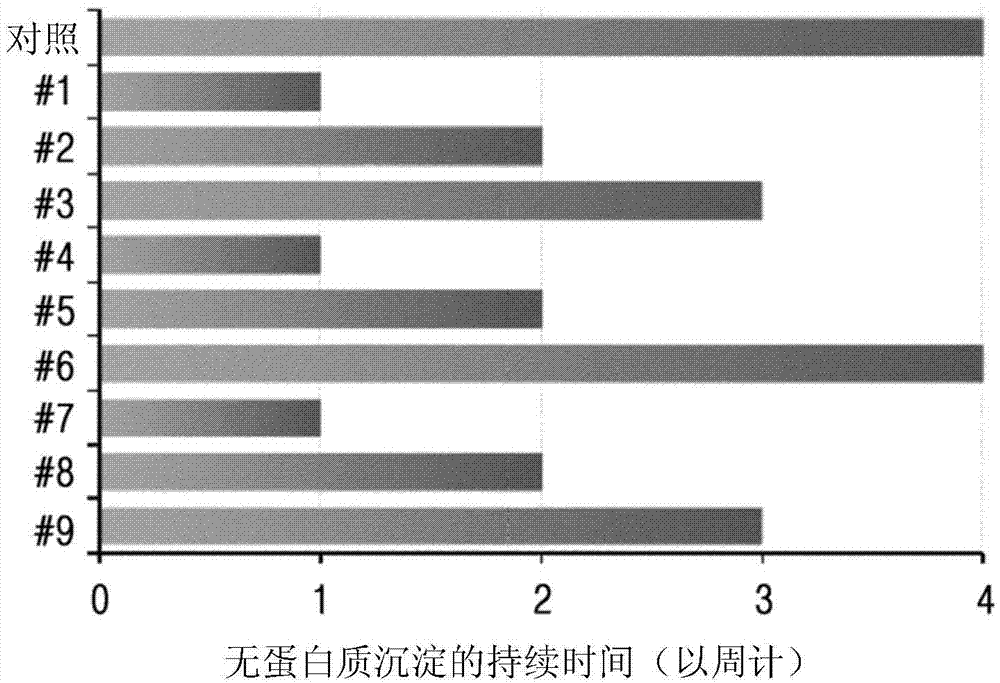
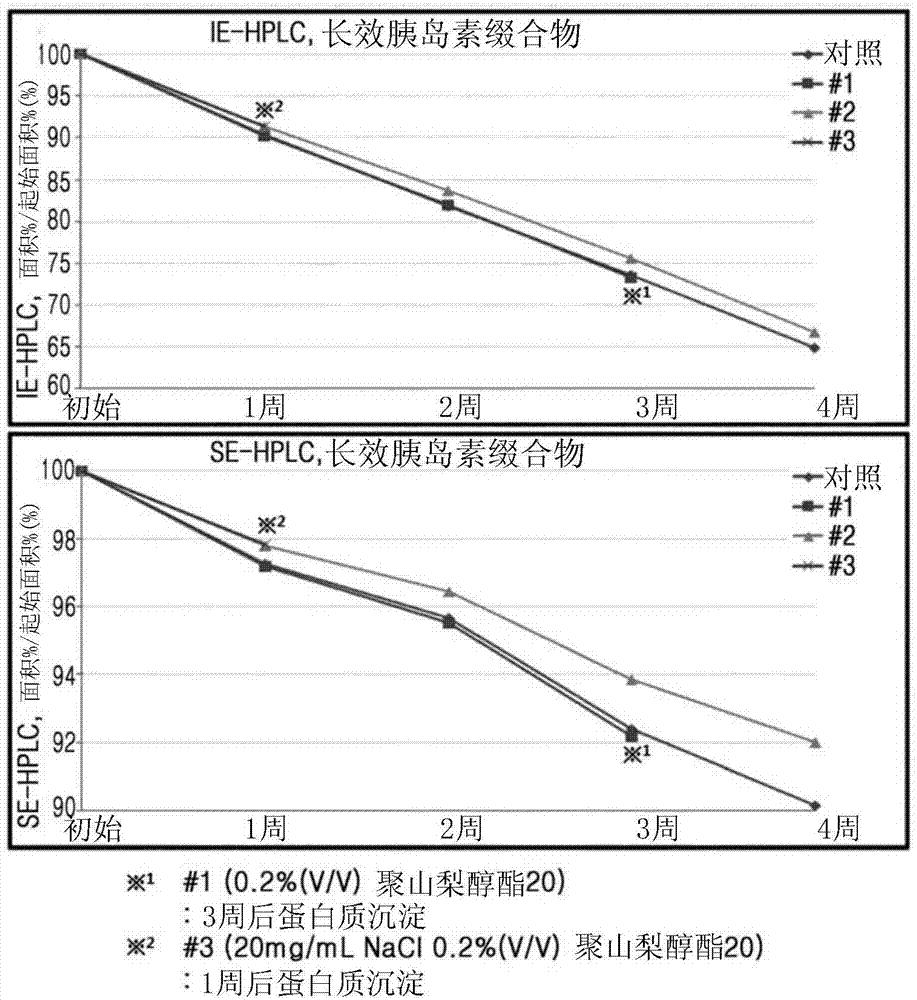
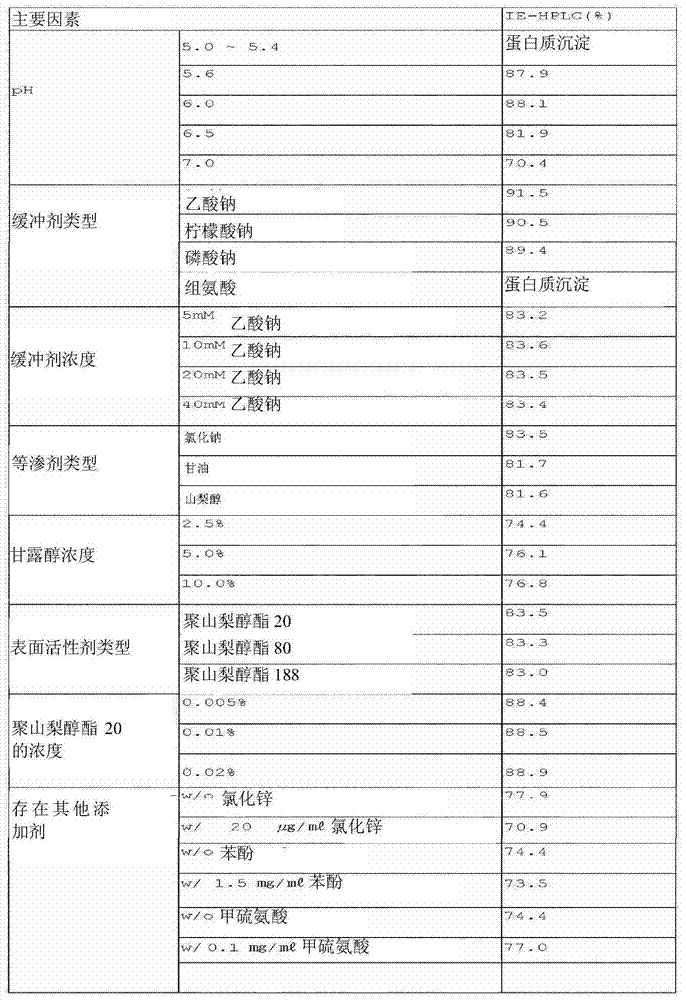
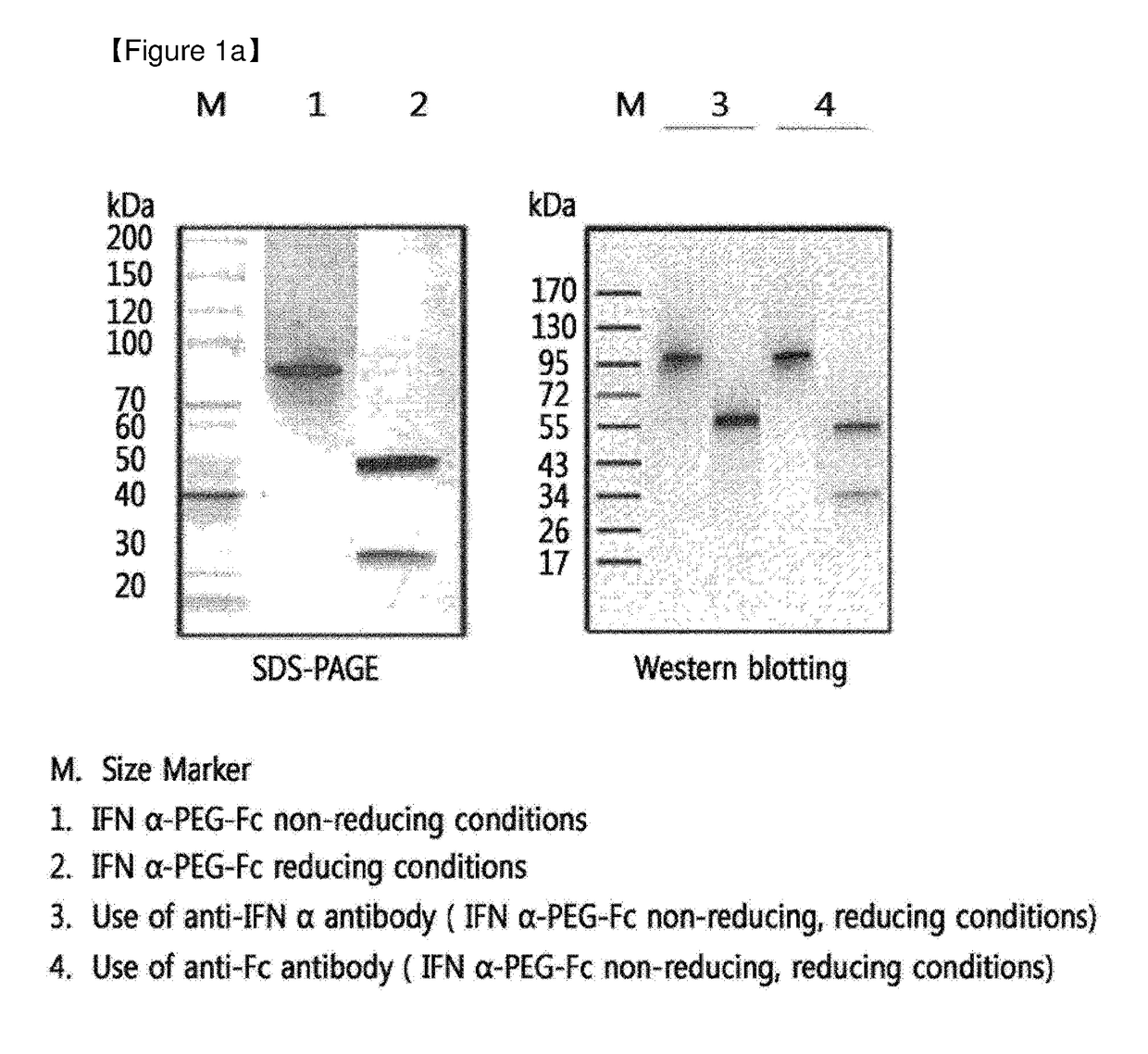
Patents
Literature
64 results about "Immunoglobulin region" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
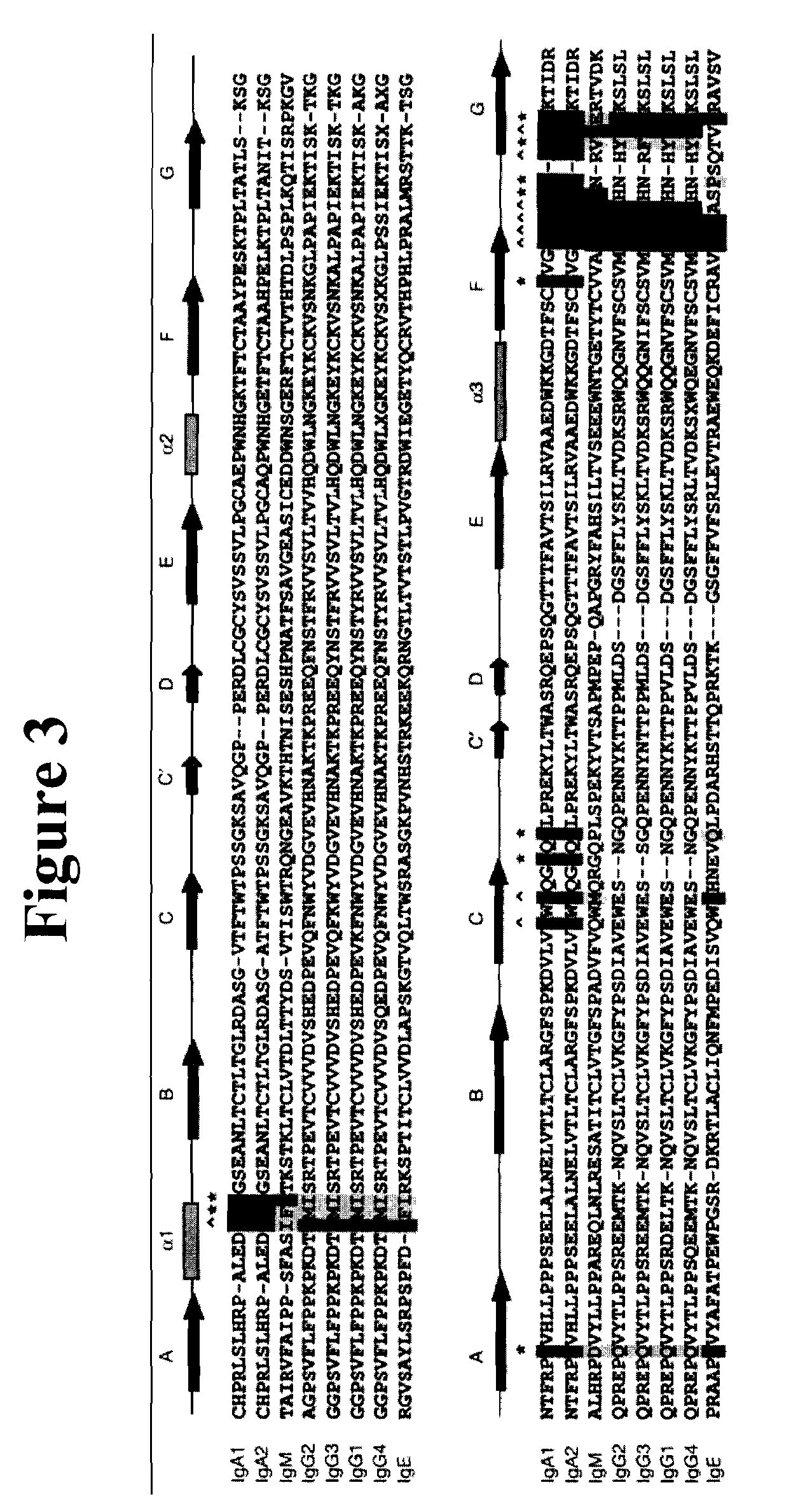
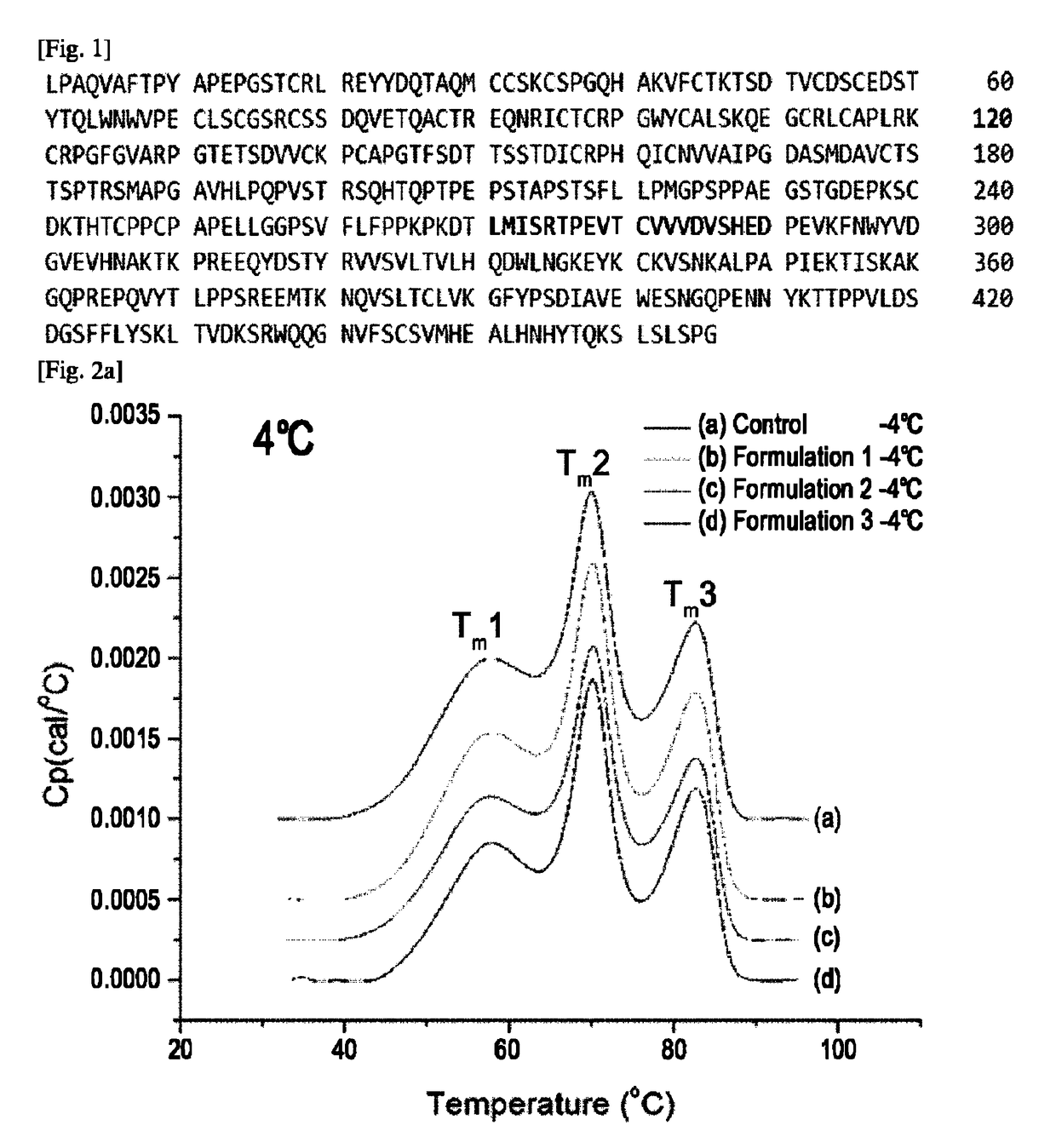
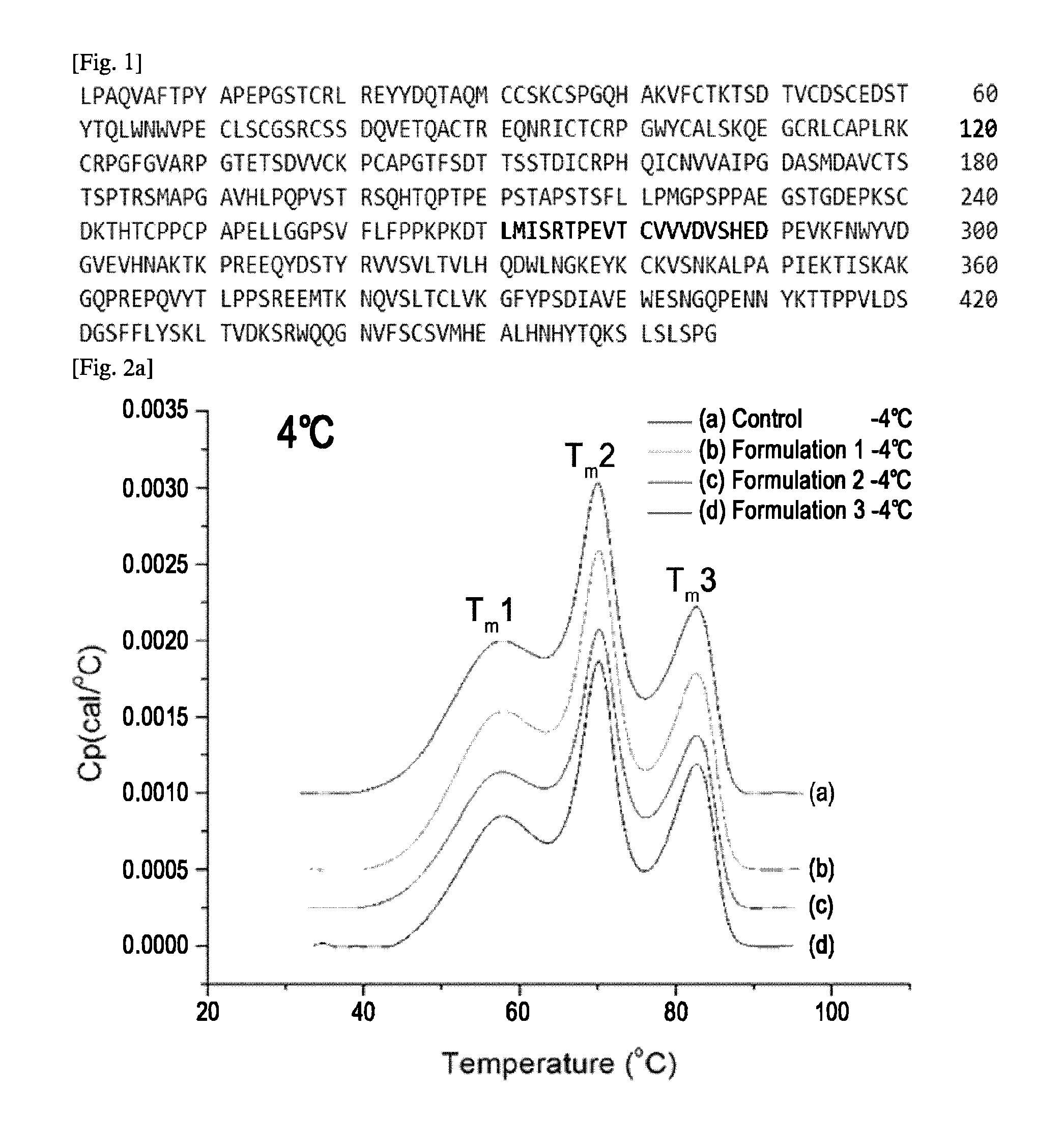
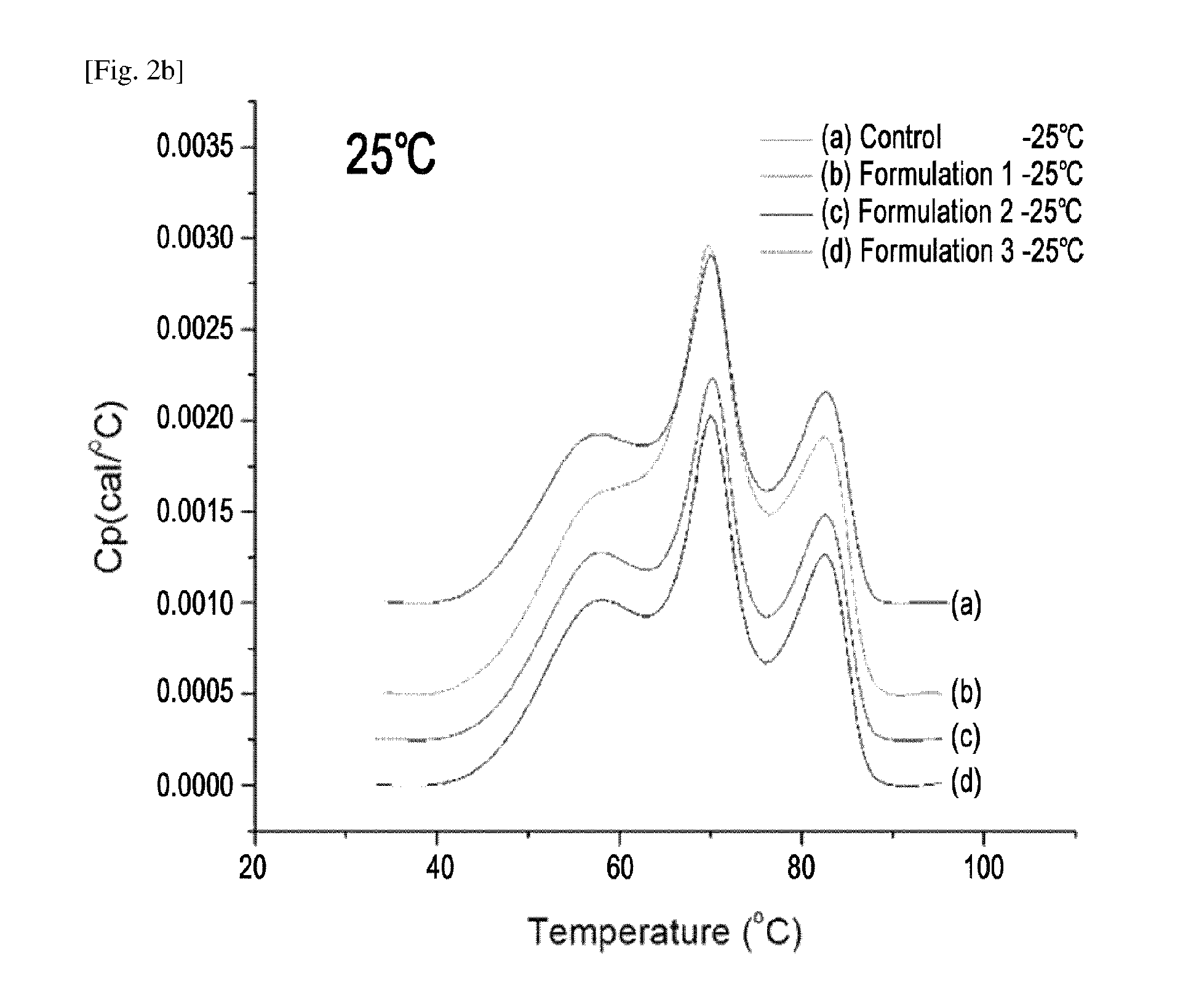
The variable region is the region of the immunoglobulin (Ig) which varies greatly in amino acid sequence among different immunoglobulins of the same class. Each immunoglobulin molecules is a tetramer of two identical light chains and two identical heavy chains linked by disulfide bonds.
Heterodimer Binding Proteins and Uses Thereof
InactiveUS20130129723A1Animal cellsFused cellsImmunoglobulin Joining RegionImmunoglobulin light chain
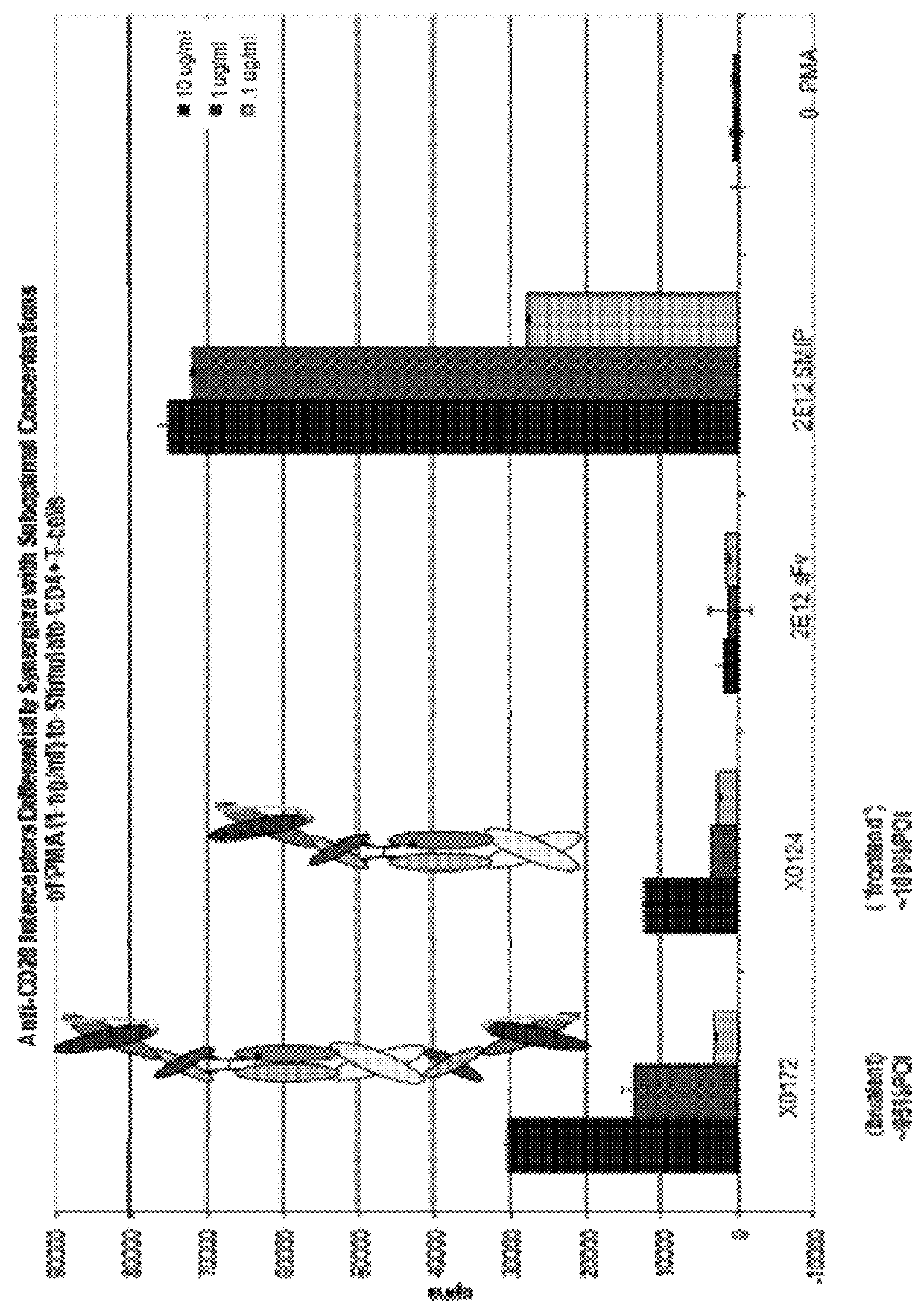
The present disclosure provides polypeptide heterodimers formed between two different single chain fusion polypeptides via natural heterodimerization of an immunoglobulin CH1 region and an immunoglobulin light chain constant region (CL). The polypeptide heterodimer comprises two or more binding domains that specifically bind one or more targets (e.g., a receptor). In addition, both chains of the heterodimer further comprise an Fc region portion. The present disclosure also provides nucleic acids, vectors, host cells and methods for making polypeptide heterodimers as well as methods for using such polypeptide heterodimers, such as in directing T cell activation, inhibiting solid malignancy growth, and treating autoimmune or inflammatory conditions.
Owner:EMERGENT PRODUCTS DEVELOPMENT SEATTLE LLC
Binding proteins comprising immunoglobulin hinge and fc regions having altered fc effector functions
Provided herein are binding proteins comprising one or more immunoglobulin Fc region hinge, CH2, and / or CH3 domain wherein one or more hinge and / or constant region CH2 and / or CH3 domain is modified to alter the binding protein's binding affinity and / or specificity for a cognate receptor (e.g., an Fc receptor) and / or to impart one or more new binding specificity(ies) to the hinge and / or constant region that the corresponding unmodified immunoglobulin does not possess (e.g., affinity for distinct class of cognate receptor distinct from the class of cognate receptor to which the unmodified binding protein specifically binds). Binding proteins according to the present invention include, for example, modified antibodies, antibody fragments, recombinant binding proteins, and molecularly engineered binding domain-immunoglobulin fusion proteins, including small modular immunopharmaceutical products (SMIP™ products).
Owner:TRUBION PHARM INC
OB fusion protein compositions and methods
InactiveUS6936439B2Stability advantageStable rateObesity gene productsBacteriaImmunoglobulin regionOb Protein
Owner:AMGEN INC
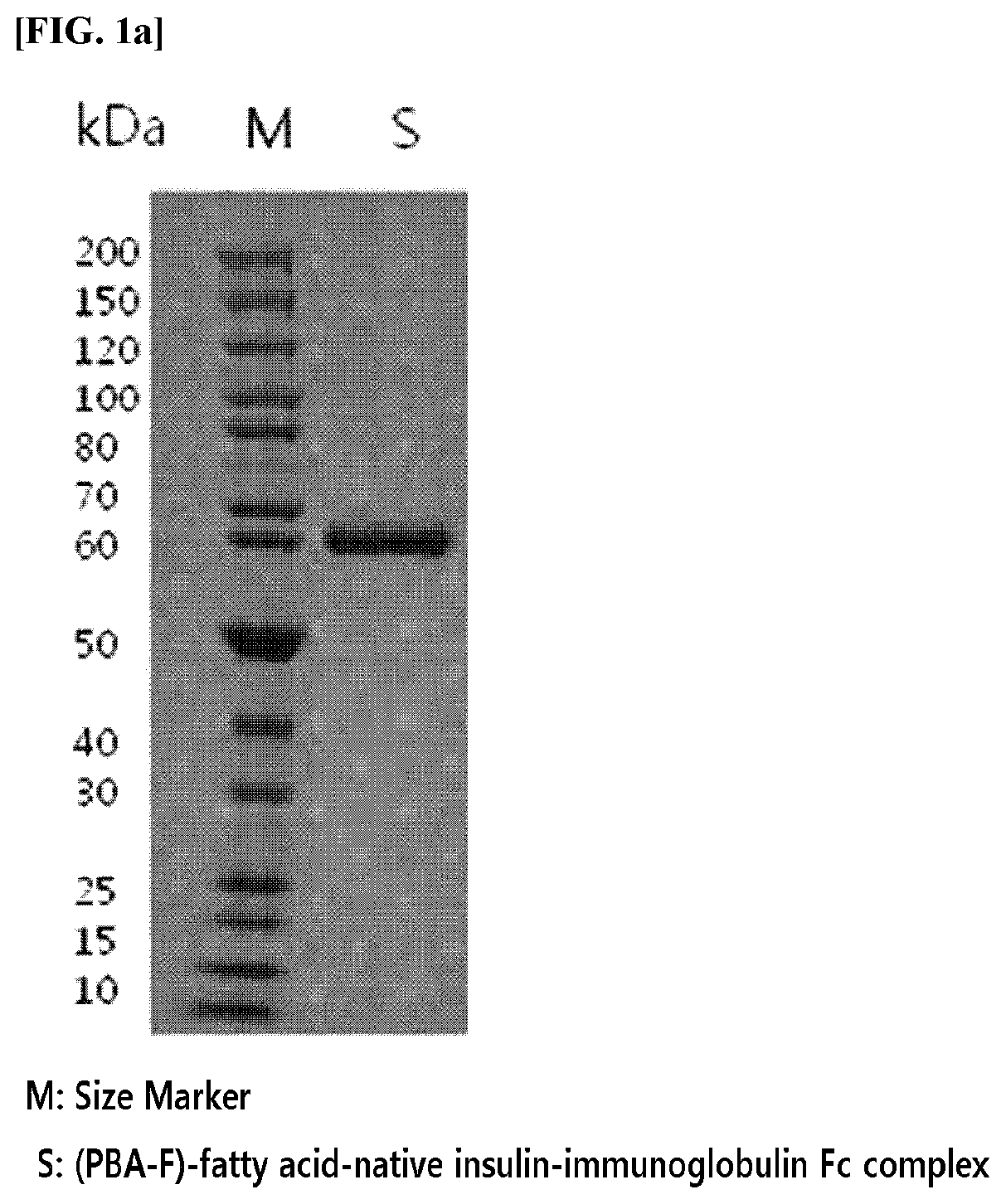
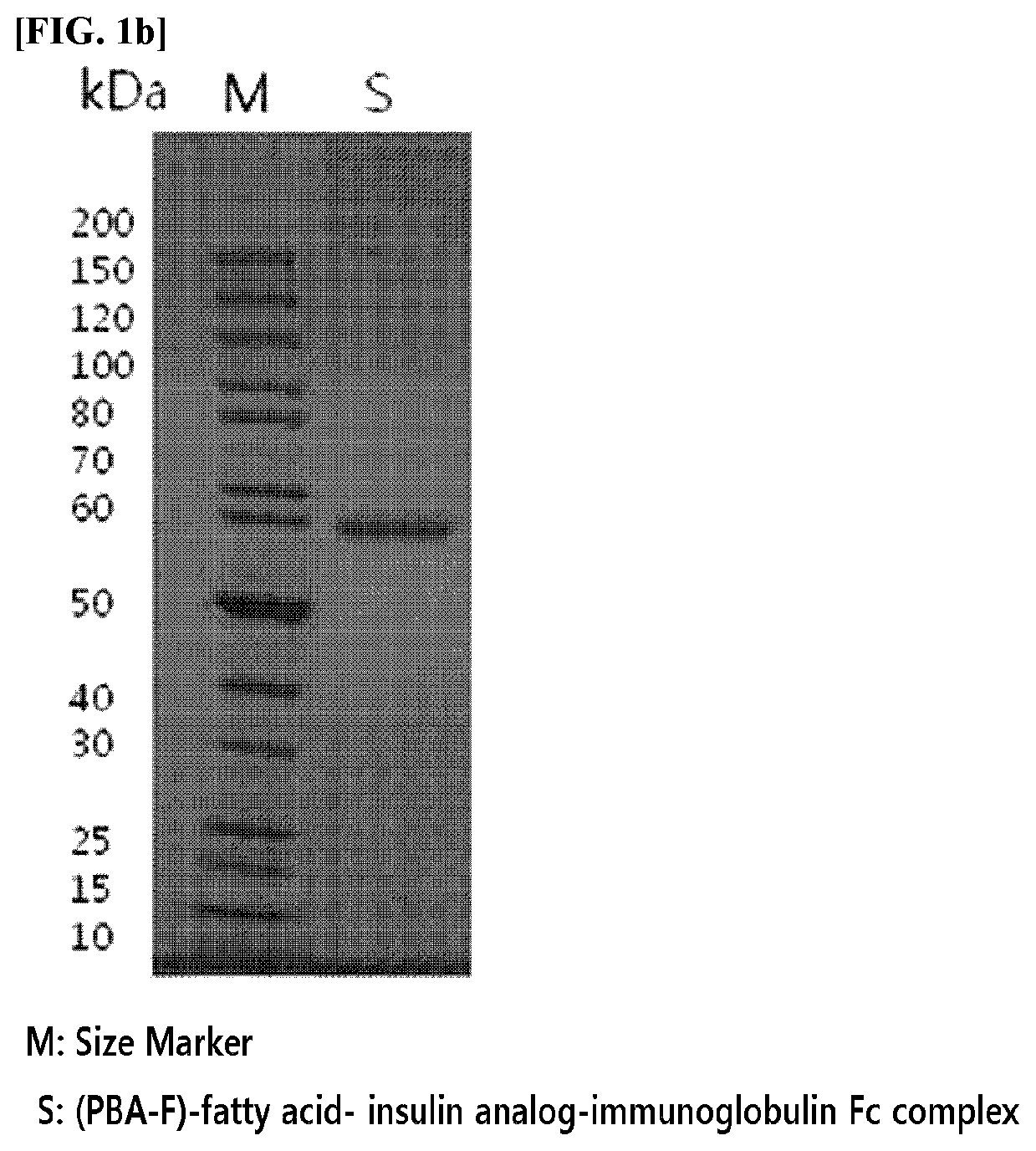
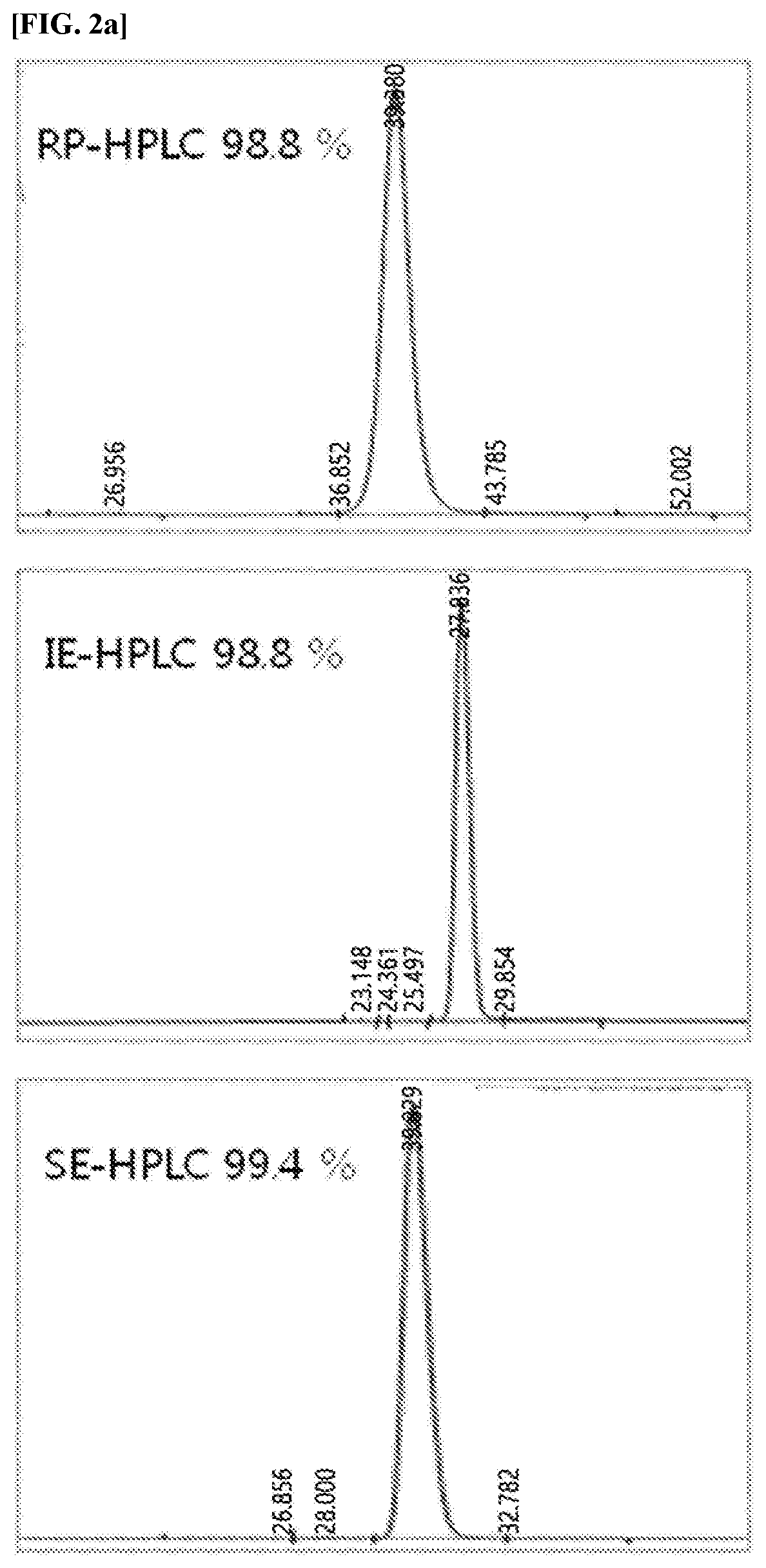
An insulinotropic complex using an immunoglobulin fragment
ActiveUS20100105877A1Improve efficacyProlong lifePeptide/protein ingredientsVasoactive intestinal peptidePeptide drugHalf-life
The present invention relates to an insulinotropic peptide conjugate having improved in-vivo duration of efficacy and stability, comprising an insulinotropic peptide, a non-peptide polymer and an immunoglobulin Fc region, which are covalently linked to each other, and a use of the same. The insulinotropic peptide conjugate of the present invention has the in-vivo activity which is maintained relatively high, and has remarkably increased blood half-life, and thus it can be desirably employed in the development of long acting formulations of various peptide drugs.
Owner:HANMI SCI CO LTD
Heteromultimer constructs of immunoglobulin heavy chains with mutations in the fc domain
InactiveUS20160257763A1Immunoglobulins against cell receptors/antigens/surface-determinantsImmunological disordersImmunoglobulin heavy chainImmunoglobulin light chain
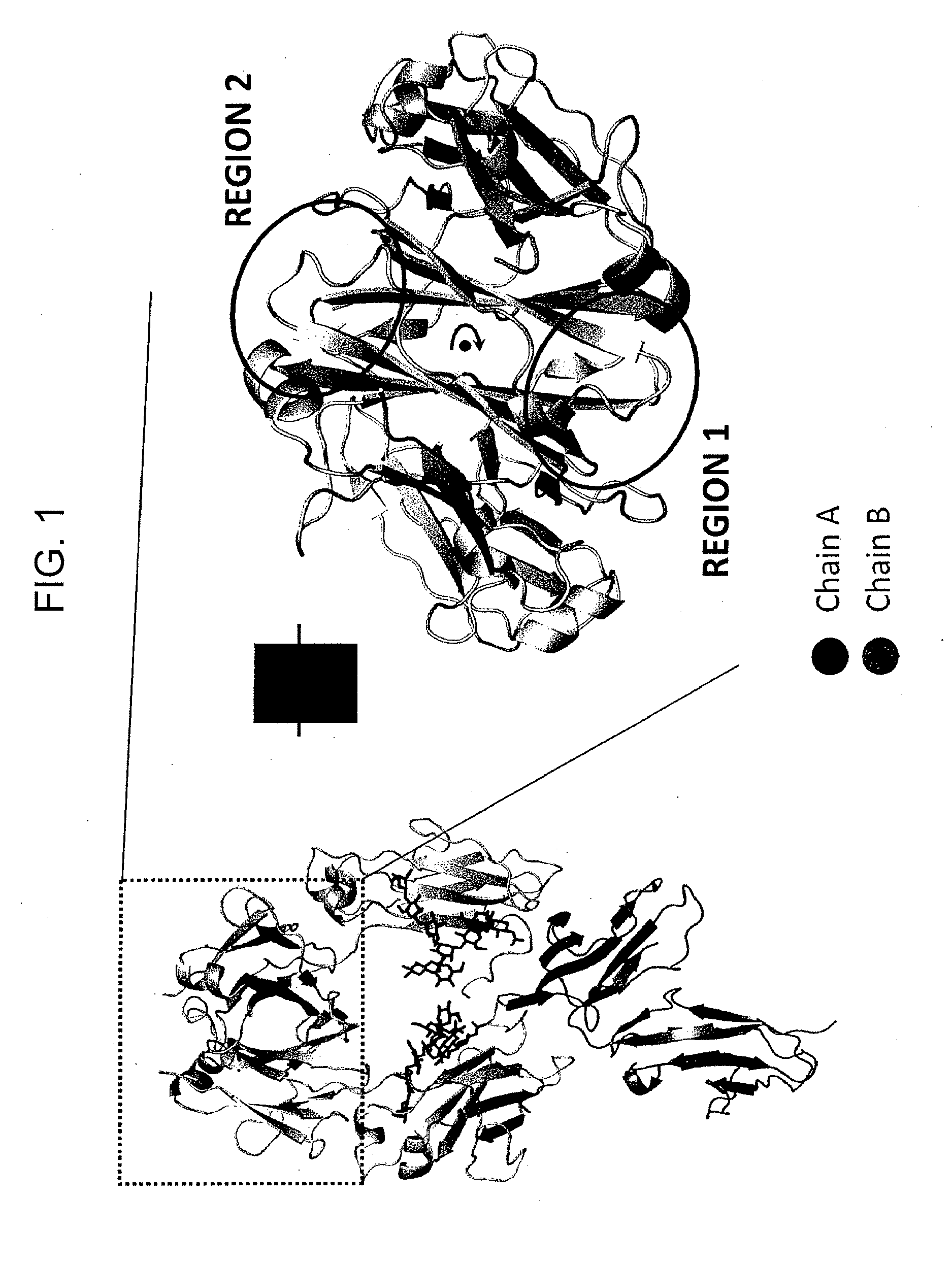
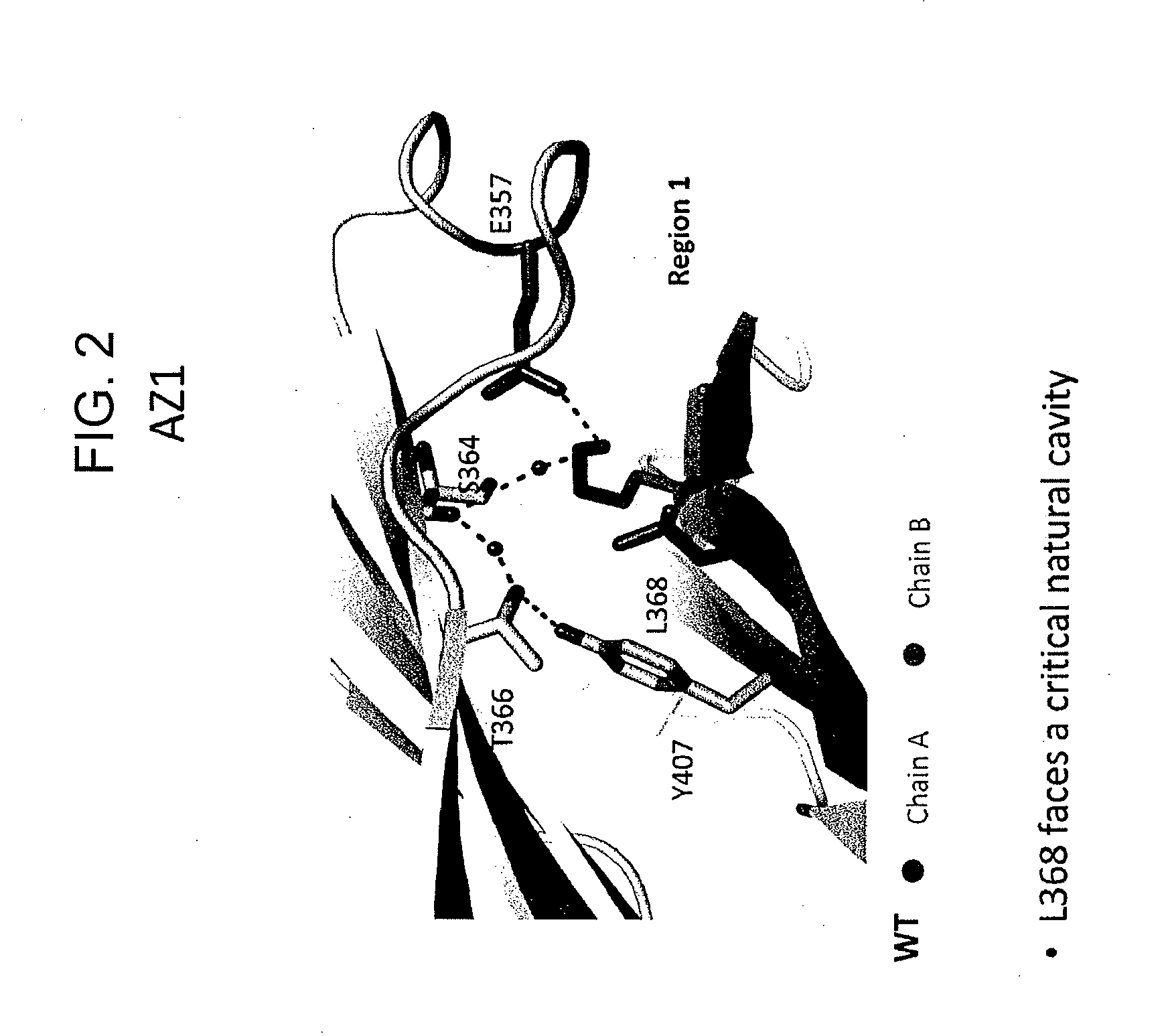
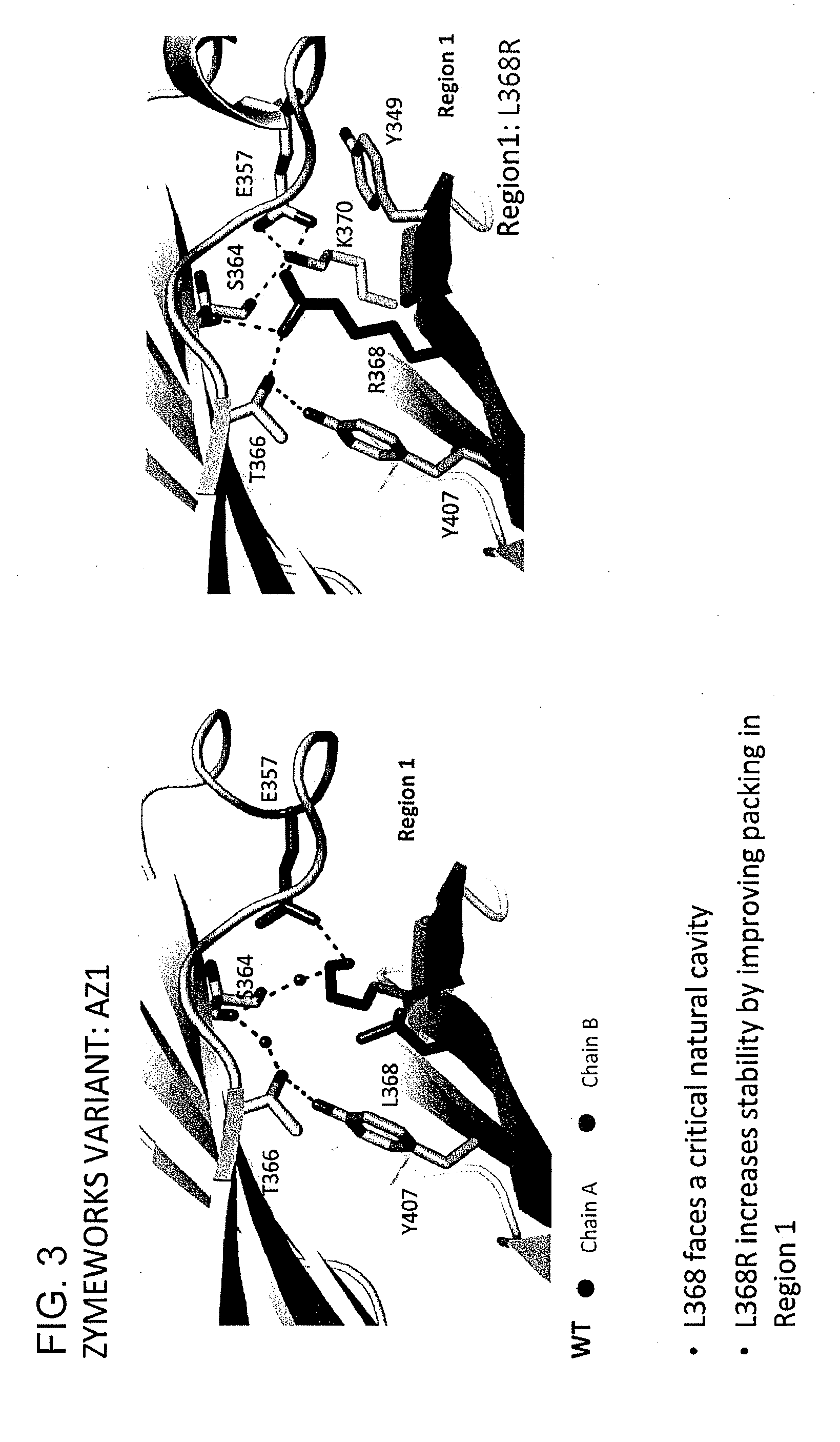
Provided herein are isolated heteromultimers comprising: at least one single domain antigen-binding construct attached to at least one monomer of a heterodimer Fc region; wherein the heterodimer Fc region comprises a variant CH3 domain comprising amino acid mutations that promote the formation of said heterodimer with stability comparable to that of a native Fc homodimer; and wherein said isolated heteromultimer is devoid of immunoglobulin light chains and optionally devoid of immunoglobulin CH1 region. These novel molecules comprise complexes of heterogeneous components designed to alter the natural way antibodies behave and that find use in therapeutics.
Owner:ZYMEWORKS INC
Transgenic animals and methods of use
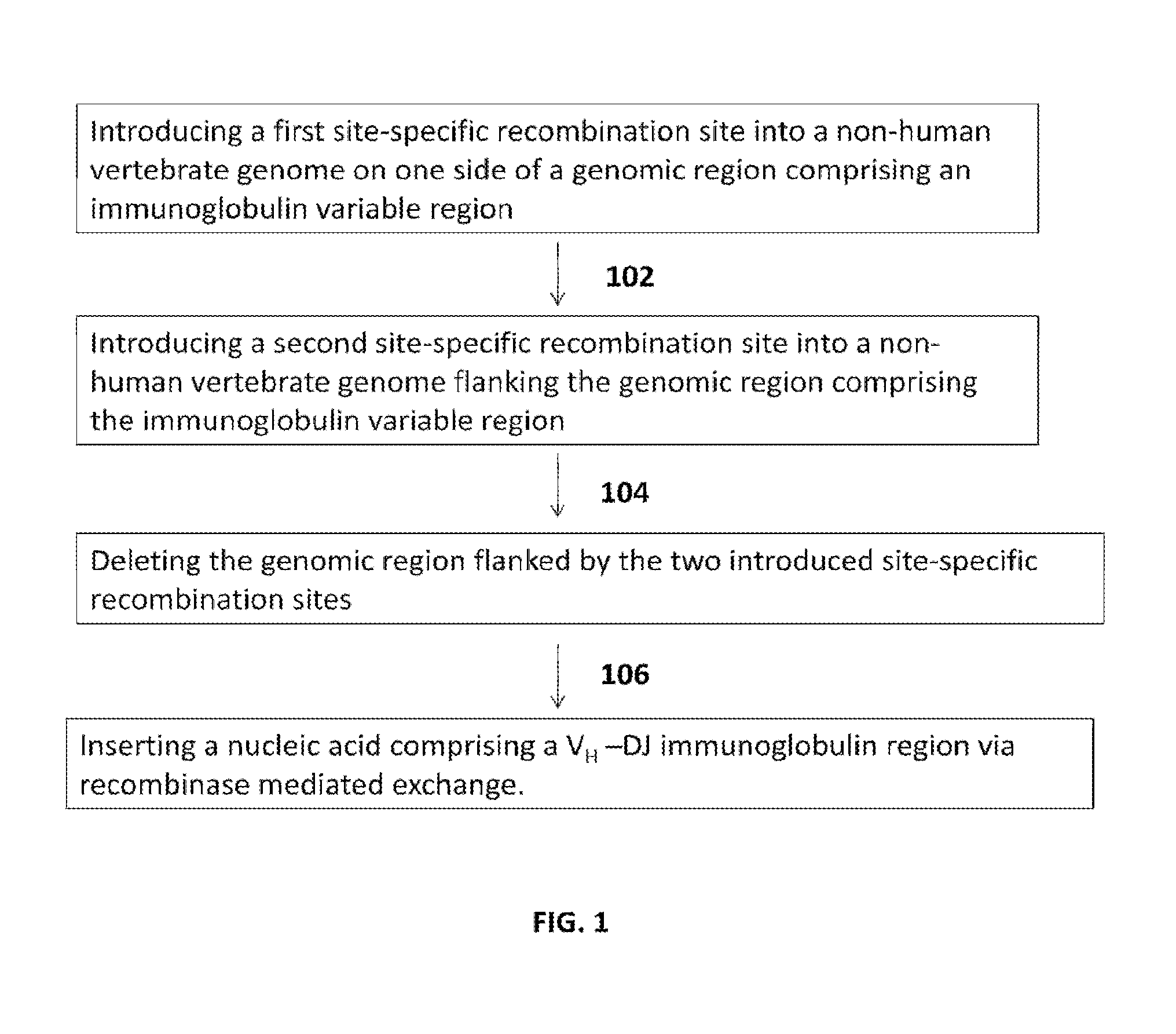
The present invention comprises non-human vertebrate cells and non-human mammals having a genome comprising an introduced partially human immunoglobulin region, said introduced region comprising human VH coding sequences and non-coding VH sequences based on the endogenous genome of the non-human mammal.
Owner:TRIANNI INC
Kind of mutated proteins a with high alkali resistance feature and application thereof
ActiveUS20160237124A1Maintain chemical stabilityBonded firmlyAntibody mimetics/scaffoldsPeptide preparation methodsComplementarity determining regionImmunoglobulin region
A series of protein A mutants having high alkali resistance, and methods of using the protein A mutants are provided. The protein A mutants have a high binding affinity for regions of immunoglobulin proteins other than the complementarity determining regions. The protein A mutants can be coupled to a solid support for immunoglobulin isolation, or conjugated to a label for immunoglobulin detection. This series of protein A mutants has high chemical stability under alkaline conditions of pH 13-14, and can also be used as chromatography ligands for purification procedures that use alkaline solutions under harsh conditions, such as Clean-In-Place (CIP). Also provided are methods of immunoglobulin separation and purification, and alkali regeneration of affinity chromatography medium that uses protein A as a ligand.
Owner:NANJING GENSCRIPT BIOTECH CO LTD
OB fusion protein compositions and methods
InactiveUS20050163799A1Stability advantageStability clearance rateObesity gene productsPeptide/protein ingredientsImmunoglobulin regionOb Protein
The present invention relates to Fc-OB fusion protein compositions, methods of preparation of such compositions and uses thereof. In particular, the present invention relates to a genetic or chemical fusion protein comprising the Fc immunoglobulin region, derivative or analog fused to the N-terminal portion of the OB protein, derivative or analog.
Owner:AMGEN INC
Insulinotropic complex using an immunoglobulin fragment
ActiveUS8476230B2Improve efficacyProlong lifePeptide/protein ingredientsVasoactive intestinal peptidePeptide drugHalf-life
The present invention relates to an insulinotropic peptide conjugate having improved in-vivo duration of efficacy and stability, comprising an insulinotropic peptide, a non-peptide polymer and an immunoglobulin Fc region, which are covalently linked to each other, and a use of the same. The insulinotropic peptide conjugate of the present invention has the in-vivo activity which is maintained relatively high, and has remarkably increased blood half-life, and thus it can be desirably employed in the development of long acting formulations of various peptide drugs.
Owner:HANMI SCI CO LTD
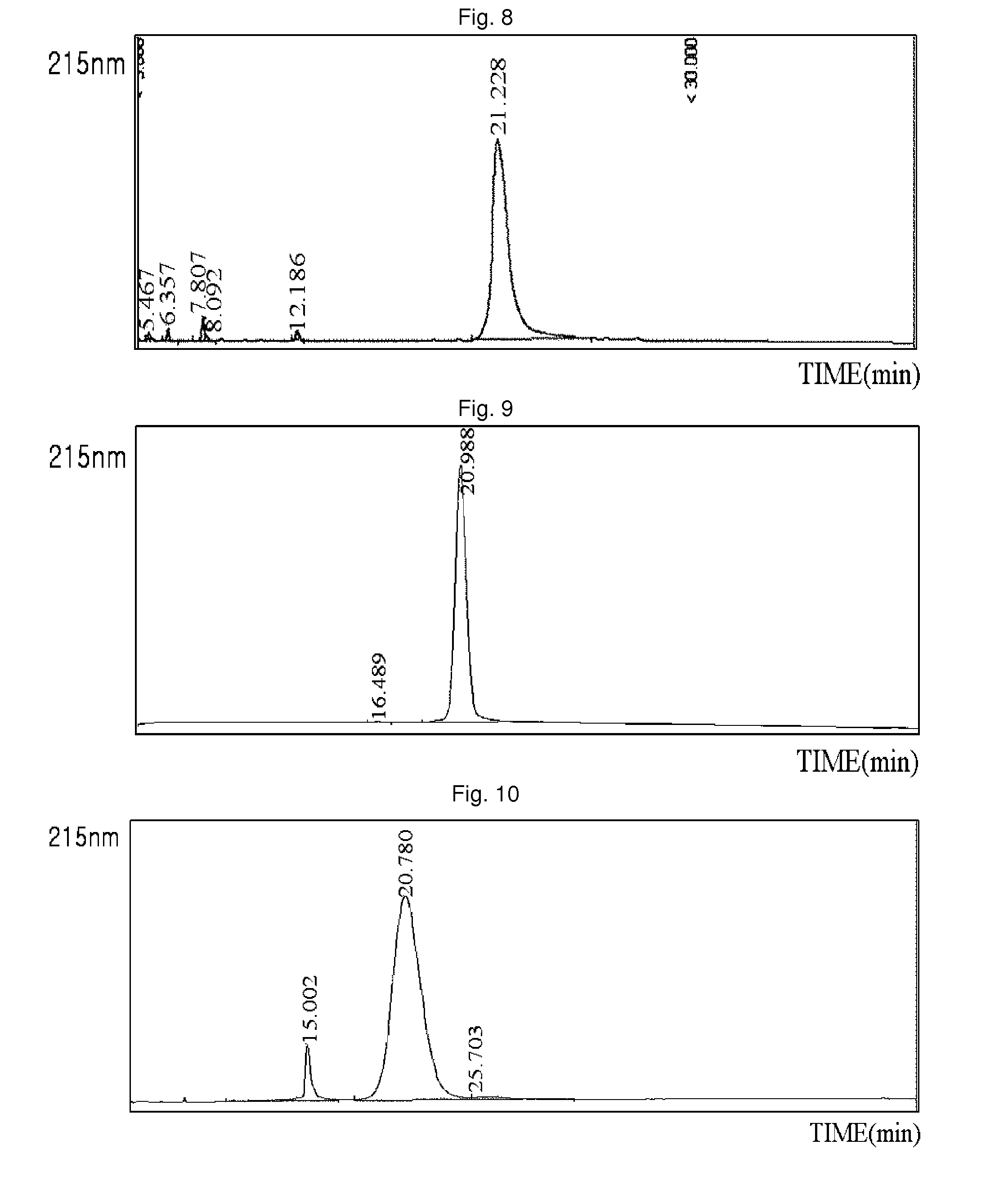
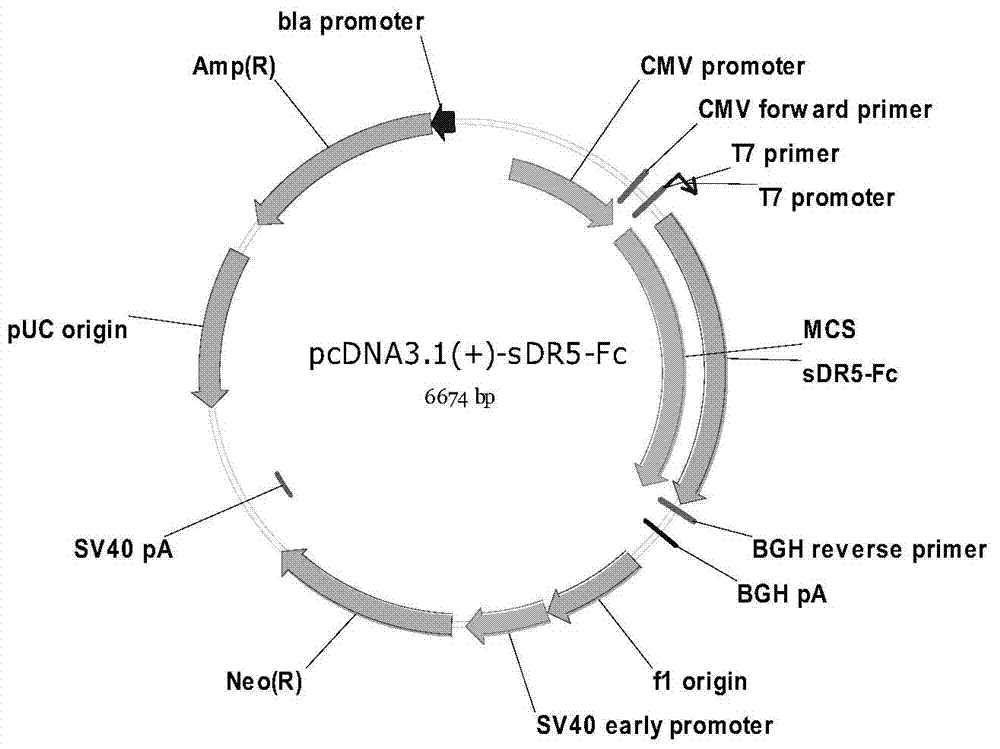
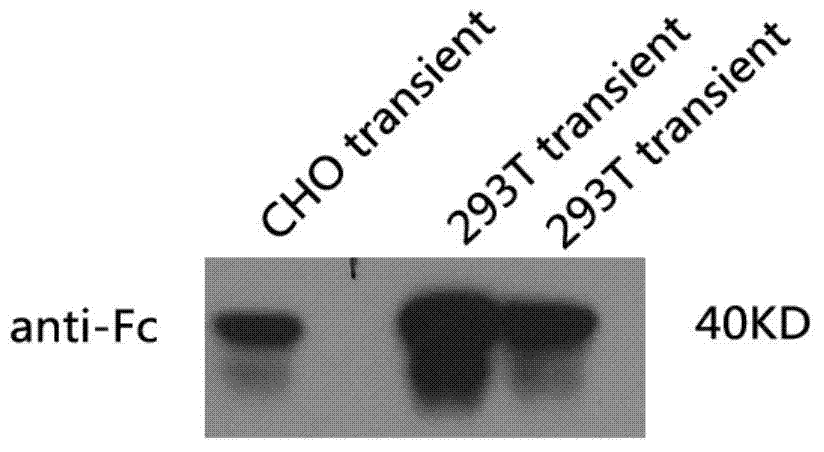
Sdr5-fc fusion protein and uses thereof
InactiveCN104710533ARelieve painLow cost of treatmentPeptide/protein ingredientsGenetic material ingredientsHalf-lifeIn vivo
The present invention belongs to the technical field of biology, and relates to disease treatment, particularly to sDR5-Fc fusion protein and uses thereof. The sDR5-Fc fusion protein comprises an sDR5 functional sequence part and an immunoglobulin Fc region, wherein the sDR5 functional sequence part and the immunoglobulin Fc region are directly connected and / or are connected through a connection fragment. According to the present invention, the natural protein DR5 is adopted to fuse with the antibody Fc fragment, such that the in vivo half-life is prolonged on the base of provision of the biological activity of the functional protein, the frequent administration is not required, and the pain and the treatment cost of patients are reduced.
Owner:SHENZHEN INST OF ADVANCED TECH CHINESE ACAD OF SCI
Multiple cytokine protein complexes
InactiveUS20070258944A1Maintains balance of activityIncreases duration of activityOrganic active ingredientsFungiImmunoglobulin regionProtein-protein complex
Owner:MERCK PATENT GMBH
Mutated protein of protein a having reduced affinity in acidic region and antibody-capturing agent
ActiveUS20140179898A1Reduced ability to bindEasy to eluteBacteriaPeptide/protein ingredientsMutated proteinProteome
A modified protein of an extracellular domain of protein A, which has the reduced ability to bind to immunoglobulin in an acidic region, compared with the wild-type extracellular domain of protein A, without impairing a selective antibody-binding activity in a neutral region. On the basis of three-dimensional structure coordinate data on a complex of the extracellular domain of protein A bound with the Fc region of immunoglobulin G, the modified protein is obtained by the substitution of amino acid residues that are located within the range of 10 angstroms from the Fc region and have a 20% or more ratio of exposed surface area, by histidine residues. Preferably, the modified protein is obtained by the substitution of amino acid residues at sites identified from the analysis of sequences selected from a library constituted by the protein group, by histidine residues. These substitutions may be combined.
Owner:NAT INST OF ADVANCED IND SCI & TECH
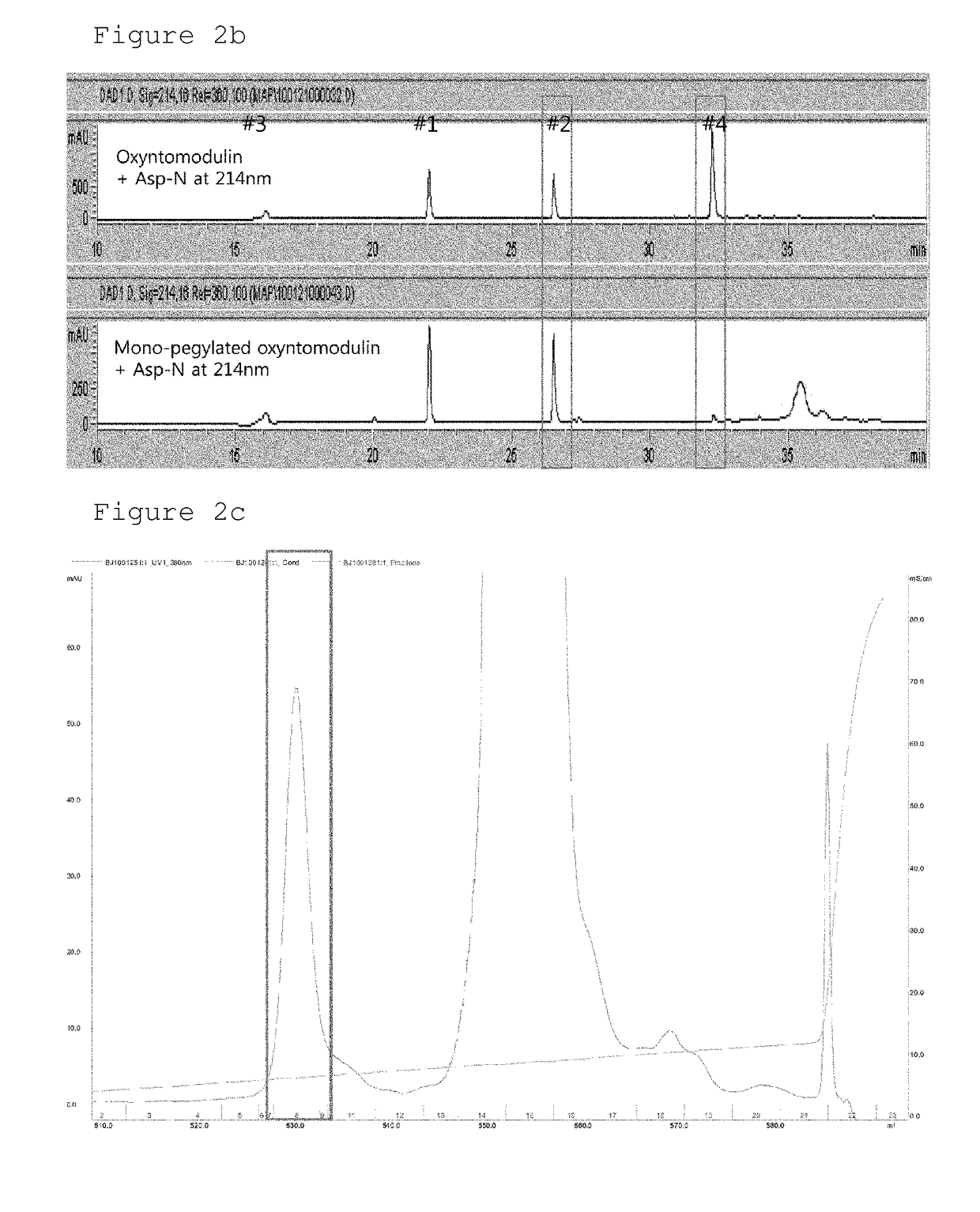
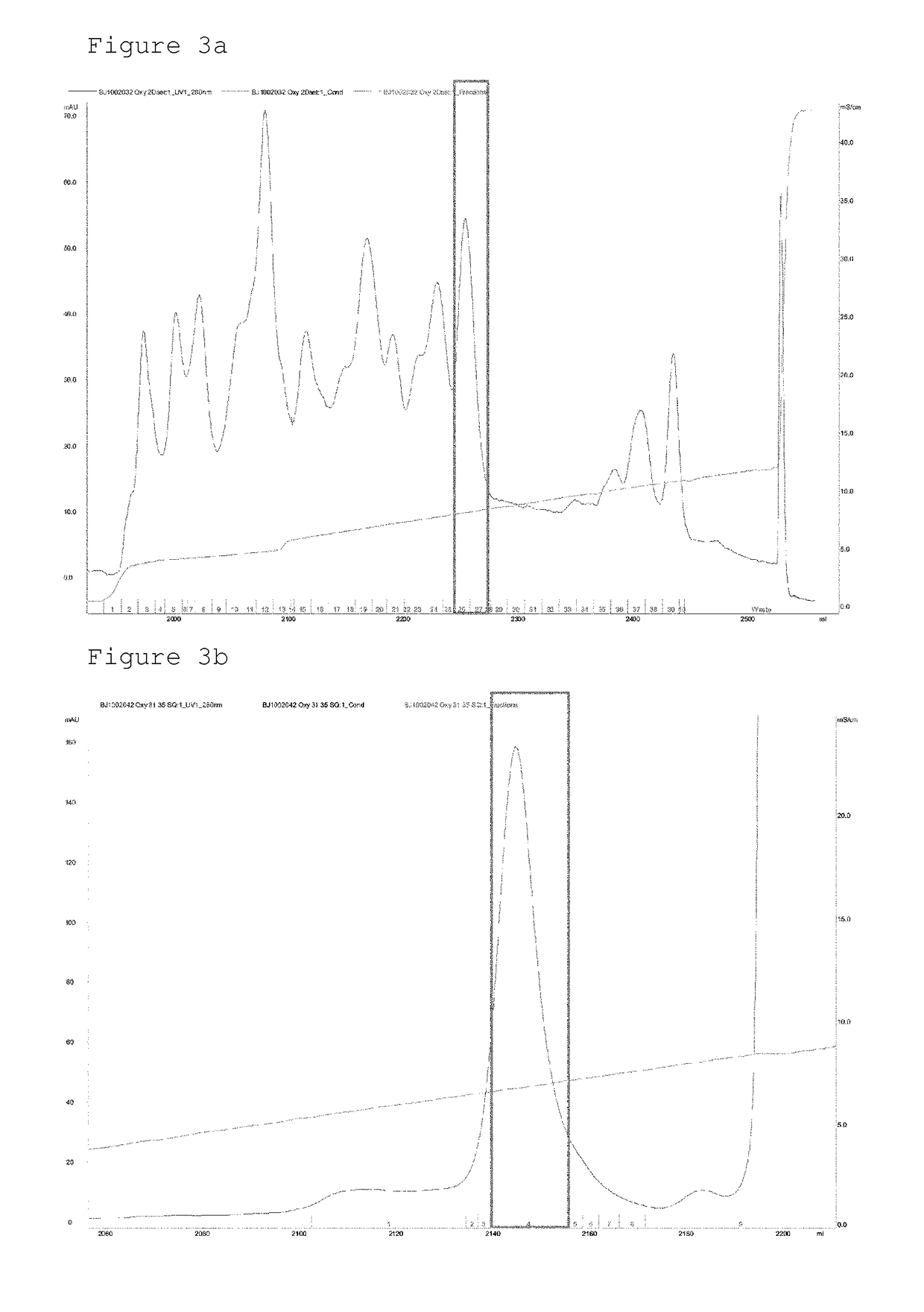
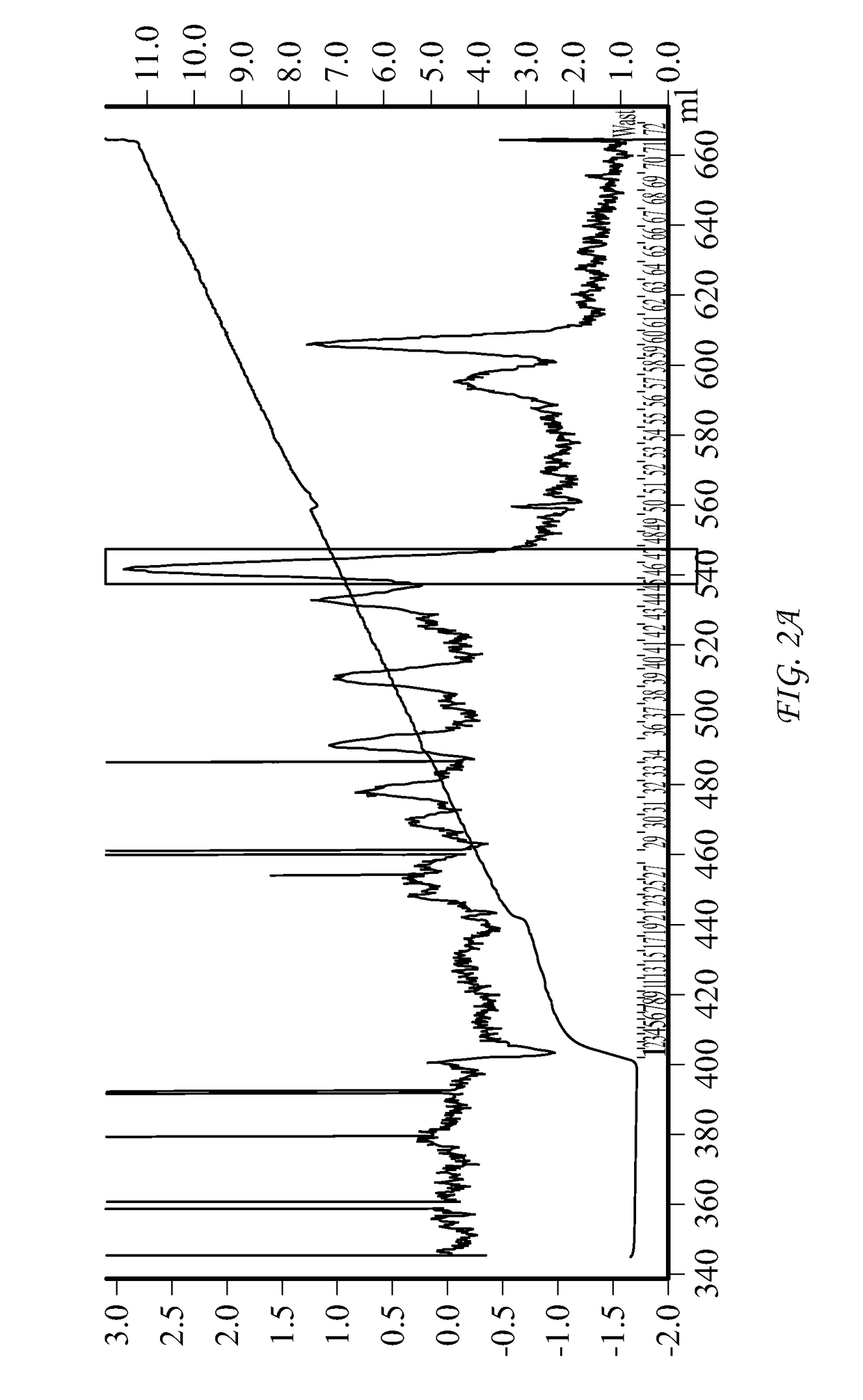
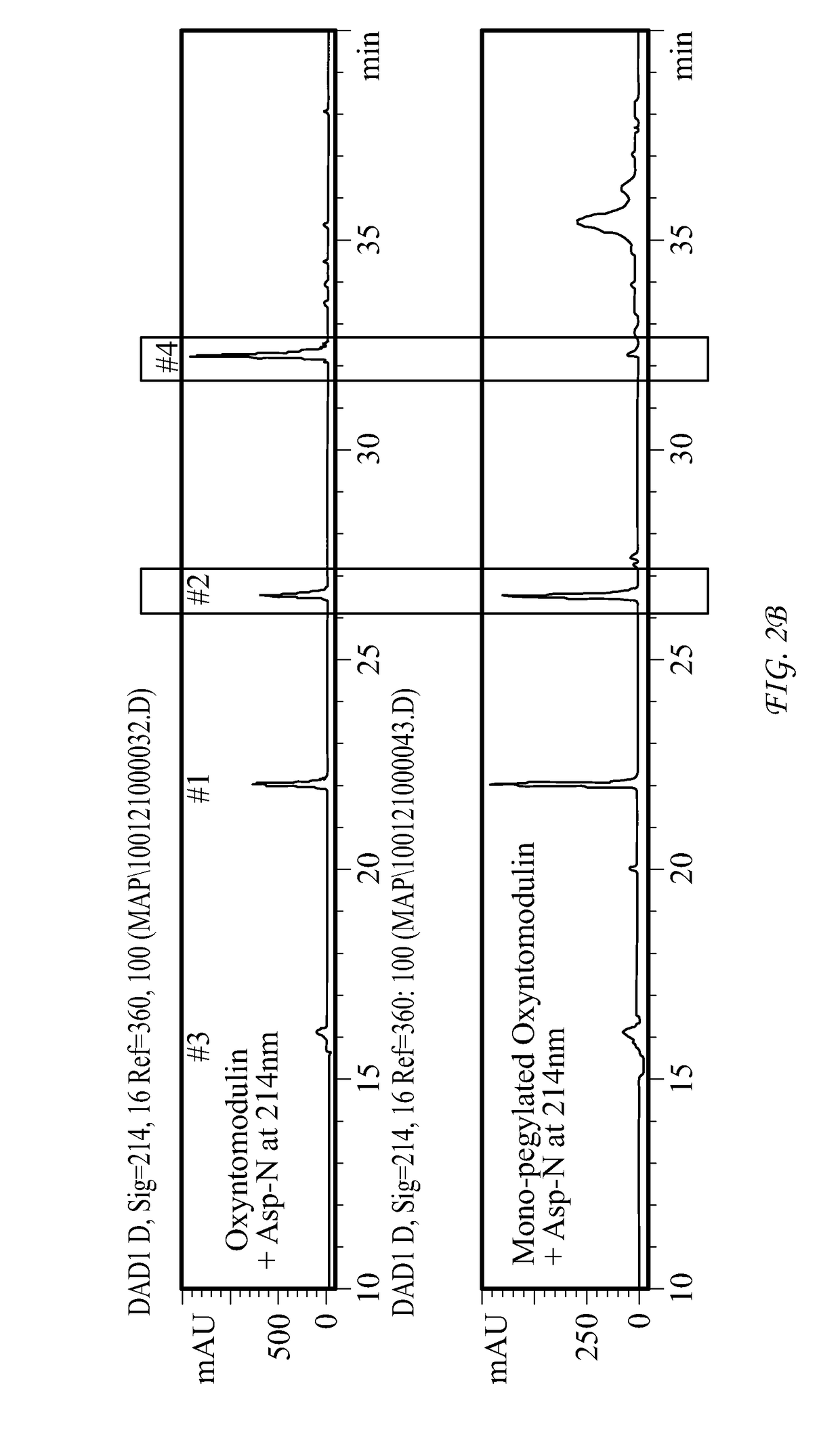
Conjugate comprising oxyntomodulin and an immunoglobulin fragment, and use thereof
ActiveUS9731031B2Reduces food intakeSuppresses gastric emptyingPeptide/protein ingredientsMetabolism disorderSide effectReceptor activation
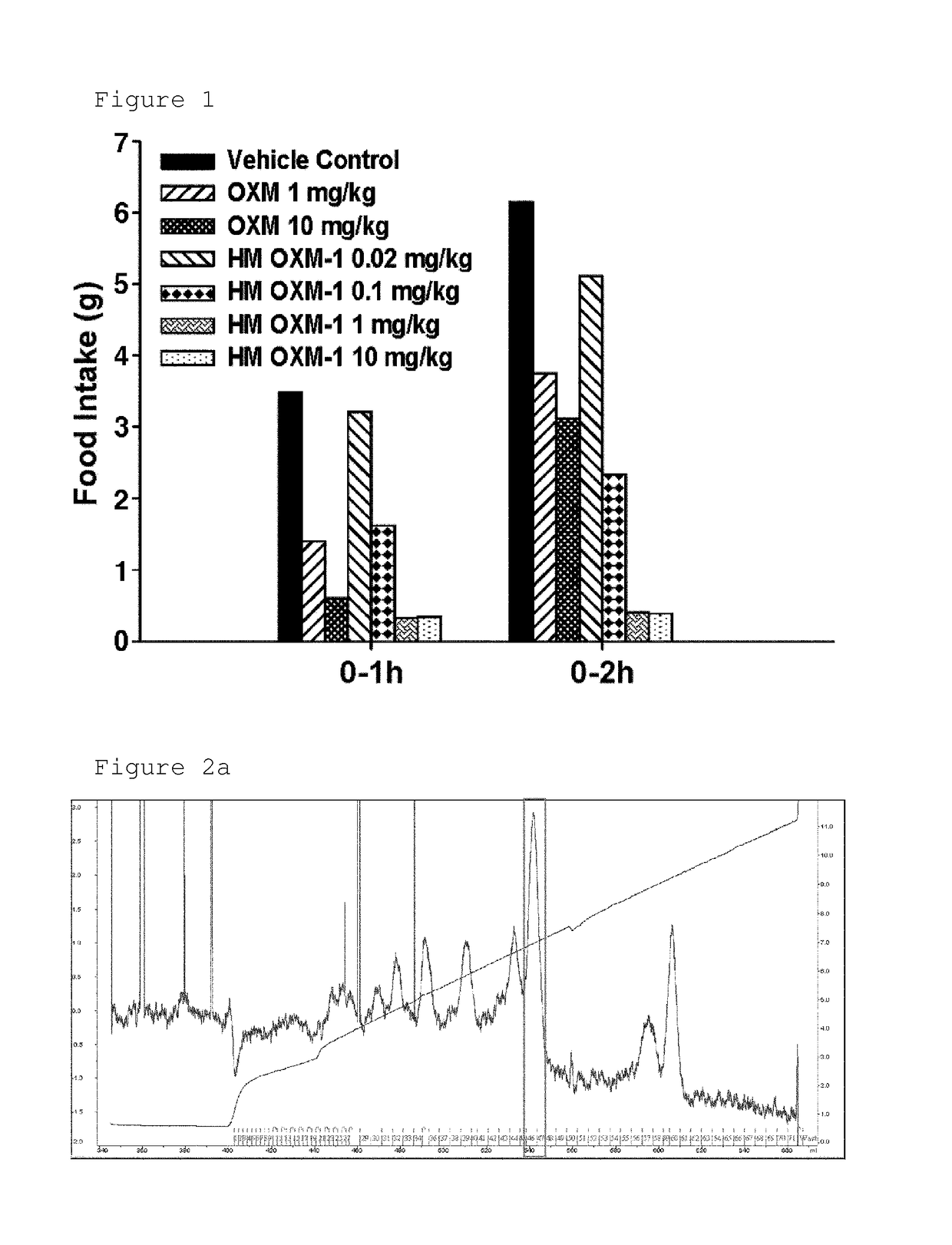
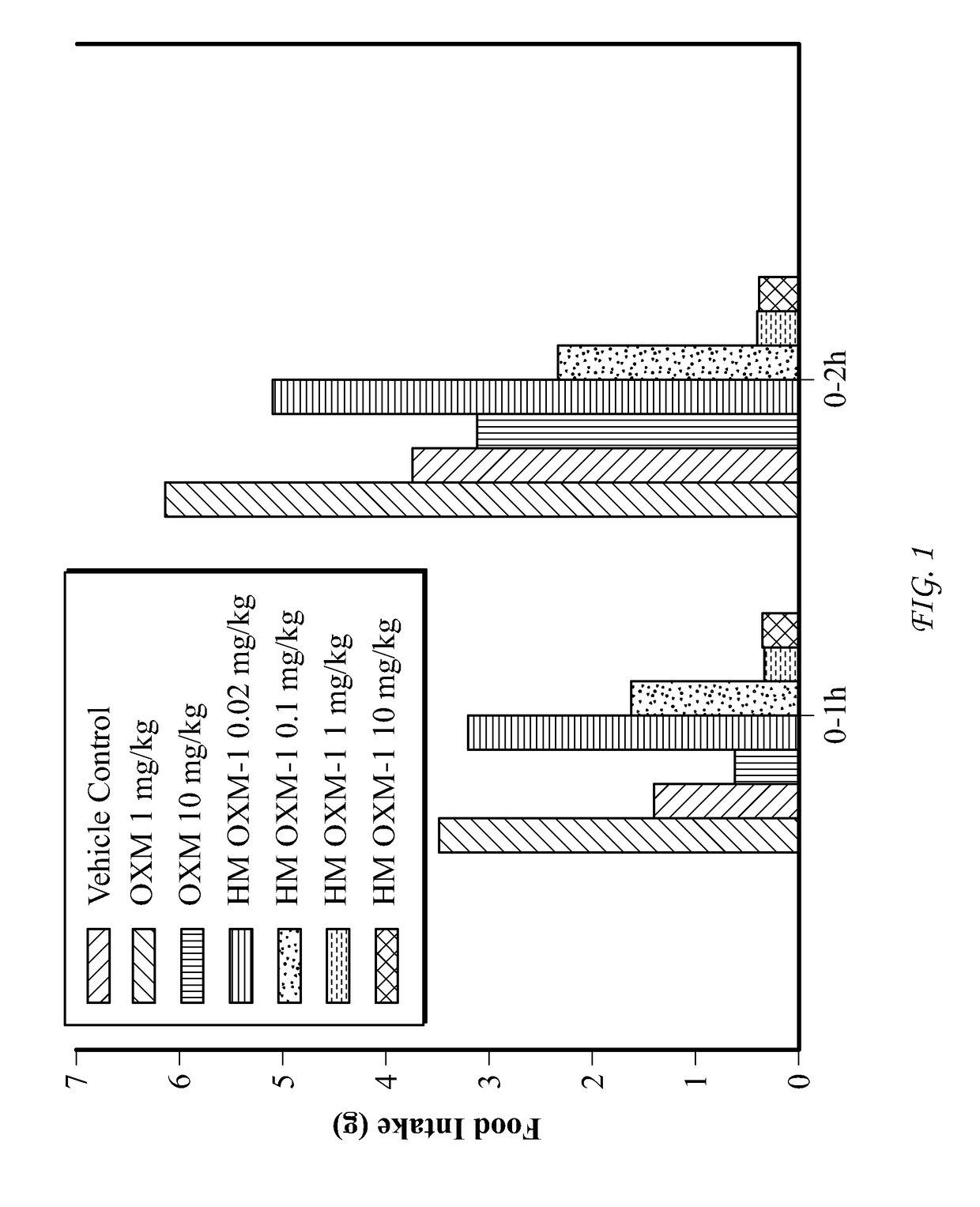
The present invention relates to a conjugate comprising oxyntomodulin, an immunoglobulin Fc region, and non-peptidyl polymer wherein the conjugate being obtainable by covalently linking oxyntomodulin to immunoglobulin Fc region via non-peptidyl polymer, and a pharmaceutical composition for the prevention or treatment of obesity comprising the conjugates. The conjugate comprising oxyntomodulin and the immunoglobulin Fc of the present invention reduces food intake, suppresses gastric emptying, and facilitates lipolysis without side-effects, unlike native oxyntomodulin, and also shows excellent receptor-activating effects and long-term sustainability, compared to native oxyntomodulin. Thus, it can be widely used in the treatment of obesity with safety and efficacy.
Owner:HANMI SCI CO LTD
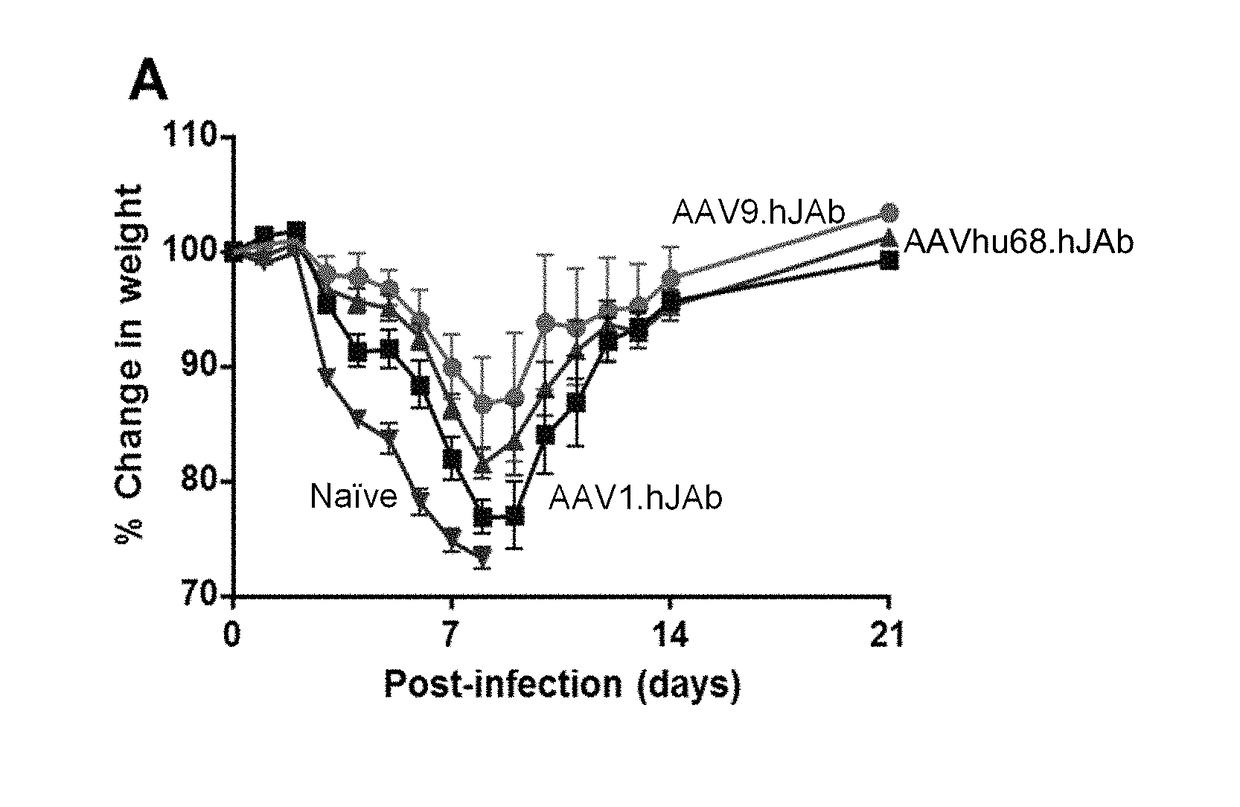
Novel aav mediated influenza vaccines
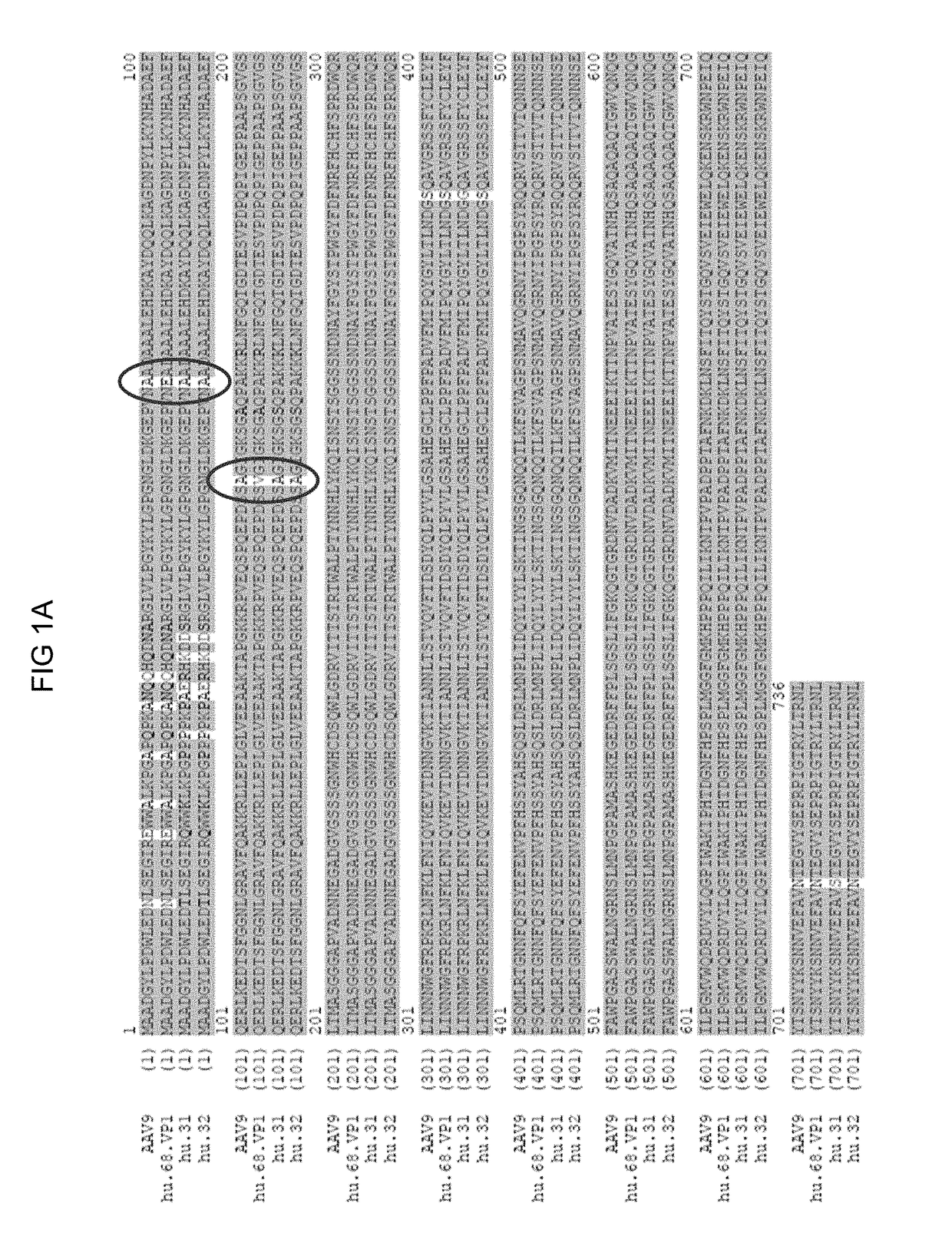
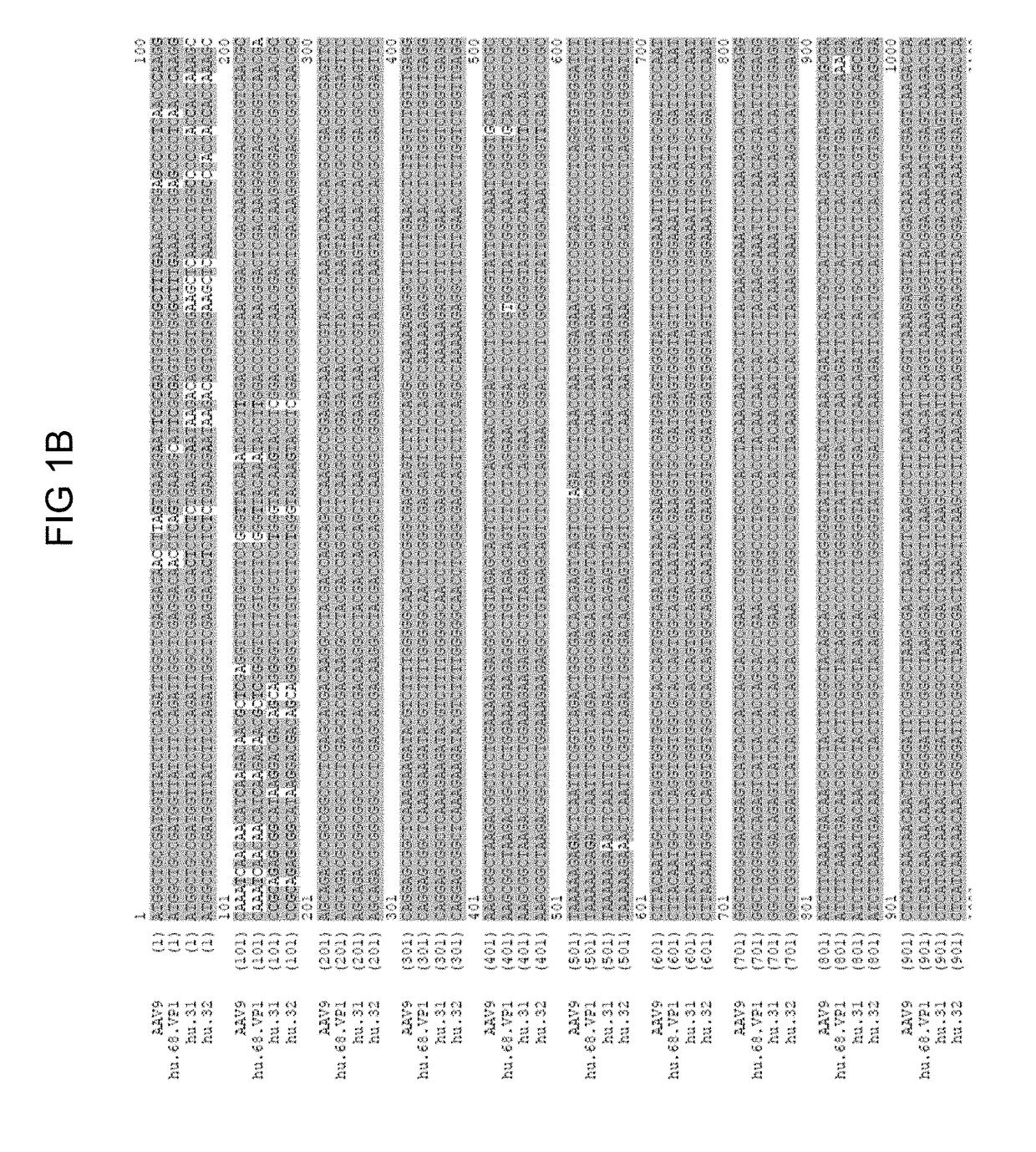
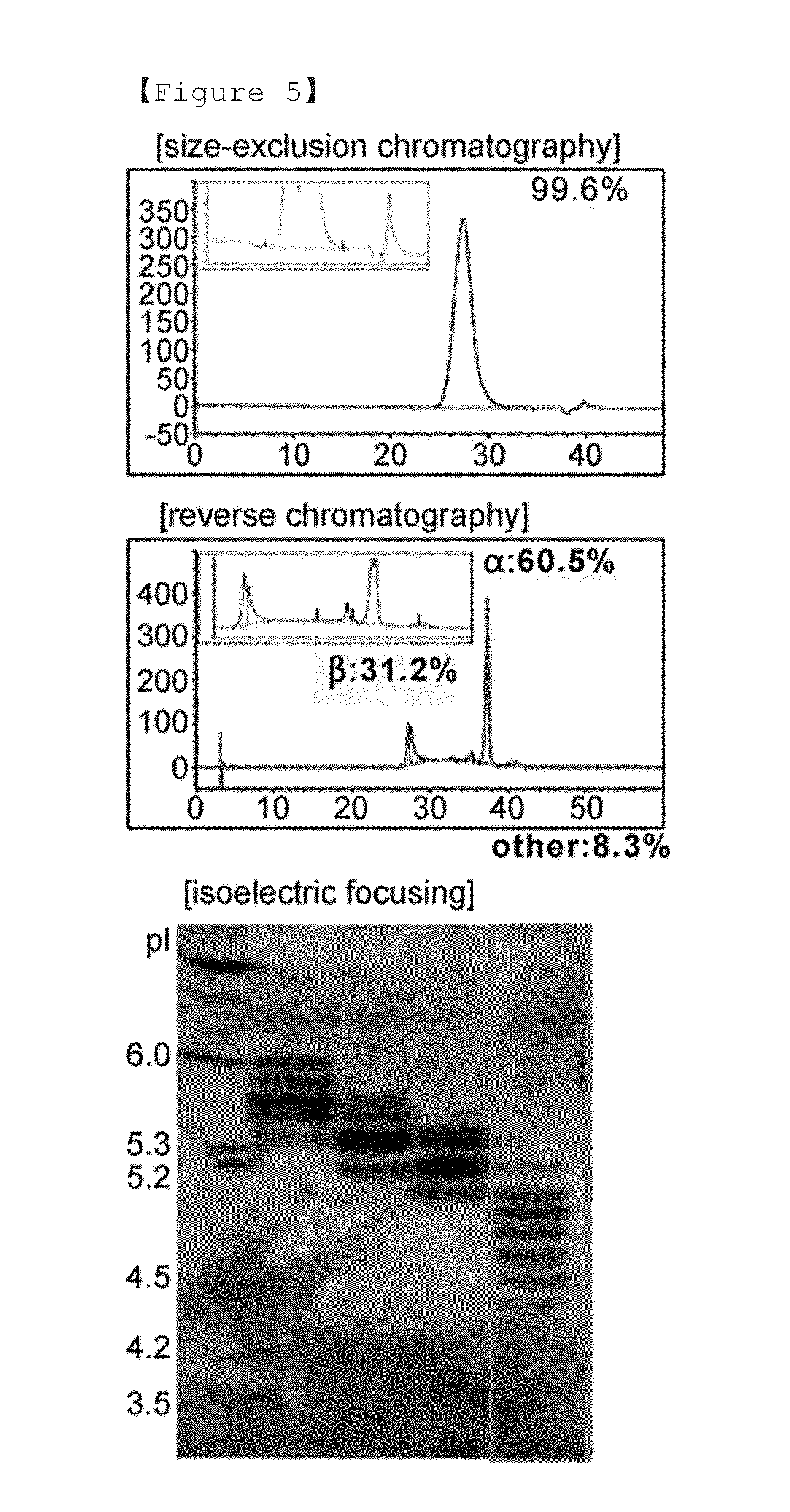
A non-replicating recombinant adeno-associated associated virus (rAAV) having an AAV capsid having packaged therein a vector genome which comprises AAV inverted terminal repeat sequences and at least one nucleic acid sequence encoding four different immunoglobulin regions (a), (b), (c) and (d) is provided. The rAAV-expressed immunoglobulins are useful for providing passive immunization against influenza A and influenza B. Also described herein are compositions containing the rAAV. Methods of vaccinating patients against influenza are provided.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA +1
Method for the mass production of immunoglobulin Fc region deleted initial methionine residues
Disclosed is a method for the mass production of a monomeric or dimeric immunoglobulin Fc region, free of initial methionine residues, using a recombinant expression vector comprising a nucleotide sequence coding for a recombinant immunoglobulin Fc region comprising an immunoglobulin Fc region linked at the N-terminus thereof to an immunoglobulin Fc region via a peptide bond.
Owner:HANMI SCI CO LTD
Insulin conjugate using an immunoglobulin fragment
ActiveUS9492507B2Extended half-lifeImprove Medication AdherencePeptide/protein ingredientsMetabolism disorderHalf-lifeIn vivo
Owner:HANMI SCI CO LTD
Liquid formulation of a fusion protein comprising TNFR and Fc region
ActiveUS9700595B2Good storage stabilityEasy to usePeptide/protein ingredientsAntibody mimetics/scaffoldsCrystallographySucrose
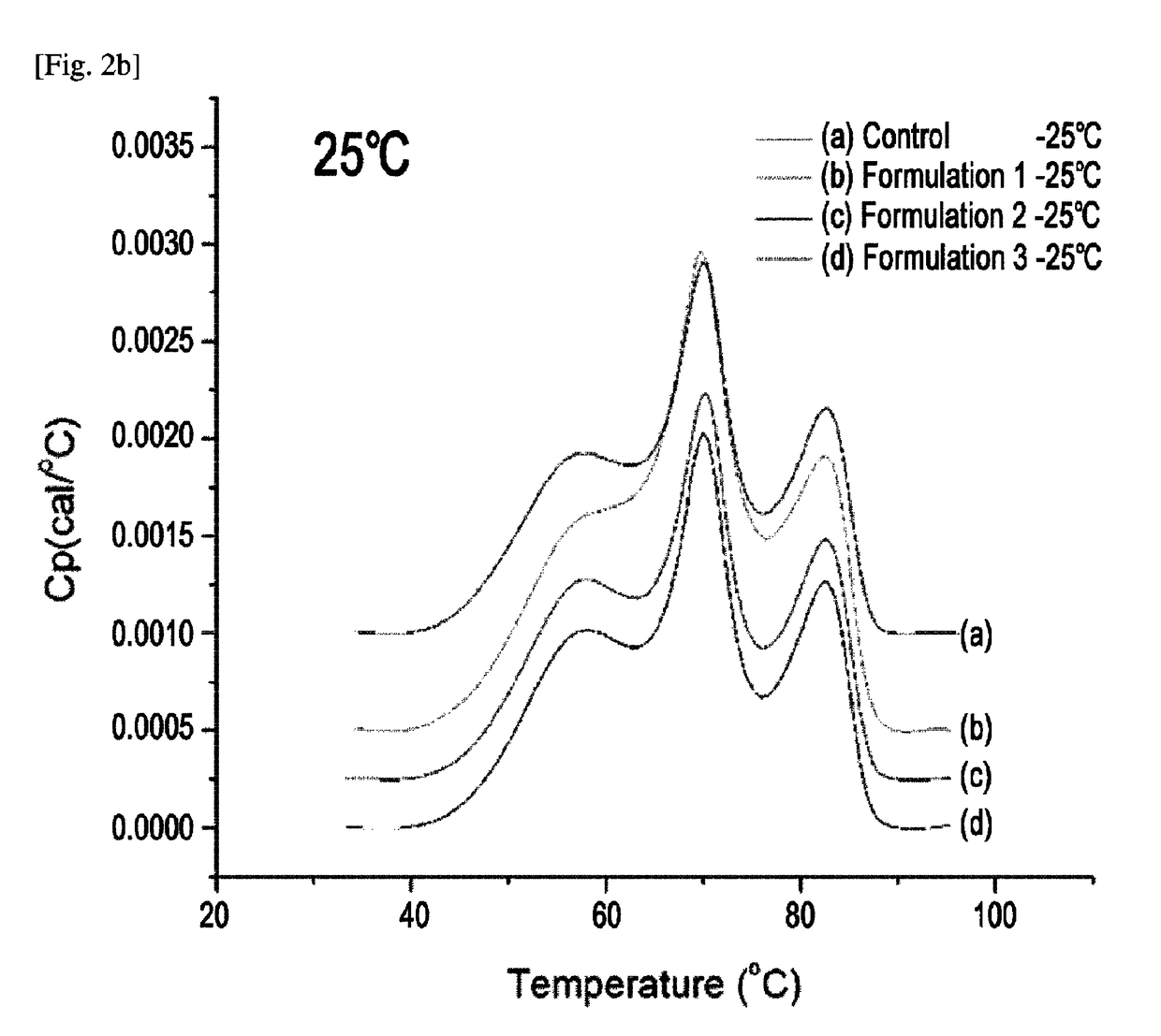
The present invention relates to a liquid formulation comprising a TNFR-Fc fusion protein and a stabilizer, in which the fusion protein comprises TNFR (tumor necrosis factor receptor) or a fragment thereof and an immunoglobulin Fc region, and the stabilizer comprises one or more amino acids selected from the group consisting of proline and histidine, a buffer solution, and an isotonic agent containing sodium chloride (NaCl) and sucrose, and a preparation method of the liquid formulation. The liquid formulation according to the present invention provides excellent storage stability because long-term storage of TNFR-Fc fusion protein (etanercept) is possible and particular storage conditions are not needed. Since the liquid formulation of the present invention shows excellent storage stability even though the formulation is simple, it is more economical than other stabilizers or lyophilized formulations, and thus the formulation can be effectively applied for uses wherein treatment of TNFR-Fc fusion protein (etanercept) is beneficial.
Owner:ARES TRADING SA
Heterodimer binding proteins and uses thereof
InactiveUS20180273642A1Peptide/protein ingredientsAntibody mimetics/scaffoldsReceptorImmunoglobulin light chain
The present disclosure provides polypeptide heterodimers formed between two different single chain fusion polypeptides via natural heterodimerization of an immunoglobulin CH1 region and an immunoglobulin light chain constant region (CL). The polypeptide heterodimer comprises two or more binding domains that specifically bind one or more targets (e.g., a receptor). In addition, both chains of the heterodimer further comprise an Fc region portion. The present disclosure also provides nucleic acids, vectors, host cells and methods for making polypeptide heterodimers as well as methods for using such polypeptide heterodimers, such as in directing T cell activation, inhibiting solid malignancy growth, and treating autoimmune or inflammatory conditions.
Owner:APTEVO RES & DEV LLC
Antibody-binding peptide
ActiveUS20150353608A1High binding activityLow ratePeptide-nucleic acidsBacteriaBinding peptideChemistry
The invention relates to polypeptides consisting of amino acid sequences represented by the following formulas 1 to 4, which have binding activity to an Fc region of immunoglobulin G and can be favorably used in detecting, purifying, immobilizing or removing an antibody, immunoglobulin G or a protein containing an Fc region of immunoglobulin G,(SEQ ID NO: 1)Y-D-P-x-T-G-T-W-R-S-x-[IL] (1)(SEQ ID NO: 10)R-[QRS]-x-x-[GS]-Y-D-P-R-T-G-T-W-R-S-S-I-A-Y-G-G(2)(SEQ ID NO: 20)G-V-V-R-Q-W-S-G-x-x-x-x-x-x-x-x-R-S-S-I-A-Y-G-G(3)(SEQ ID NO: 23)D-A-A-W-H-L-G-E-L-V-W-A-T-Y-Y-D-P-E-T-G-T-W-x-P-D-W-x-x-M (4)where x represents any amino acid residue; and amino acid residues within the square brackets indicate that any one of the amino acid residues is selected.
Owner:NAT INST OF ADVANCED IND SCI & TECH
A liquid formulation of long-acting insulin conjugate
ActiveCN104519871AGood storage stabilityNo risk of contaminationPeptide/protein ingredientsMetabolism disorderActive agentAlcohol sugars
The present invention relates to a liquid formulation of long-acting insulin conjugate, comprising a pharmaceutically effective amount of a long-acting insulin conjugate, wherein a physiologically active peptide, which is an insulin, is linked to an immunoglobulin Fc region; and an albumin-free stabilizer, wherein the stabilizer comprises a buffer, a sugar alcohol, a non-ionic surfactant, and an isotonic agent, and a method for preparing the formulation. For preventing microbial contamination in multiple uses, a preservative can be added to the formulation. The liquid formulation of the present invention does not comprise a human serum albumin and potentially hazardous factors to body, and thus it has excellent storage stability for insulin conjugate without a risk of viral infection.
Owner:HANMI PHARMA
Protein complex by use of a specific site of an immunoglobulin fragment for linkage
ActiveUS20180326013A1Improved in vivo durationImproving in vivo durationPeptide/protein ingredientsSolid sorbent liquid separationPeptide drugBinding site
Provided is a complex composition, of which positional isomers are minimized by using a N-terminus of an immunoglobulin Fc region as a binding site when the immunoglobulin Fc region is used as a carrier. Also provided are a protein complex which is prepared by N-terminal-specific binding of immunoglobulin Fc region, thereby prolonging blood half-life of the physiologically active polypeptide, maintaining in vivo potency at a high level, and having no risk of immune responses, a preparation method thereof, and a pharmaceutical composition including the same for improving in vivo duration and stability of the physiologically active polypeptide. The protein complex prepared by the present invention may be usefully applied to the development of long-acting formulations of various physiologically active polypeptide drugs.
Owner:HANMI PHARMA
Long-acting human follicle-stimulating hormone formulation using immunoglobulin fragment
ActiveUS20130022626A1Improve the level ofExtended half-lifePeptide/protein ingredientsAntibody ingredientsSerum igeHalf-life
The present invention relates to a long-acting human follicle-stimulating hormone formulation having improved in vivo duration and stability, comprising a human follicle-stimulating hormone conjugate that is prepared by covalently linking human follicle-stimulating hormone with an immunoglobulin Fc region via a non-peptidyl polymer, and a preparation method thereof. The long-acting human follicle-stimulating hormone formulation of the present invention maintains in vivo activity of human follicle-stimulating hormone at a relatively high level and remarkably increases the serum half-life thereof.
Owner:HANMI SCI CO LTD
AAV mediated influenza vaccines
A non-replicating recombinant adeno-associated associated virus (rAAV) having an AAV capsid having packaged therein a vector genome which comprises AAV inverted terminal repeat sequences and at least one nucleic acid sequence encoding four different immunoglobulin regions (a), (b), (c) and (d) is provided. The rAAV-expressed immunoglobulins are useful for providing passive immunization against influenza A and influenza B. Also described herein are compositions containing the rAAV. Methods of vaccinating patients against influenza are provided.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA +1
Conjugate Comprising Oxyntomodulin And An Immunoglobulin Fragment, And Use Thereof
ActiveUS20170340753A1Maintain activityExtended half-lifePeptide/protein ingredientsAntibody mimetics/scaffoldsSide effectReceptor activation
Owner:HANMI SCI CO LTD
Protein a mutants having high alkali resistance and methods of use thereof
ActiveUS20190048046A1Maintain chemical stabilityAntibody mimetics/scaffoldsPeptide preparation methodsHigh resistanceComplementarity determining region
A series of protein A mutants having high alkali resistance, and methods of using the protein A mutants are provided. The protein A mutants have a high binding affinity for regions of immunoglobulin proteins other than the complementarity determining regions. The protein A mutants can be coupled to a solid support for immunoglobulin isolation, or conjugated to a label for immunoglobulin detection. This series of protein A mutants have high chemical stability under alkaline conditions of pH 13-14, and can also be used as chromatography ligands for purification procedures that use alkaline solutions under harsh conditions, such as Clean-In-Place (CIP). Also provided are methods of immunoglobublin separation and purification, and alkali regeneration of affinity chromatography medium that uses protein A as a ligand.
Owner:NANJING GENSCRIPT BIOTECH CO LTD
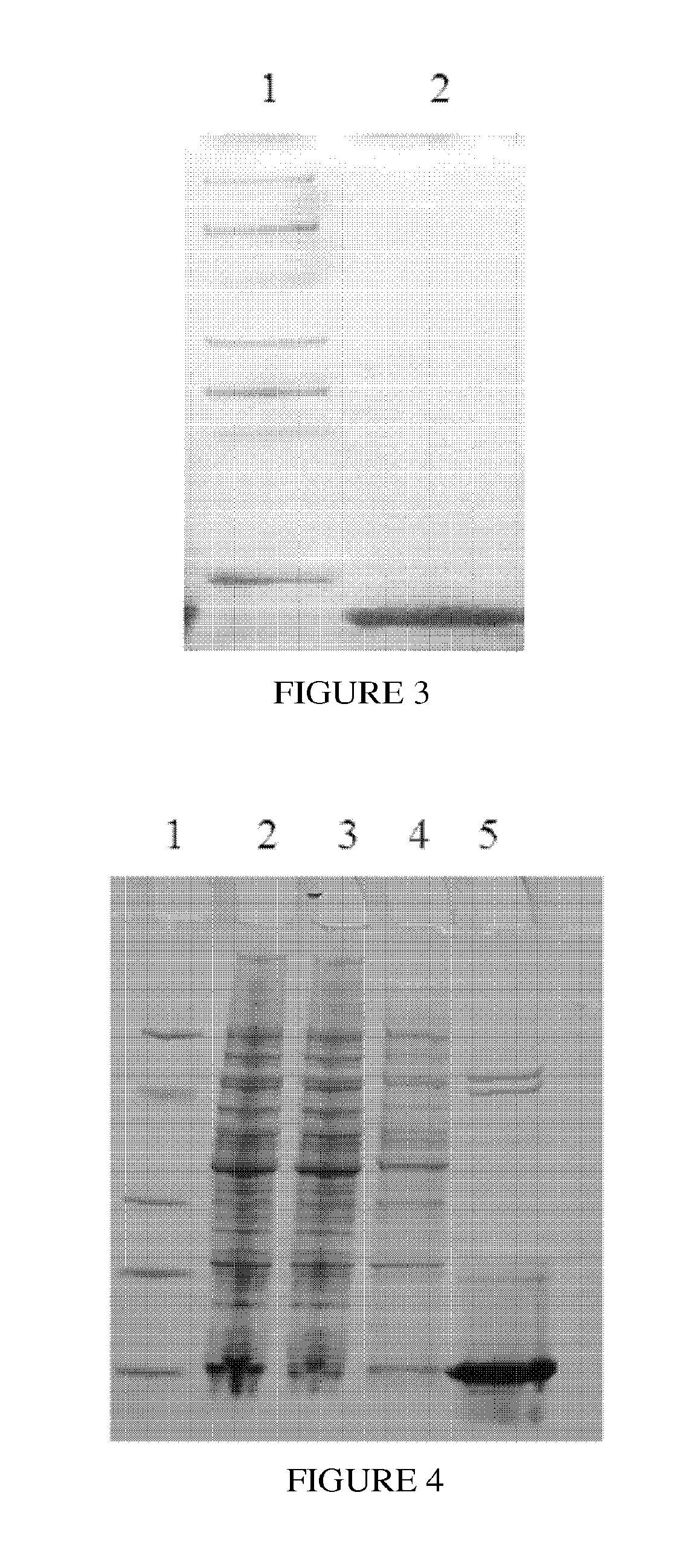
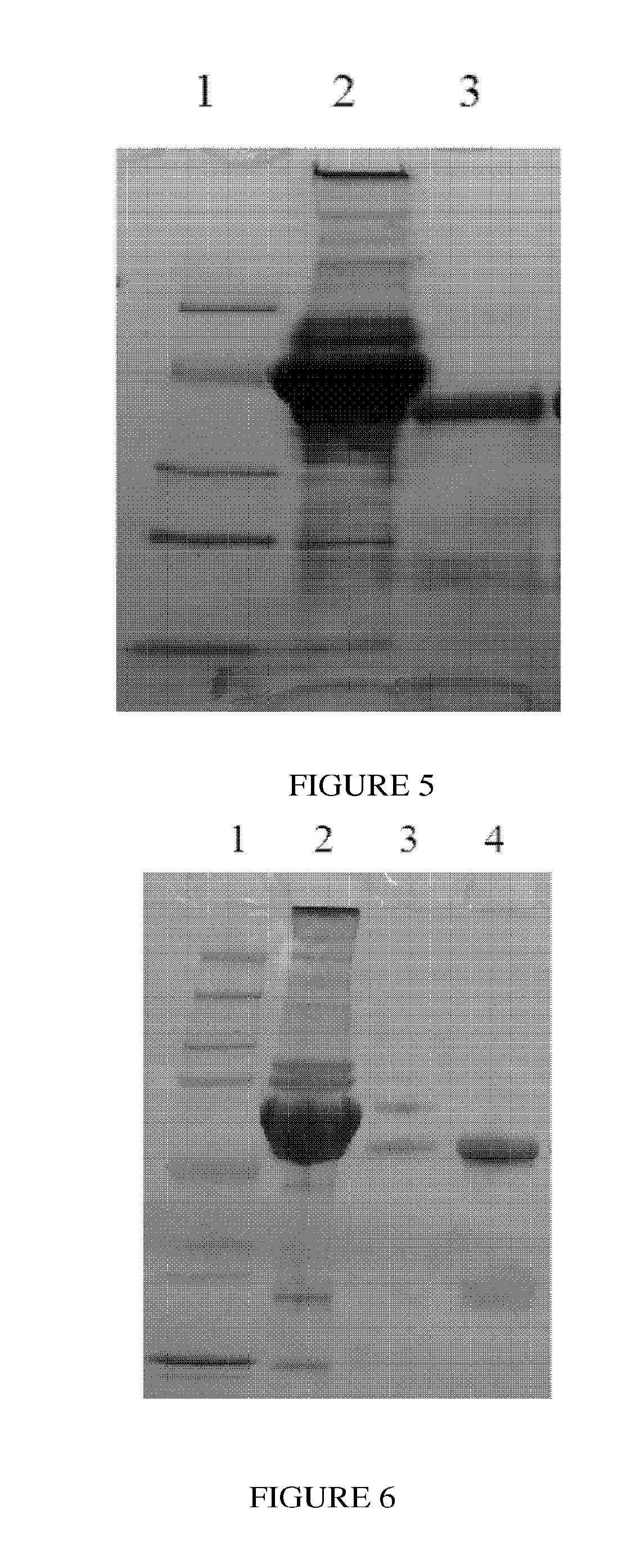
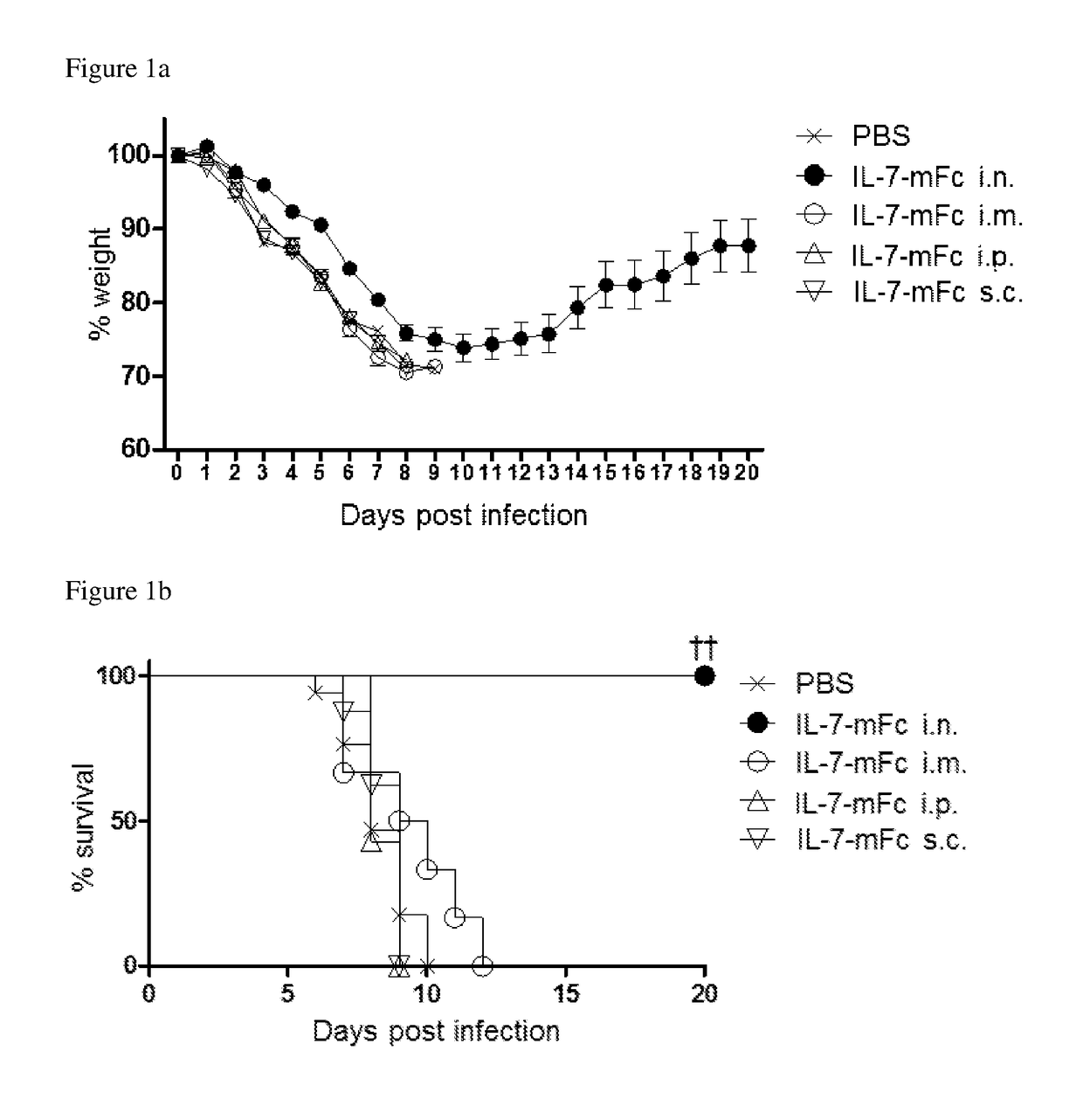
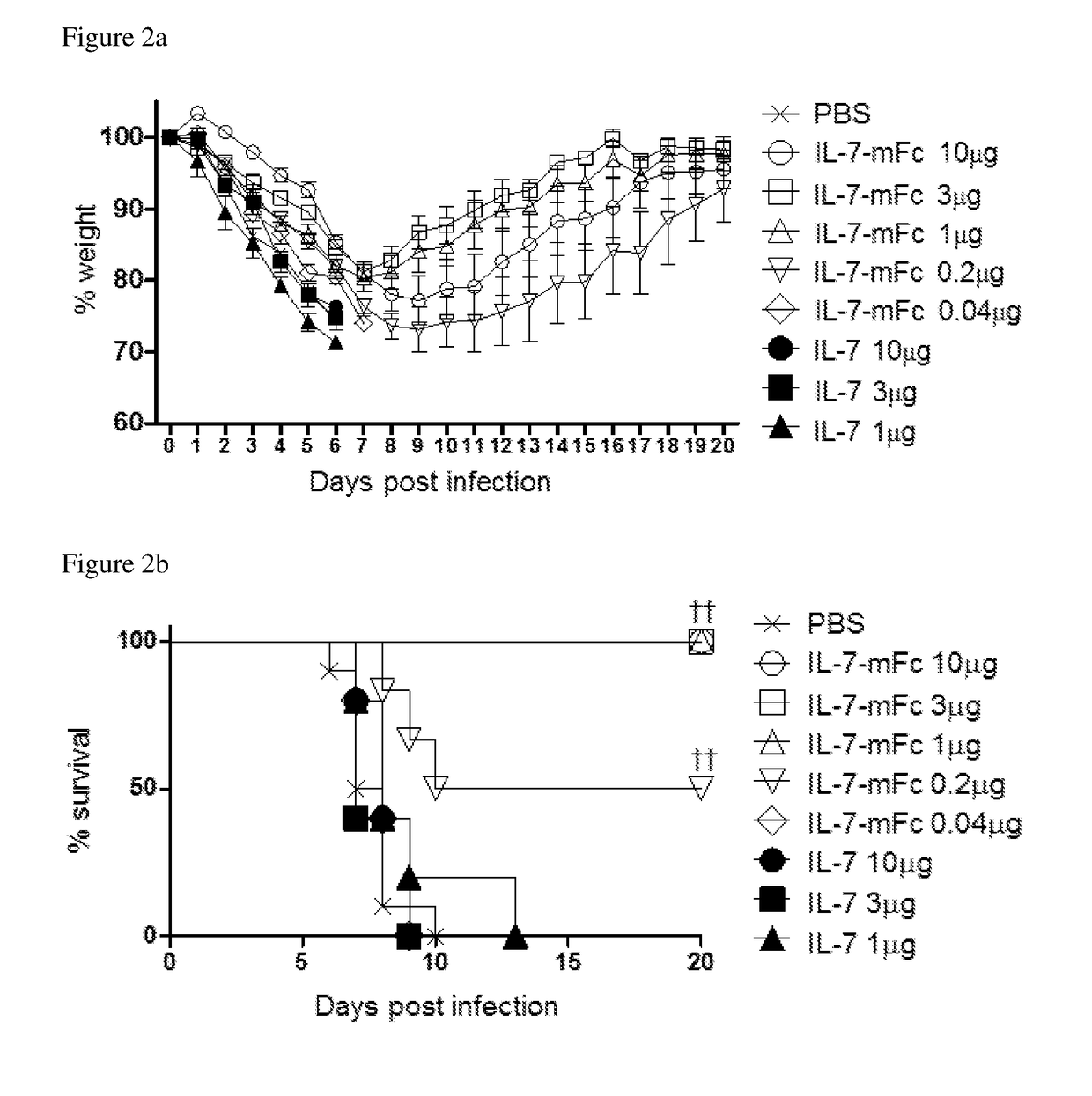
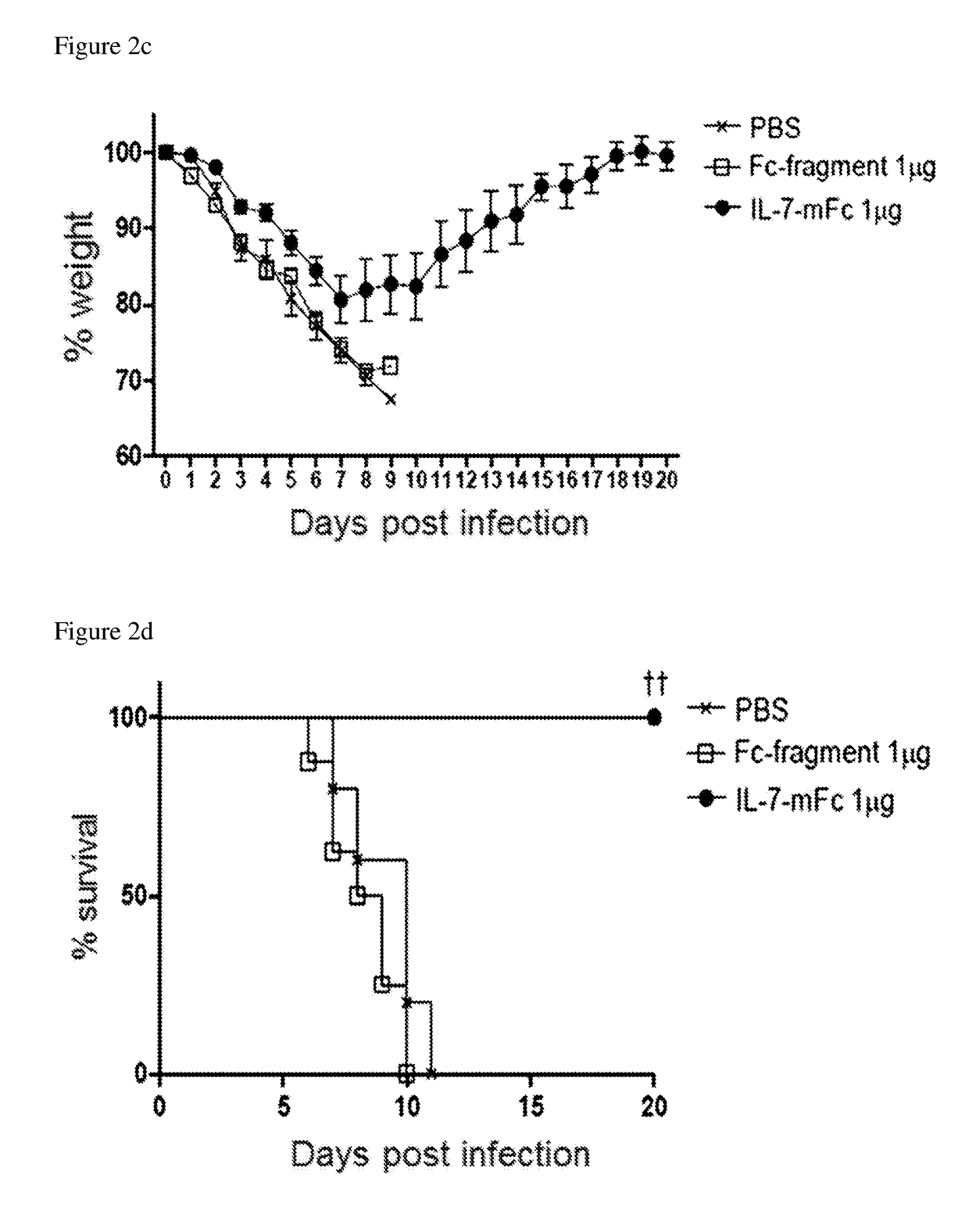
Pharmaceutical composition comprising immunoglobulin fc-fused interleukin-7 fusion protein for preventing or treating infection from influenza virus
ActiveUS20180353573A1Avoid infectionPeptide/protein ingredientsPharmaceutical delivery mechanismImmunoglobulin regionVirus
The present invention relates to a pharmaceutical composition comprising an interleukin-7 fusion protein to which an immunoglobulin Fc region has been fused for preventing or treating diseases caused by influenza virus A. The fusion protein comprising the immunoglobulin Fc region and IL-7 according to the present invention protects the body from infection due to influenza virus A and thus can treat diseases which can be caused by the virus.
Owner:GENEXINE INC
Long acting protein complex having an enhanced efficiency
PendingUS20200283492A1Excellent in vivo durability and activityUseful for developmentPeptide/protein ingredientsAntibody mimetics/scaffoldsBinding siteIntravenous gammaglobulin
The present invention relates to a protein complex, in which a physiologically active polypeptide modified with a fatty acid molecule and an immunoglobulin Fc region are linked through a non-peptide polymer linker, a pharmaceutical composition comprising the same, and a method for preparing the protein complex. The protein complex extends the blood half-life of the physiologically active polypeptide through the linkage of the binding site of immunoglobulin. Fc and the intramolecular bonding of the fatty acid molecule; has an excellent in vivo activity; and has no risk of inducing an immune response.
Owner:HANMI PHARMA
Non-peptidic polymeric linker compound, conjugate comprising same linker compound, and methods for preparing same linker compound and conjugate
ActiveCN110545852AHigh yieldImprove stabilityOrganic chemistryPharmaceutical non-active ingredientsImmunoglobulin regionBiochemistry
One aspect of the present invention provides a compound in which a functional group capable of binding to a globulin Fc region or a physiologically active polypeptide is introduced at one end of a non-peptidic polymer and a functional group capable of a click reaction is introduced at the other end; a polypeptide conjugate in which a physiologically active polypeptide binds to one end of the compound; a physiologically active polypeptide conjugate in which a physiologically active polypeptide and an immunoglobulin Fc region bind to both ends thereof by using the compound as a linker; and methods for preparing the same compound, polypeptide conjugate, and physiologically active polypeptide conjugate.
Owner:HANMI PHARMA
Liquid formulation of a fusion protein comprising tnfr and fc region
ActiveUS20170028020A1Good storage stabilityEasy to usePeptide/protein ingredientsAntibody mimetics/scaffoldsCrystallographySucrose
The present invention relates to a liquid formulation comprising a TNFR-Fc fusion protein and a stabilizer, in which the fusion protein comprises TNFR (tumor necrosis factor receptor) or a fragment thereof and an immunoglobulin Fc region, and the stabilizer comprises one or more amino acids selected from the group consisting of proline and histidine, a buffer solution, and an isotonic agent containing sodium chloride (NaCl) and sucrose, and a preparation method of the liquid formulation. The liquid formulation according to the present invention provides excellent storage stability because long-term storage of TNFR-Fc fusion protein (etanercept) is possible and particular storage conditions are not needed. Since the liquid formulation of the present invention shows excellent storage stability even though the formulation is simple, it is more economical than other stabilizers or lyophilized formulations, and thus the formulation can be effectively applied for uses wherein treatment of TNFR-Fc fusion protein (etanercept) is beneficial.
Owner:ARES TRADING SA
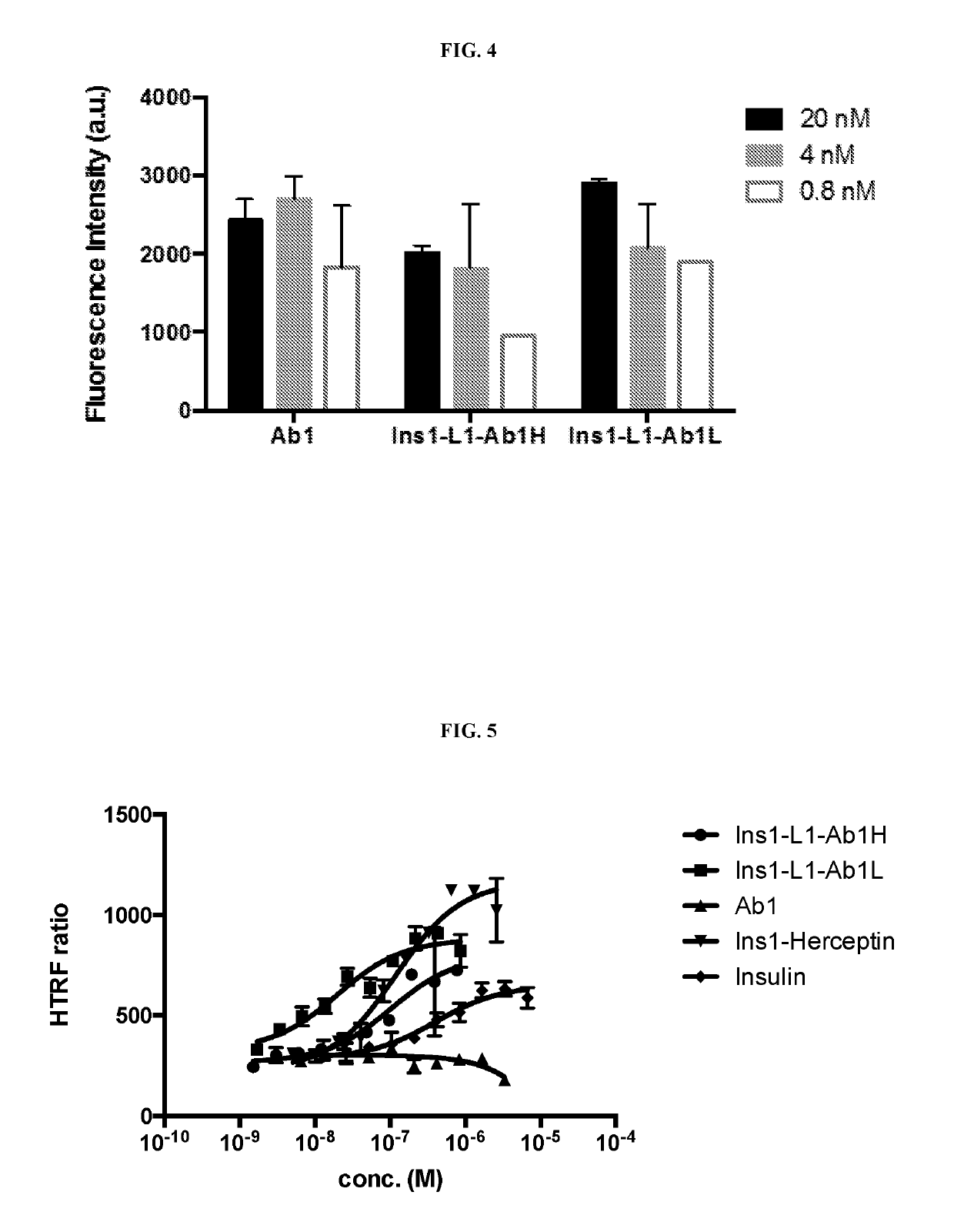
Insulin immunoglobulin fusion proteins
Owner:THE SCRIPPS RES INST
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com