementImage analysisPattern recognitionMonosomal karyotype
The invention relates to the technical field of karyotype analysis, and provides a karyotype analysis method and system based on deep learning, and the method comprises the steps: obtaining a chromosome original image; preprocessing the original chromosome image to obtain a clear image; according to the semantic class information of the clear image, performing minimum chromosome unit cluster segmentation on the foreground semantic class according to the contour number to obtain a plurality of clustered images containing a single chromosome or a plurality of chromosomes; carrying out chromosome segmentation and classification on the clustered images by adopting a chromosome instance segmentation model based on deep learning to obtain single chromosome images with classification numbers and contour information; performing polarity prediction and structural variation classification on the chromosome images by adopting a classification model, performing centroid detection on the chromosomes by adopting a feature point detection model, and finally arranging the chromosomes according to classification numbers, polarity numbers, structural variation numbers and centroid positions of the chromosome images. And obtaining a standard karyotype graph to finish chromosome karyotype analysis.
Owner:易构智能科技(广州)有限公司
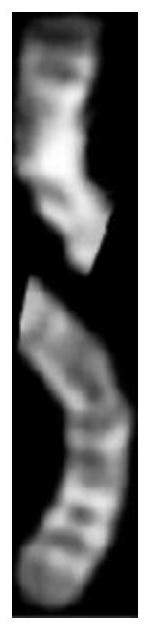
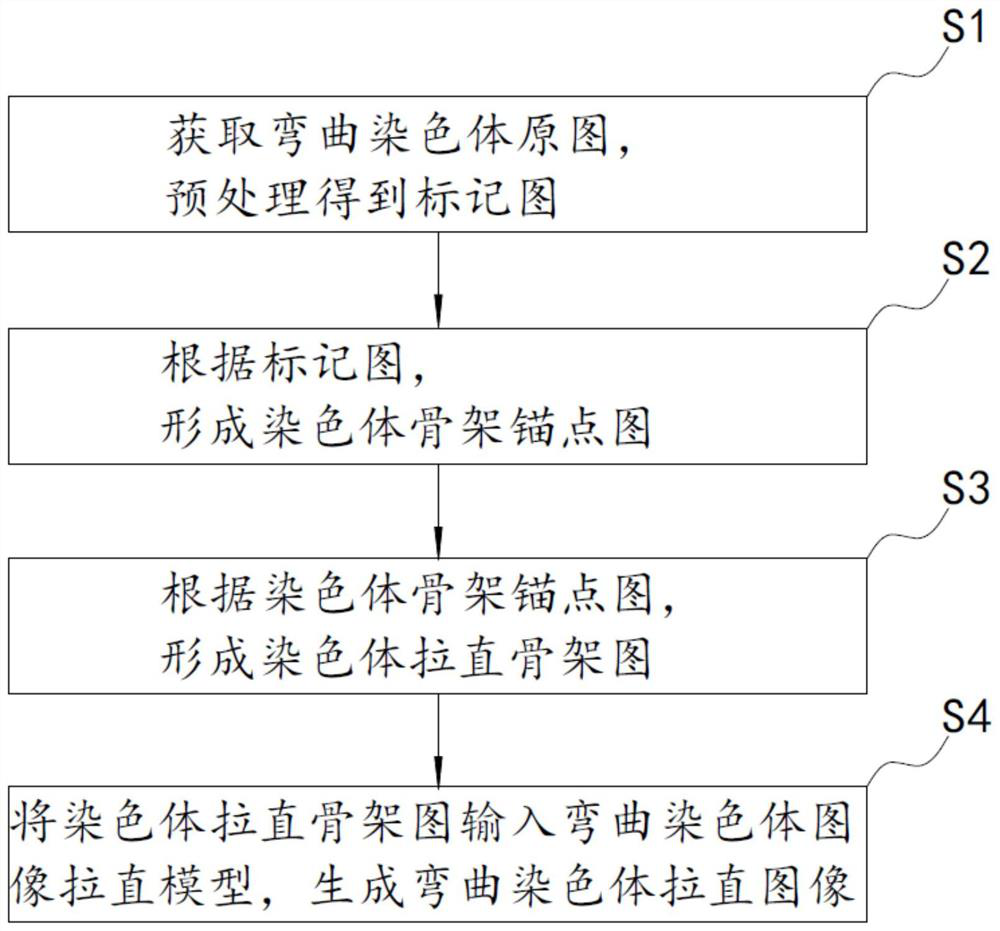
Curved chromosome image straightening method and system based on BagPix2Pix self-learning model, storage medium and device
PendingCN111612745AImprove integrityImprove continuityImage enhancementImage analysisAlgorithmGenetics
The invention discloses a curved chromosome image straightening method and system, based on a BagPix2Pix self-learning model, a storage medium and a device. The method comprises the following steps: S1, receiving a curved chromosome original image, and processing the curved chromosome original image to obtain a mark image; S2, generating a chromosome skeleton anchor point graph according to the marker image obtained in the step S1; S3, generating a chromosome straightening skeleton diagram according to the chromosome skeleton anchor point diagram; and S4, inputting the chromosome straighteningskeleton diagram into a curved chromosome image straightening model which is trained to be convergent and can receive a prediction diagram which can be output and matched by the skeleton diagram, andgenerating a curved chromosome straightening image. Compared with an existing method based on iconography, the method has the advantages that the result of the method does not contain obvious fractures and sections, the quality of the straightened chromosome image and the integrity and continuity of effective features are greatly improved, the method is not affected by the number of curves and curved parts, and the accuracy is high.
Owner:XIAN JIAOTONG LIVERPOOL UNIV +1
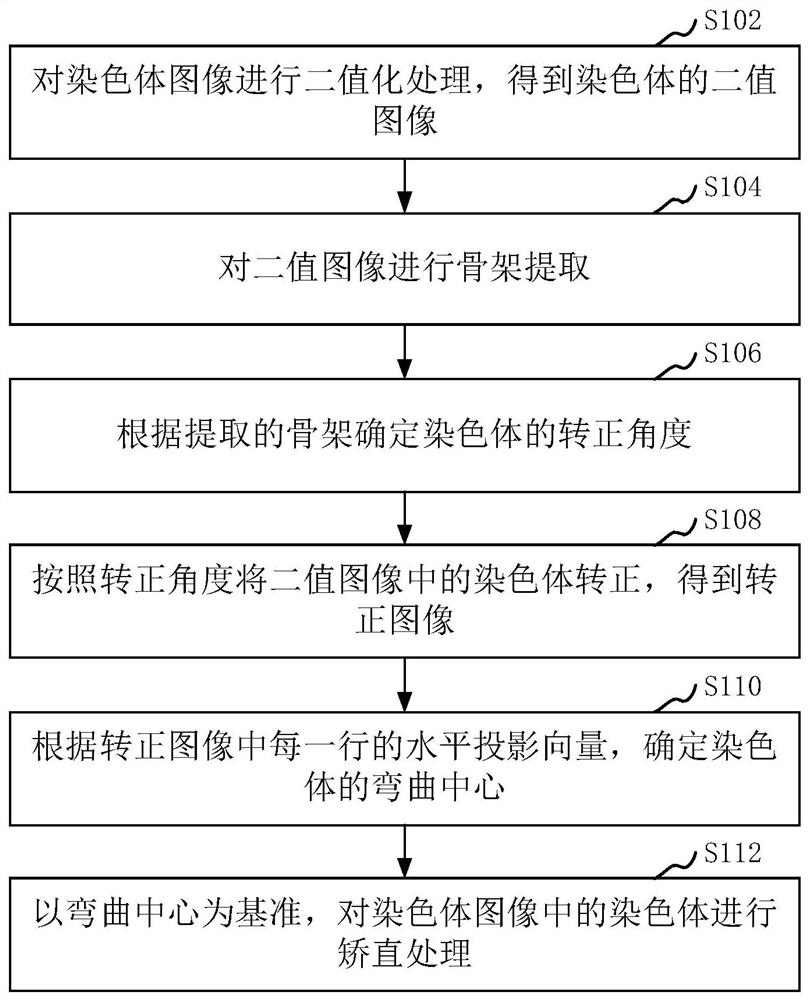
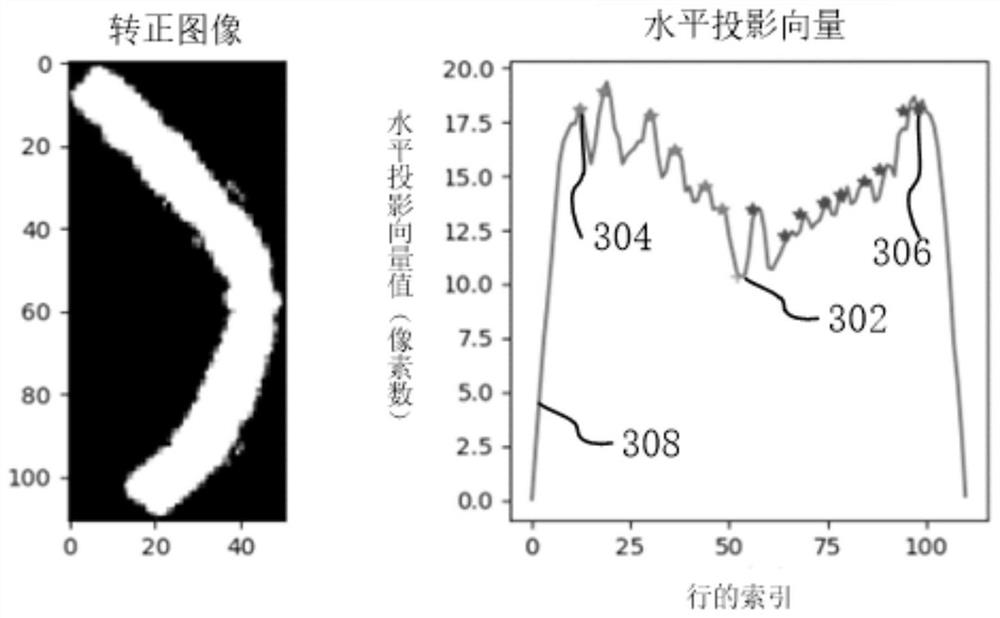
Chromosome straightening method and device, computer equipment and storage medium
The invention relates to a chromosome straightening method and device, computer equipment and a storage medium. The method comprises the following steps: carrying out binarization processing on a chromosome image to obtain a binary image of a chromosome; performing skeleton extraction on the binary image; determining a positive rotation angle of the chromosome according to the extracted skeleton; correcting the chromosome in the binary image according to the correcting angle to obtain a corrected image; determining the bending center of the chromosome according to the horizontal projection vector of each row in the corrected image; and carrying out straightening processing on the chromosome in the chromosome image by taking the bending center as a reference. By adopting the method, the accuracy of chromosome straightening can be improved.
Owner:SHENZHEN REETOO BIOTECHNOLOGY CO LTD
Chromosome stripe image enhancement method and system, intelligent terminal and storage medium
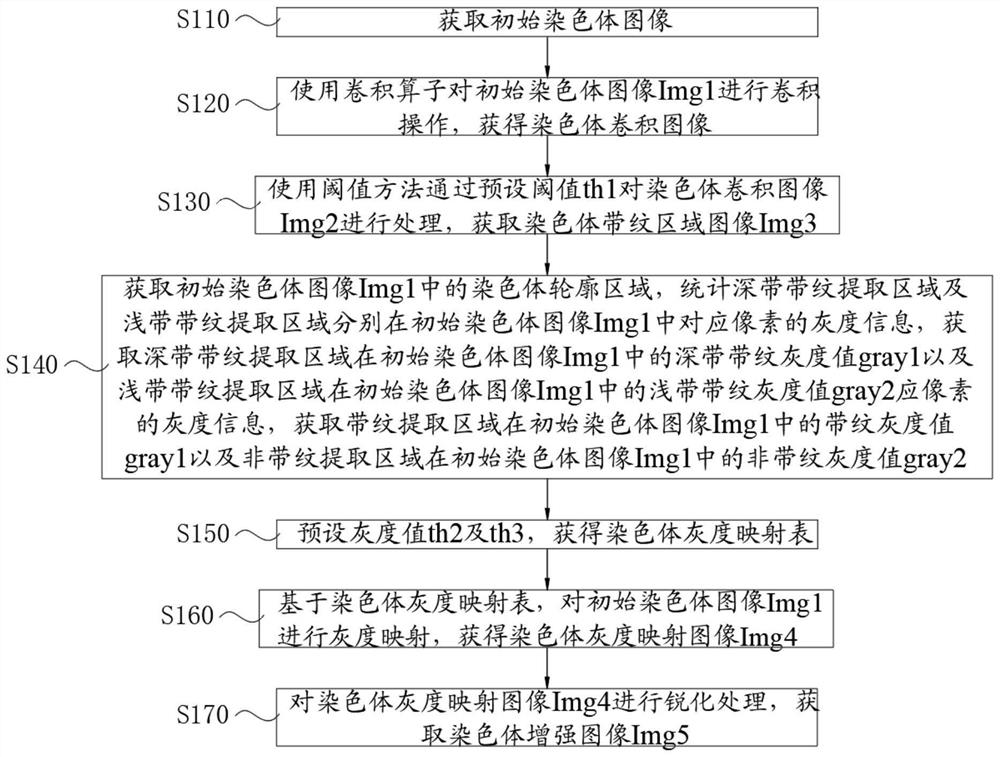
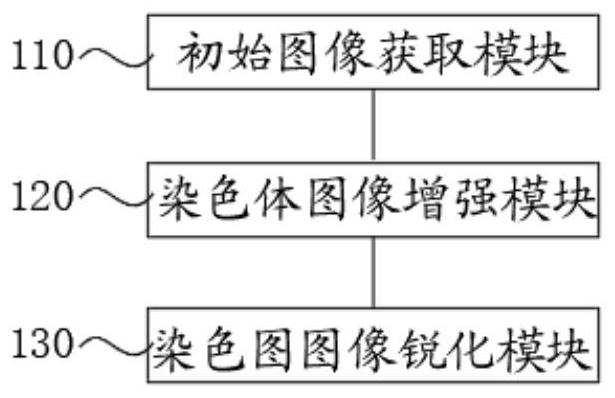
PendingCN114155162ANo loss of banding detailEfficient expressionImage enhancementImage analysisImaging analysisImaging quality
The invention discloses a chromosome stripe image enhancement method and system, an intelligent terminal and a storage medium. The method comprises the following steps: acquiring an initial chromosome image; obtaining a chromosome convolution image; obtaining a chromosome deep zone stripe region image Img3; obtaining a deep belt stripe gray value gray1 and a shallow belt stripe gray value gray2; obtaining a chromosome gray mapping table; obtaining a chromosome gray mapping image Img4; and obtaining a chromosome enhanced image Img5. According to the method, through stripe enhancement and sharpening processing, main stripes of chromosomes in the processed image are obvious, main features of each type of chromosomes can be highlighted, transition between secondary deep stripes and shallow stripes is obvious, stripe details of the chromosomes are not lost, and sharpening is carried out, so that the chromosome image becomes clearer; the chromosome image stripe expression is obvious, the features are prominent, the image analysis is facilitated, the working efficiency is improved, and the problem of large chromosome image quality floating caused by a chromosome dyeing technology and imaging hardware is solved.
Owner:HUNAN ZIXING INTELLIGENT MEDICAL TECH CO LTD
Chromosome division phase positioning and sequencing method
InactiveCN110879996AAccurate identificationEfficient identificationAcquiring/recognising microscopic objectsNeural architecturesFeature extractionAlgorithm
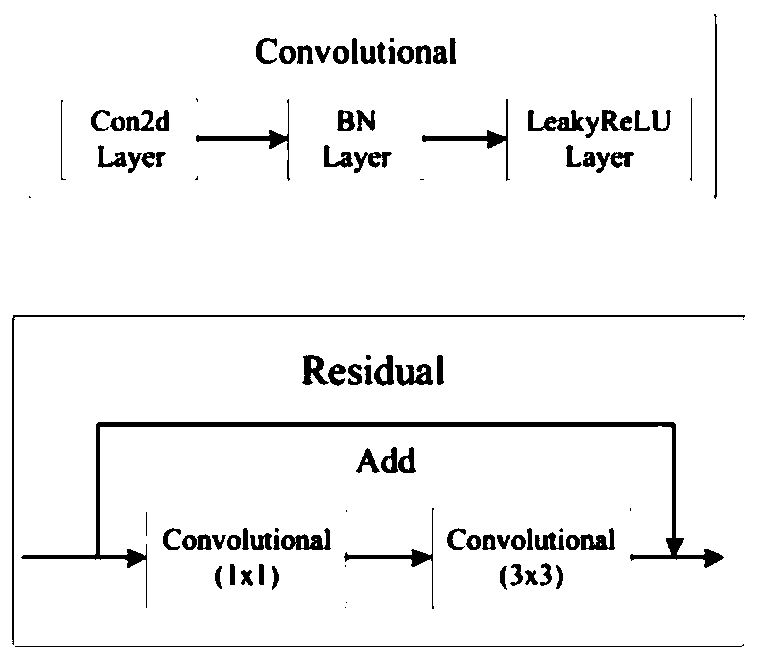
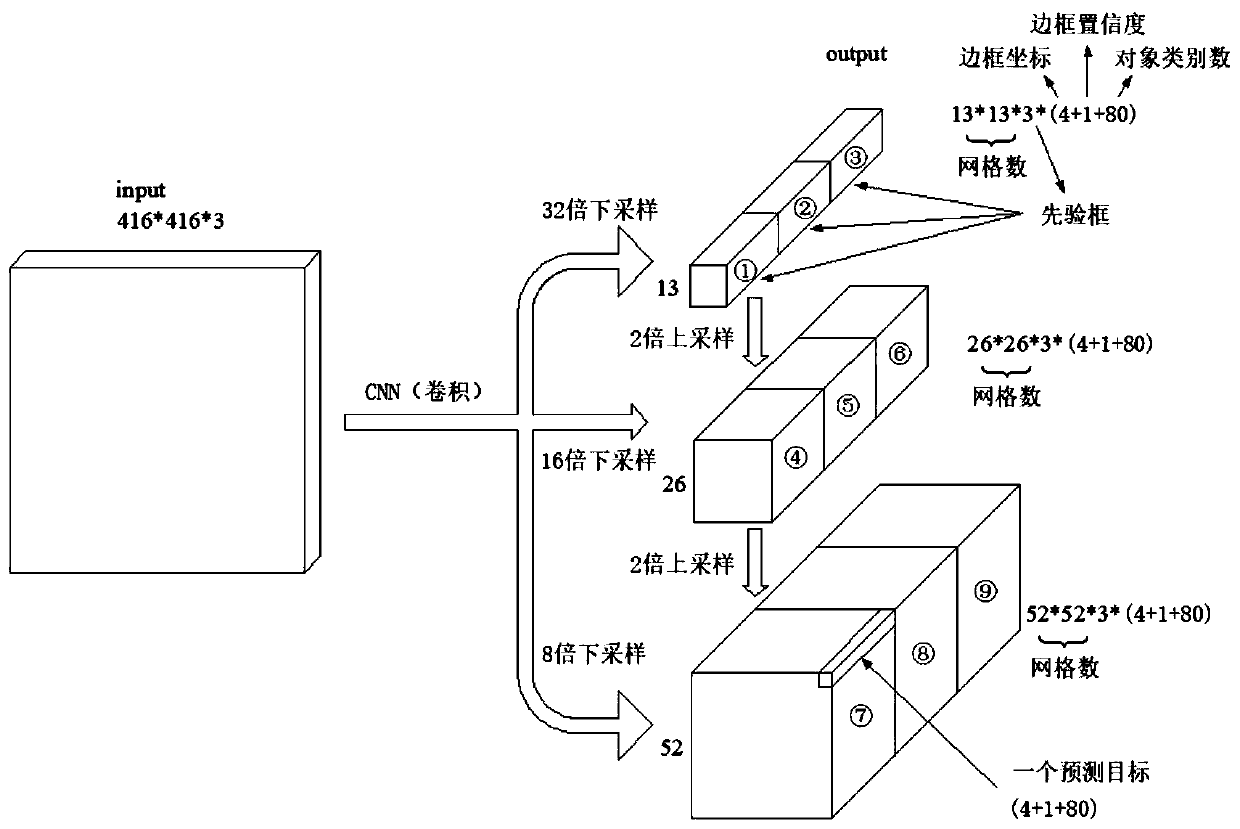
The invention discloses a chromosome division phase positioning and sequencing method, and belongs to the technical field of chromosome recognition. The chromosome division phase positioning and sequencing method comprises the following steps: 1) establishing an identification model based on a darknet53 basic network; 2) obtaining an independent chromosome image, and inputting the image accordingto a specified size; 3) performing feature extraction on the input chromosome image by using an identification model, setting three downsampling scales for the structure of the identification model, setting three priori boxes for each downsampling scale, and clustering the priori boxes with nine sizes in total; and 4) carrying out frame prediction by using the output of logistic. According to theinvention, an identification model is established based on a darknet53 basic network; characteristic extraction is performed on the input chromosome image by using the recognition model, and frame prediction is performed quickly, so that the chromosome type can be recognized accurately and efficiently, and compared with the existing recognition technology, the analysis efficiency of the chromosomekaryotype can be effectively improved, the recognition sorting time is shortened, and the chromosome sorting is completed with high accuracy.
Owner:SHANGHAI BEION MEDICAL TECH CO LTD
Chromosome recognition method and device based on deep learning, and computer equipment
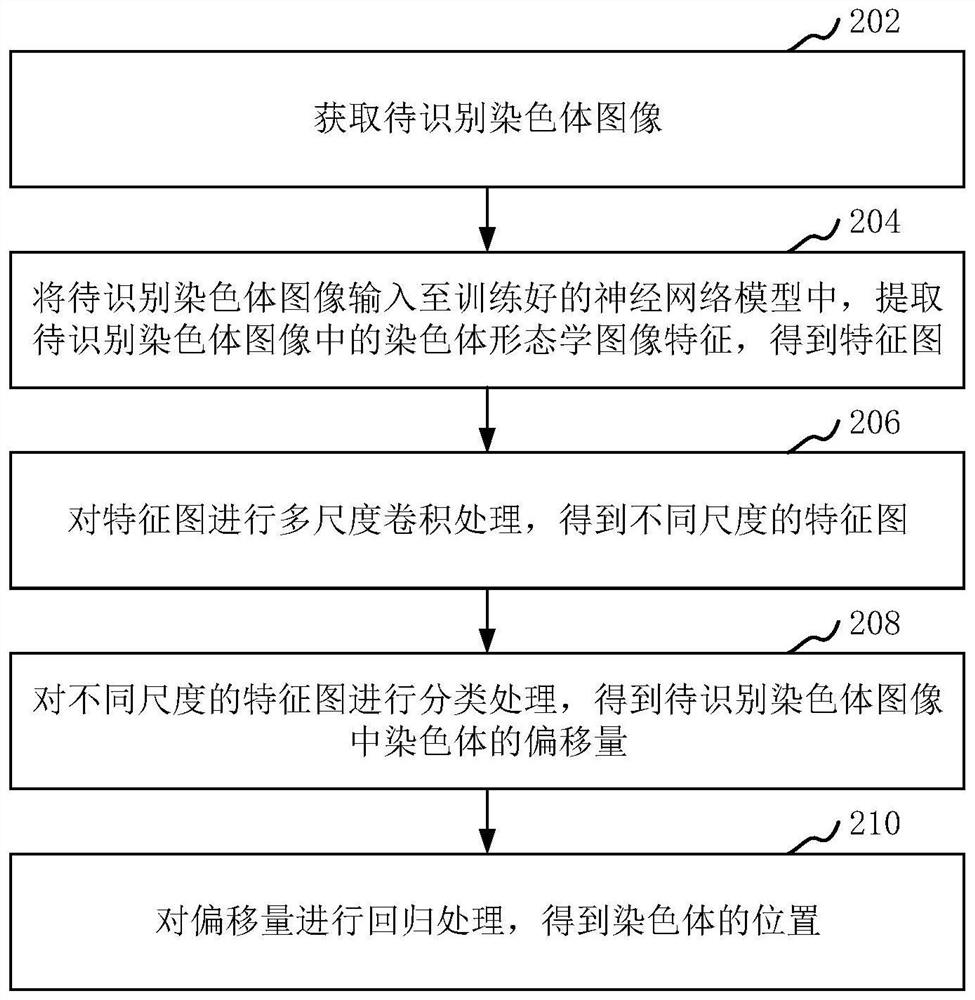
PendingCN112330652AImprove accuracyAccurate identification of chromosome positionsImage enhancementImage analysisPattern recognitionNetwork model
The invention relates to a chromosome recognition method and device based on deep learning, computer equipment and a storage medium. The method comprises the following steps of acquiring a chromosomeimage to be identified; inputting a chromosome image to be recognized into the trained neural network model, and extracting chromosome morphological image features in the chromosome image to be recognized to obtain a feature map; performing multi-scale convolution processing on the feature map to obtain feature maps of different scales; classifying the feature maps of different scales to obtain the offset of chromosomes in the to-be-identified chromosome image; and performing regression processing on the offset to obtain the position of the chromosome. By adopting the method, chromosome recognition accuracy can be improved.
Owner:SHENZHEN UNIV
Convolutional neural network-based chromosome important feature visualization method and device
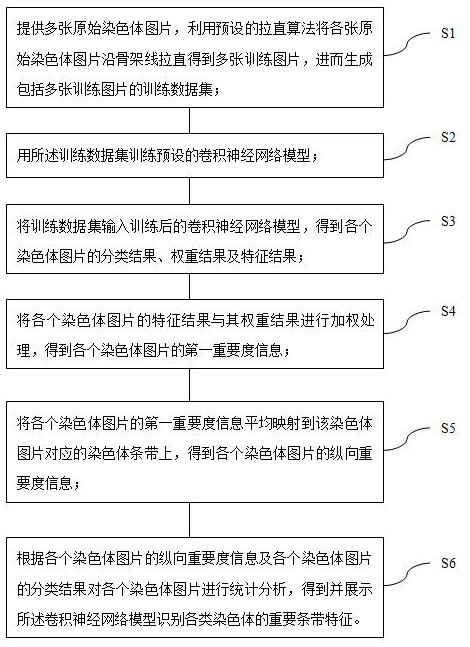
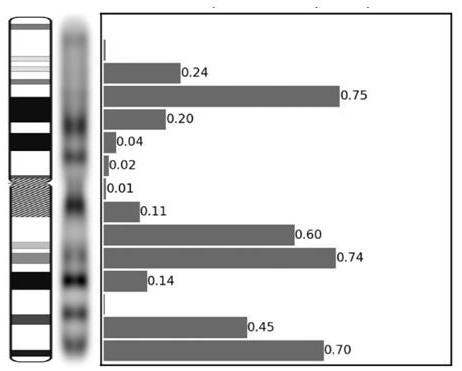
ActiveCN113724793AEasy to findRealize visual displayData visualisationBiostatisticsData setStatistical analysis
The invention provides a chromosome important feature visualization method based on a convolutional neural network. The method comprises the following steps: training a preset convolutional neural network model by using a training data set; inputting the training data set into the trained convolutional neural network model to obtain a classification result, a weight result and a feature result of each chromosome picture; multiplying the feature result and the weight result of each chromosome picture to obtain first importance information of each chromosome picture; averagely mapping the first importance information of each chromosome picture to a chromosome stripe corresponding to the chromosome picture to obtain longitudinal importance information; performing statistical analysis on each chromosome picture according to the longitudinal importance information and the classification result of each chromosome picture, obtaining and displaying the important stripe features of the convolutional neural network model for identifying various chromosomes, and realizing visual display of the chromosome important stripe features of the convolutional neural network for discriminating chromosome categories.
Owner:HUNAN ZIXING INTELLIGENT MEDICAL TECH CO LTD
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com