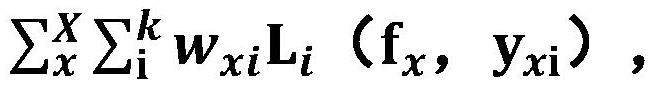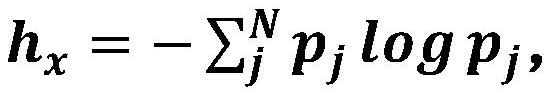Patents
Literature
35 results about "Single voxel" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
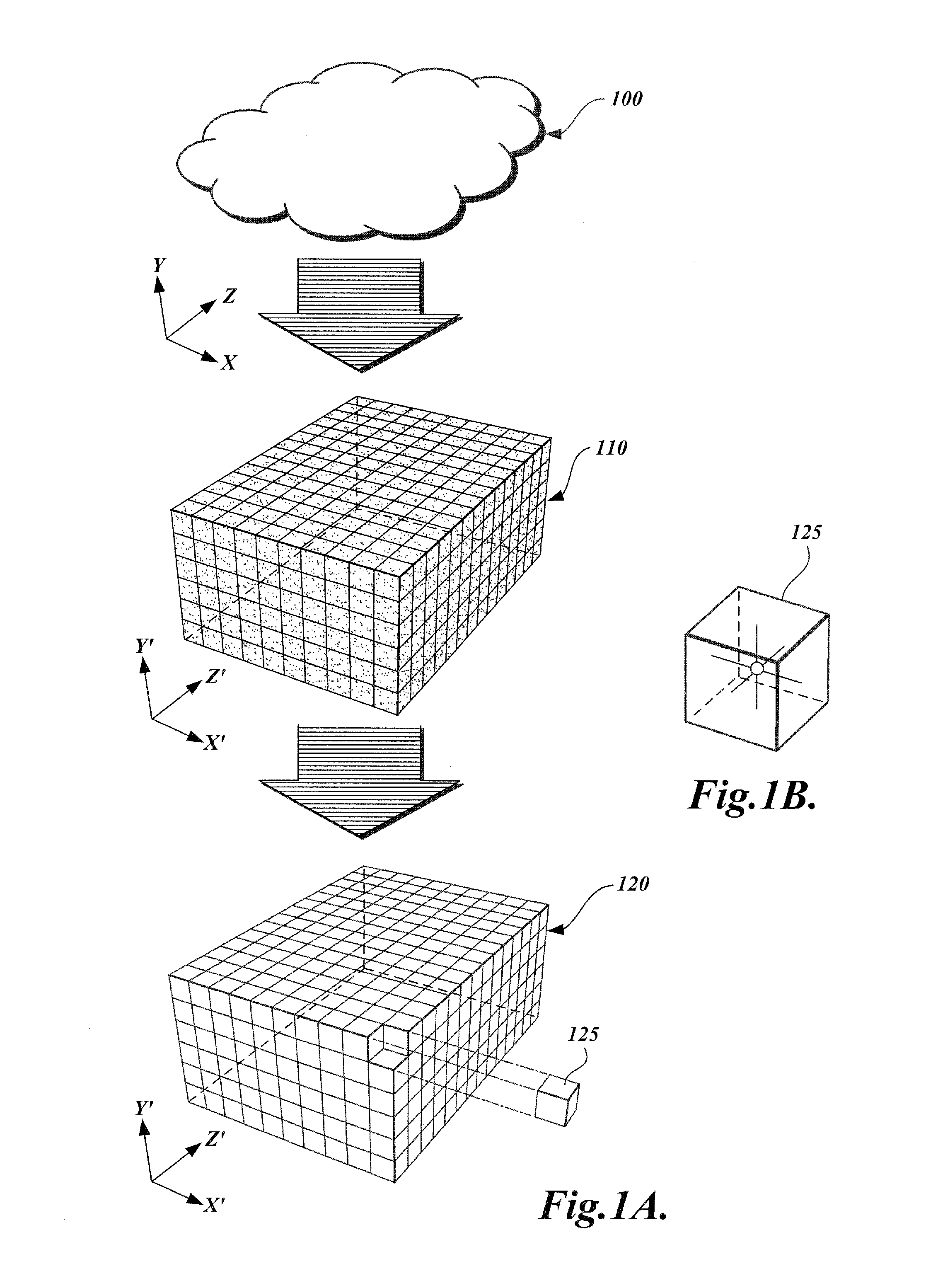
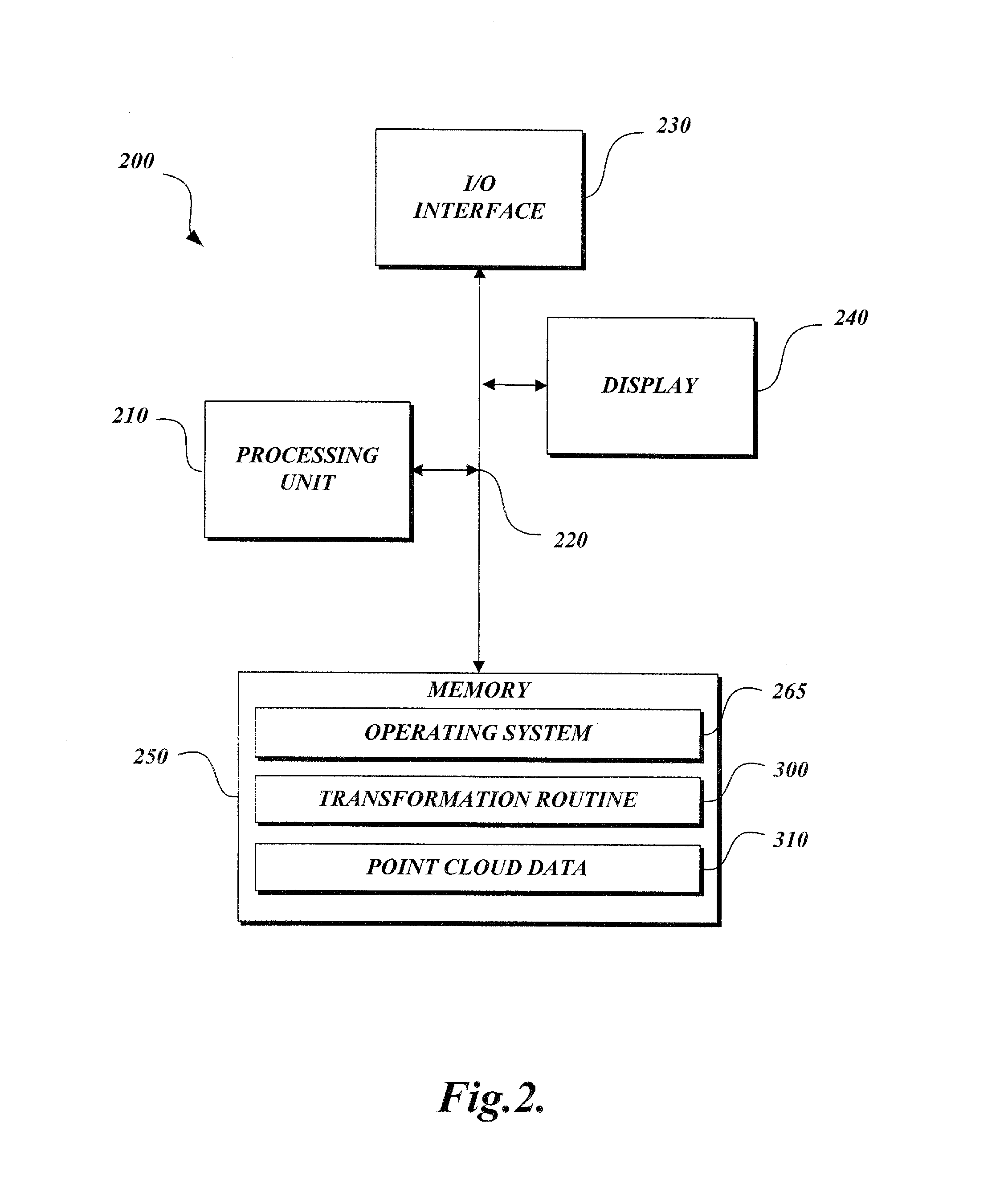
Method and apparatus for transforming point cloud data to volumetric data
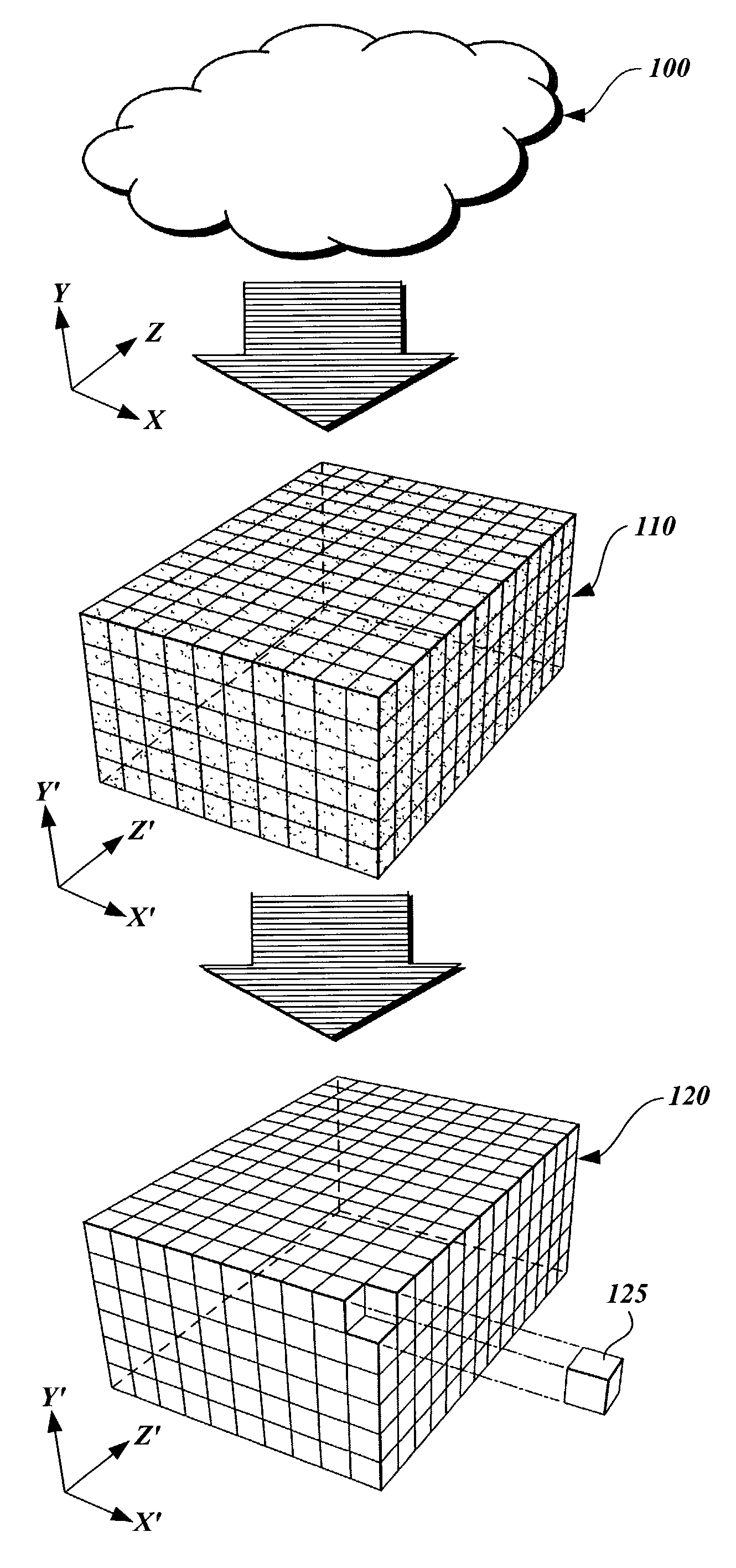
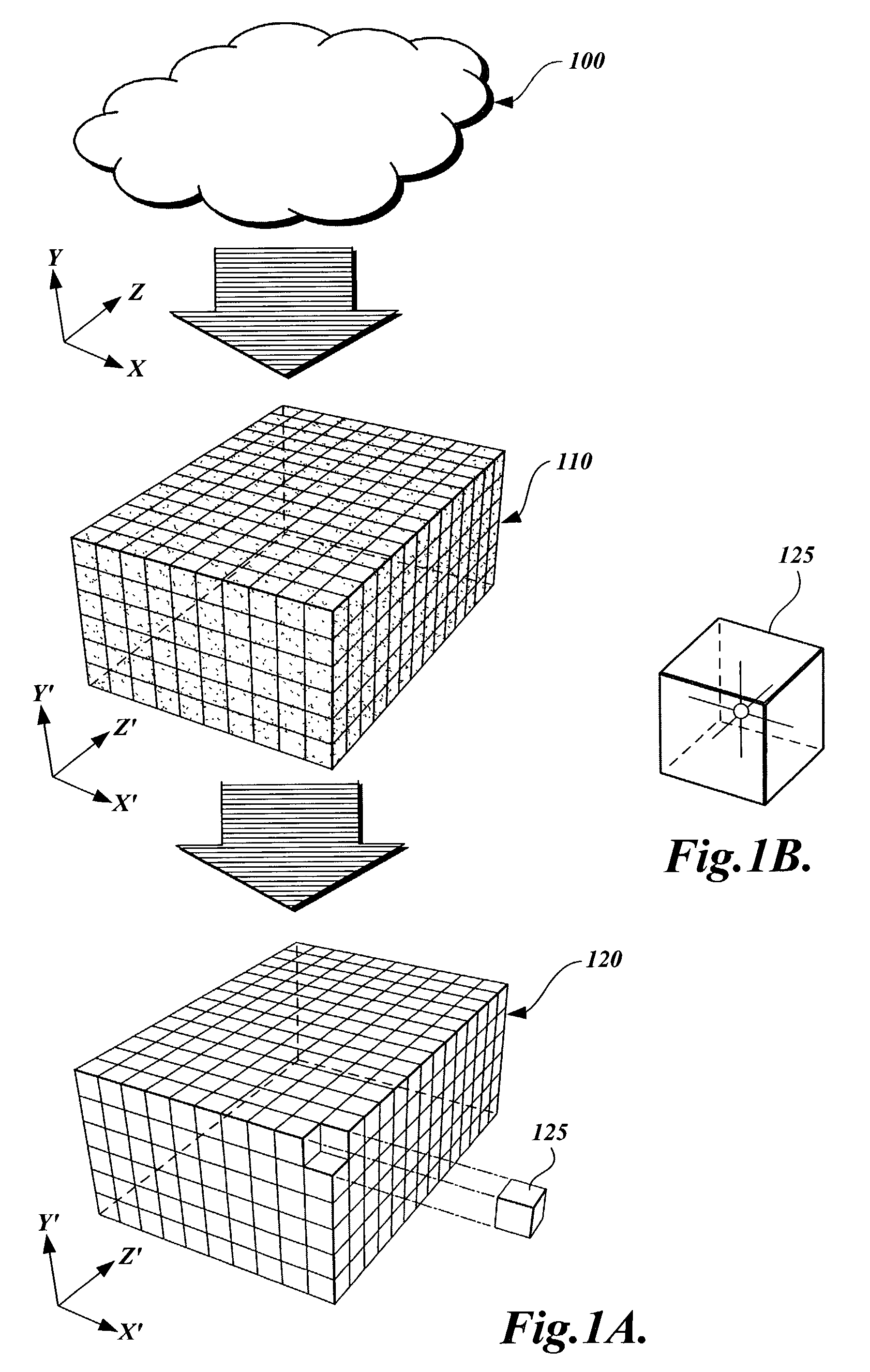
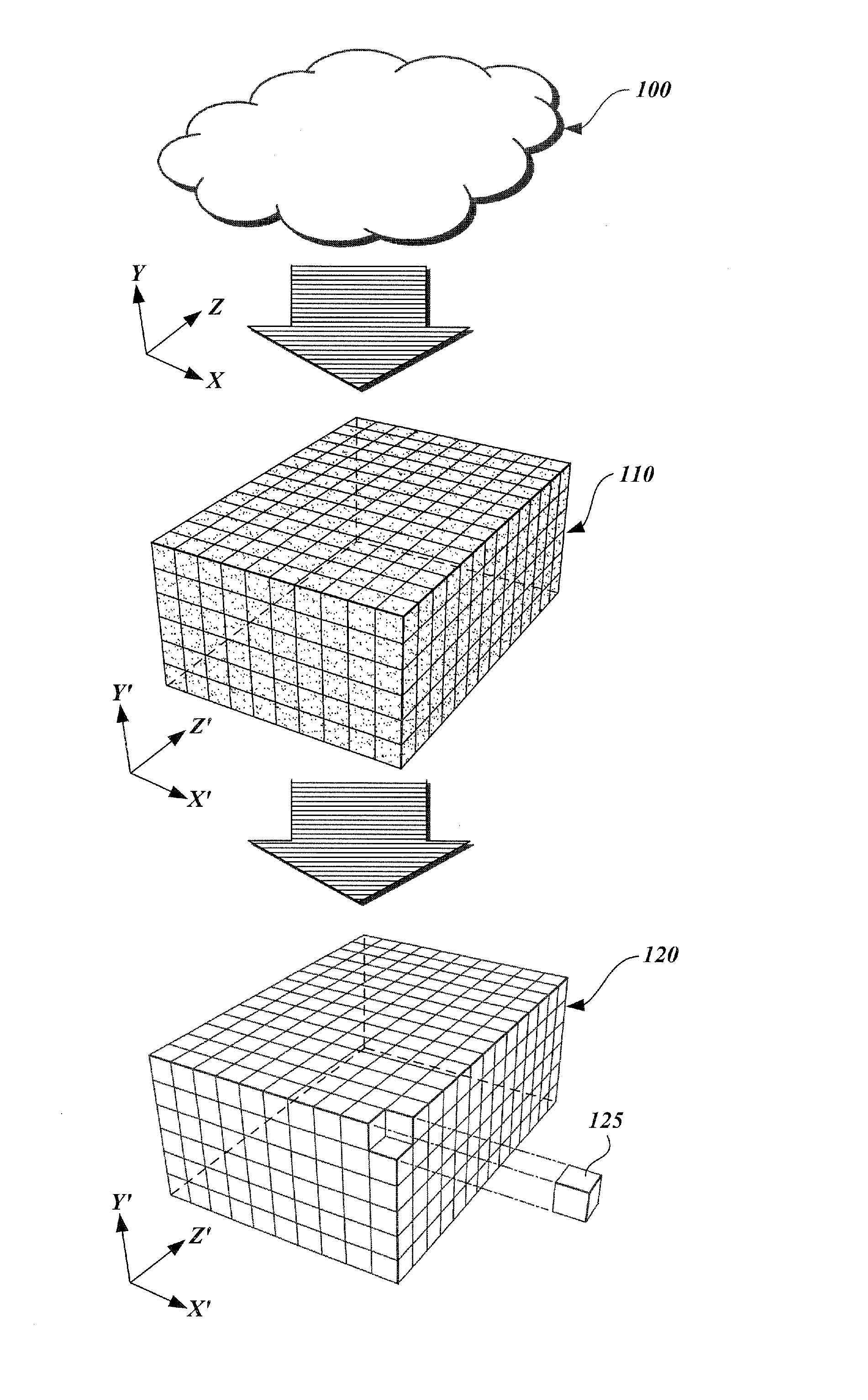
A method and apparatus are provided for transforming an irregular, unorganized cloud of data points (100) into a volumetric data or “voxel” set (120). Point cloud (100) is represented in a 3D cartesian coordinate system having an x-axis, a y-axis and a z-axis. Voxel set (120) is represented in an alternate 3D cartesian coordinate system having an x′-axis, a y′-axis, and a z′-axis. Each point P(x, y, z) in the point cloud (100) and its associated set of attributes, such as intensity or color, normals, density and / or layer data, are mapped to a voxel V(x′, y′, z′) in the voxel set (120). Given that the point cloud has a high magnitude of points, multiple points from the point cloud may be mapped to a single voxel. When two or more points are mapped to the same voxel, the disparate attribute sets of the mapped points are combined so that each voxel in the voxel set is associated with only one set of attributes.
Owner:MCLOUD TECH USA INC
Method and apparatus for transforming point cloud data to volumetric data
A method and apparatus are provided for transforming an irregular, unorganized cloud of data points (100) into a volumetric data or “voxel” set (120). Point cloud (100) is represented in a 3D Cartesian coordinate system having an x-axis, a y-axis and a z-axis. Voxel set (120) is represented in an alternate 3D Cartesian coordinate system having an x′-axis, a y′-axis, and a z′-axis. Each point P(x, y, z) in the point cloud (100) and its associated set of attributes, such as intensity or color, normals, density and / or layer data, are mapped to a voxel V(x′, y′, z′) in the voxel set (120). Given that the point cloud has a high magnitude of points, multiple points from the point cloud may be mapped to a single voxel. When two or more points are mapped to the same voxel, the disparate attribute sets of the mapped points are combined so that each voxel in the voxel set is associated with only one set of attributes.
Owner:MCLOUD TECH USA INC
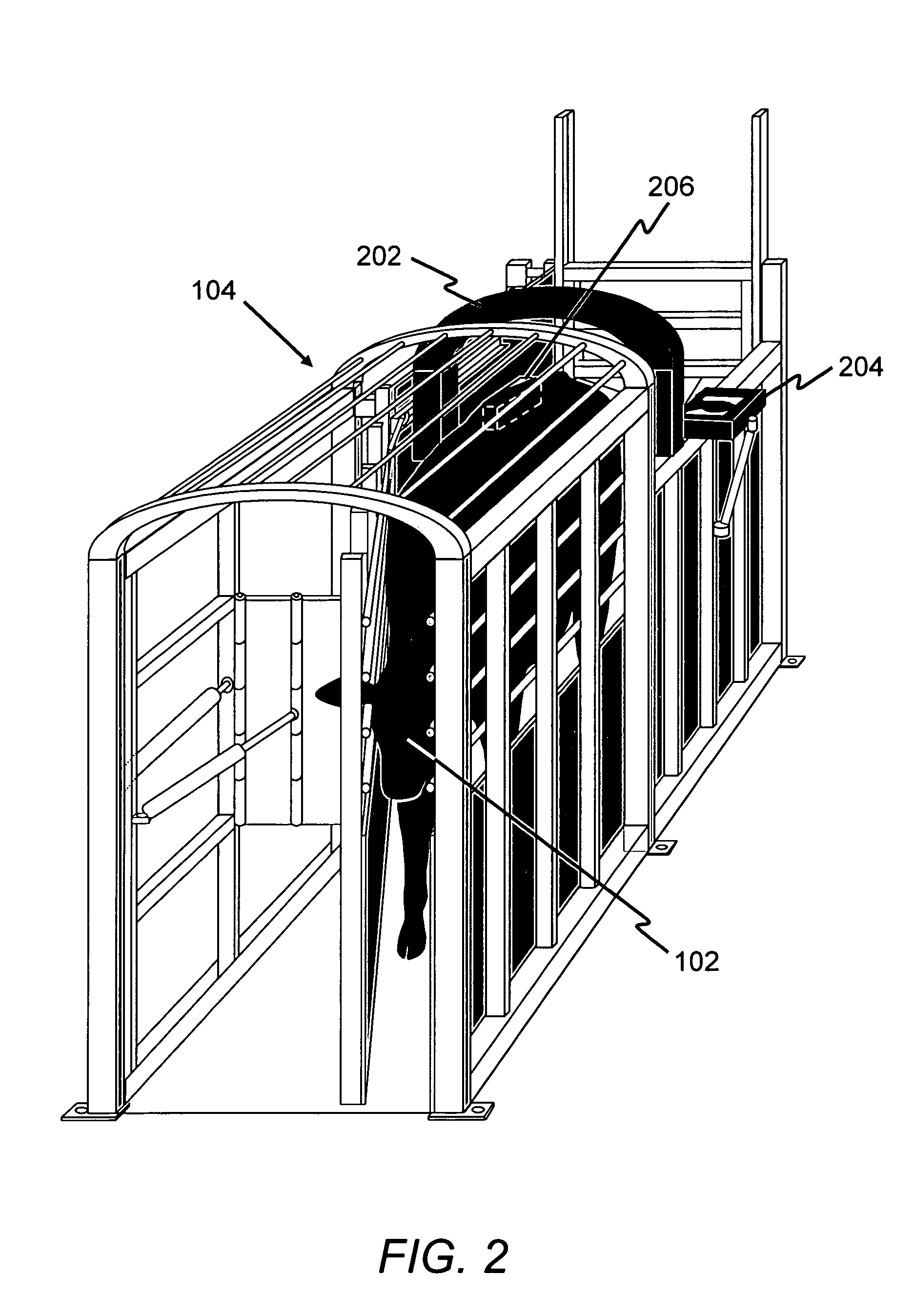
Animal sorting and grading system using an internal evaluation to predict maximum value
InactiveUS7444961B1Reduces the dollars lostAccurate measurementCharacter and pattern recognitionAnimal housingComputed tomographyEngineering
A system that compares, ranks, sorts and grades animals into groups of like kinds according to previously determined predicted maximum values. The system uses a single method such as multi-dimensional single voxel(s) of MR, NMR, MRI, ultrasound, C.A.T. scan, CT scan, P.E.T. scan, or a combination of these, or any equivalent internal measuring device, on a single occasion, to evaluate the animal and determine a number of days the animal must be fed to reach a maximum value. The system also combines internal evaluation with an external animal evaluation system to refine the number of days remaining for the animal to reach a maximum value, and the system, when used in a feedlot, will direct the animal to a feed pen based on the number of days remaining for the animal to reach maximum value.
Owner:ELLIS JAMES S
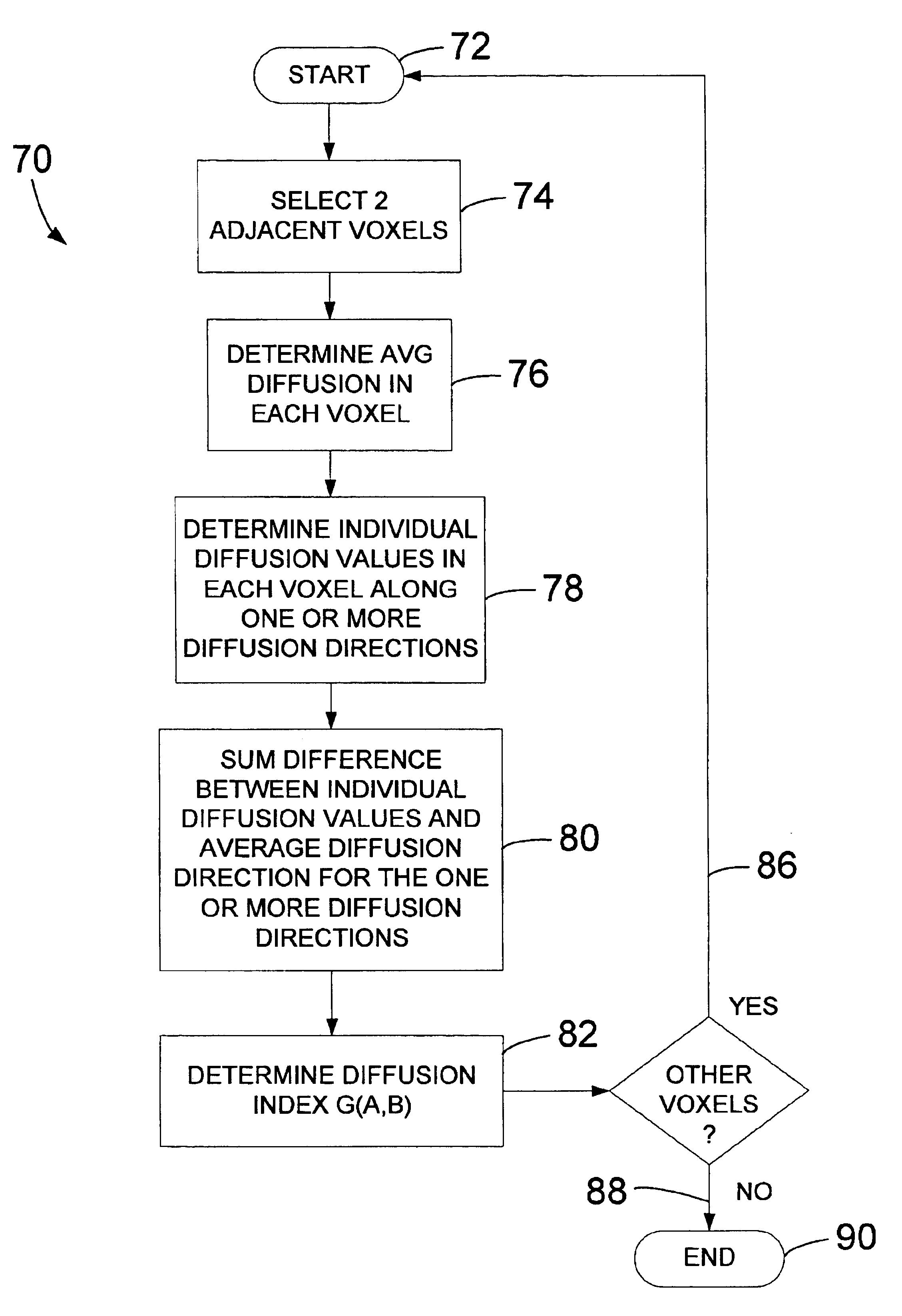
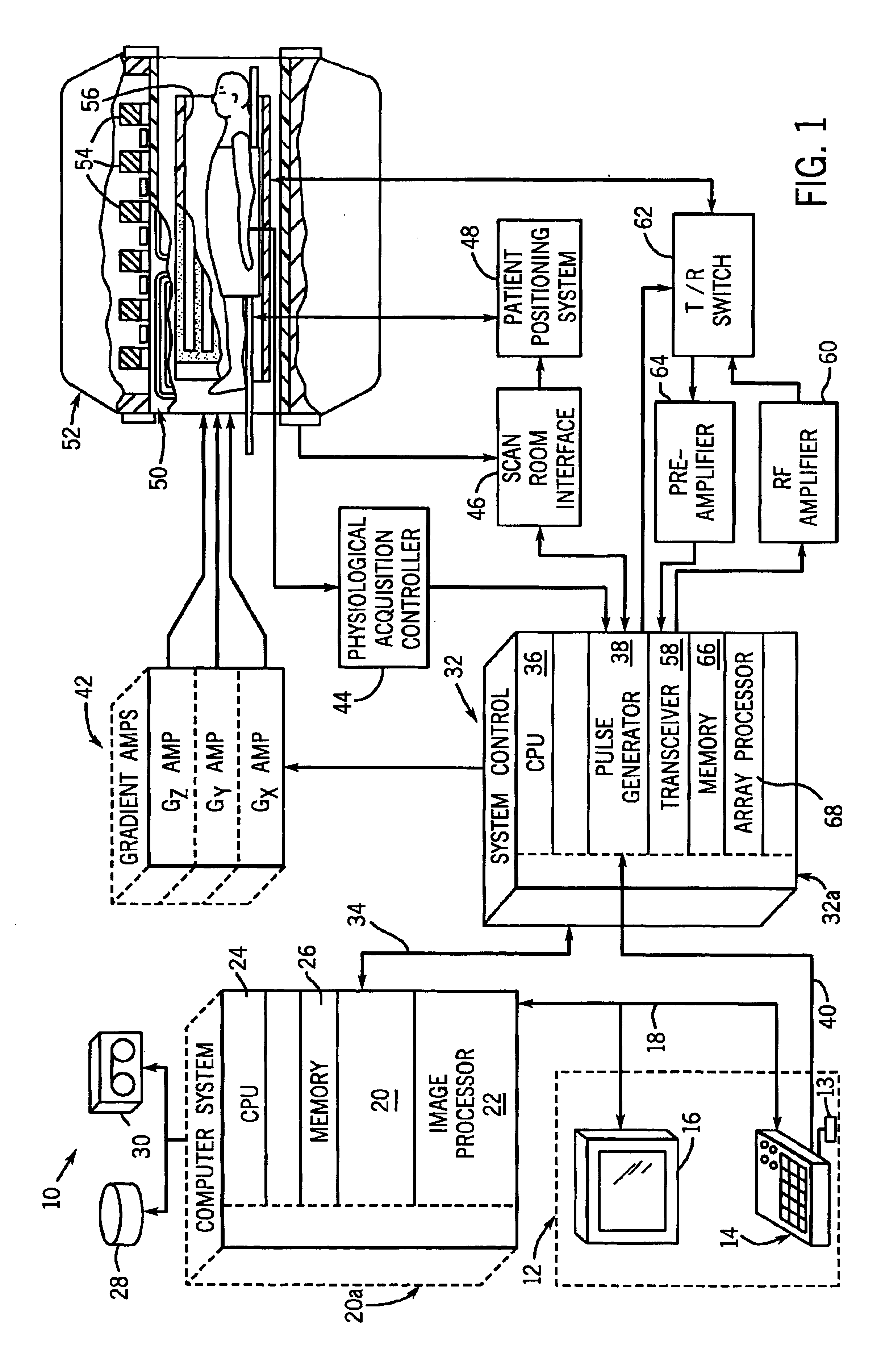
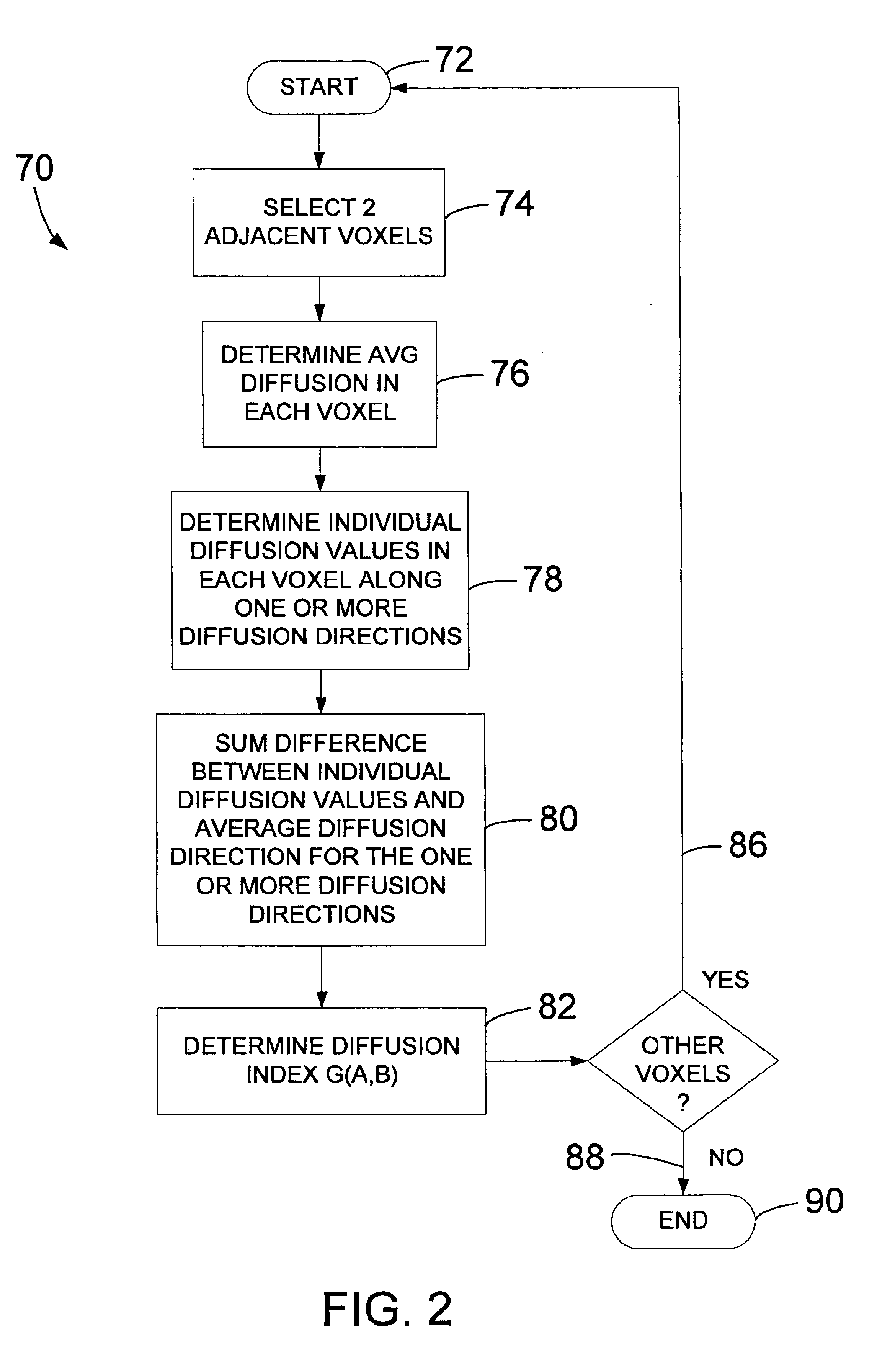
Method and system of quantitatively assessing diffusion characteristics in a tissue
InactiveUS6853189B1Quick fixTechnique is effectiveMeasurements using NMR imaging systemsElectric/magnetic detectionDiffusion AnisotropyDiffusion
A process and system to implement such a process is configured to determine diffusion properties within a region-of-interest. A metric or index of diffusion is determined from individual diffusion values corresponding to diffusion along, multiple diffusion directions relative to average diffusion at a related voxel or pair of voxels. The invention may be implemented to determine shared diffusion anisotropy between two voxels. Based on a value of a diffusion index or metric, the present invention provides an efficient and tensor-free technique of determining if diffusion in neighboring voxels is anisotropic and if the diffusion is oriented in the same direction within each voxel. The present invention may be used to rapidly determine fractional anisotropy in a single voxel or used to filter diffusion weighted imaging data. The present invention may also be used to color-code diffusion data such that diffusion and tract orientation are readily identifiable in a reconstructed image.
Owner:CATHOLIC HEALTHCARE WEST CALIFORNIA NONPROFIT PUBLIC BENEFIT D B A ST JOSEPHS HOSPITAL & MEDICAL CENT
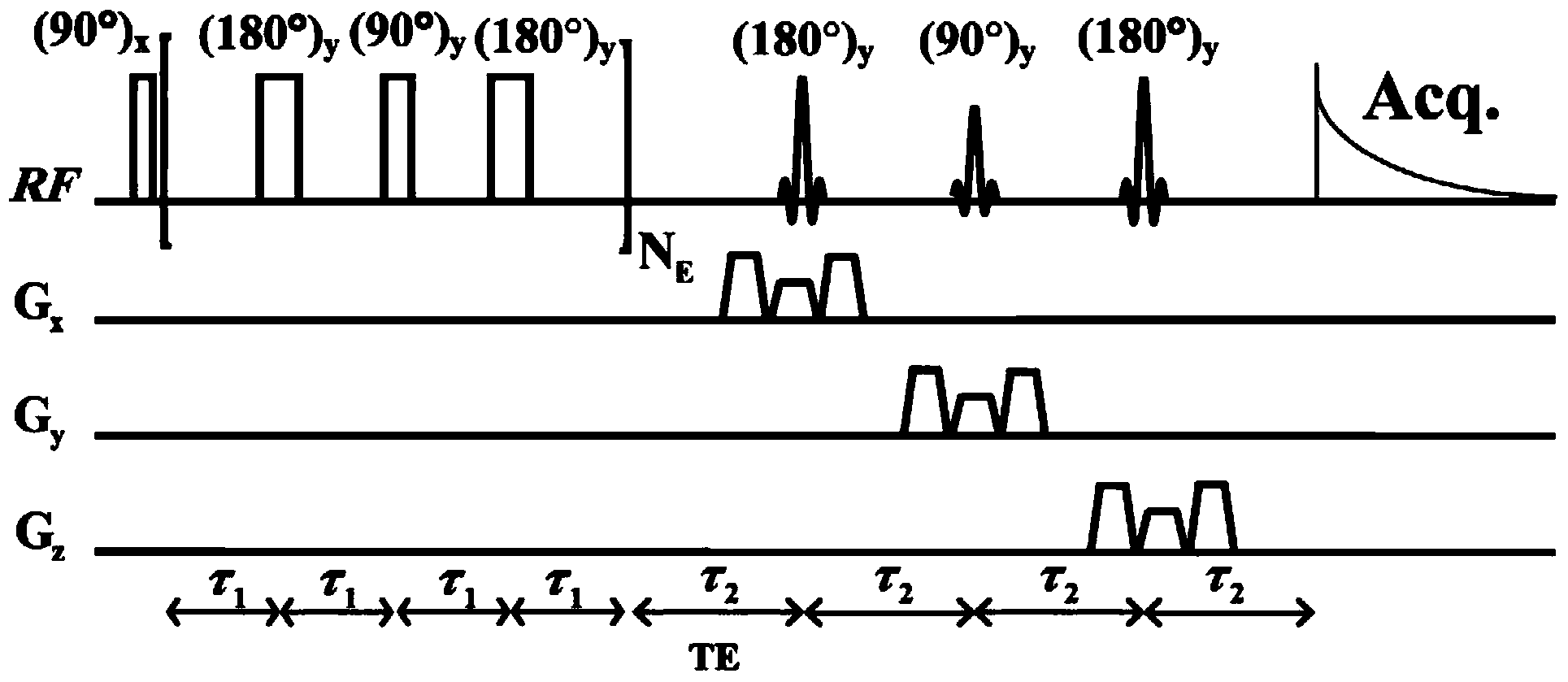
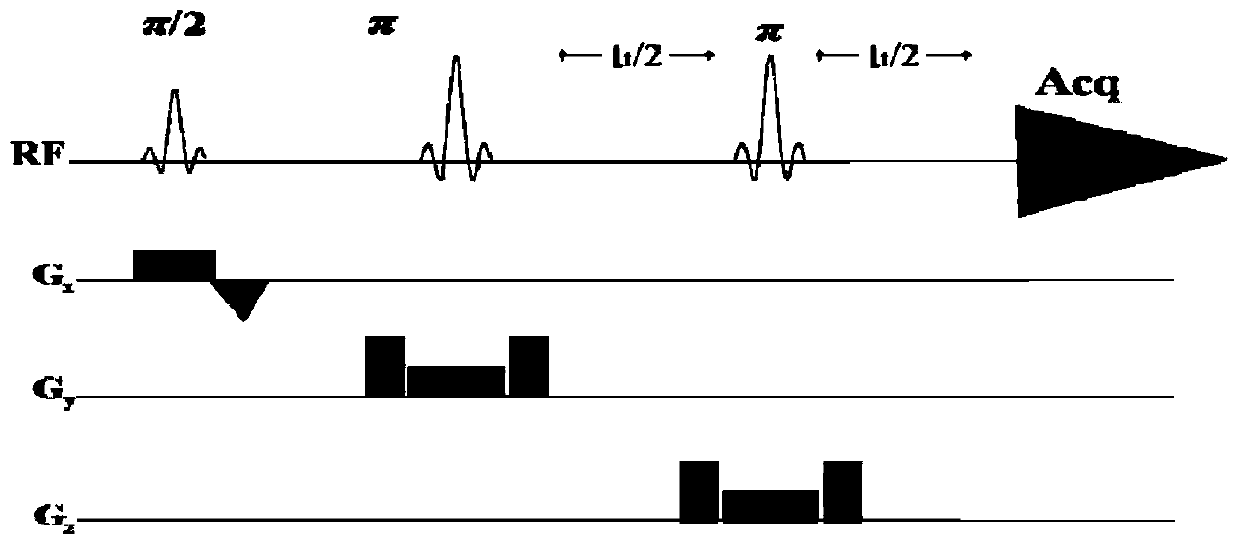
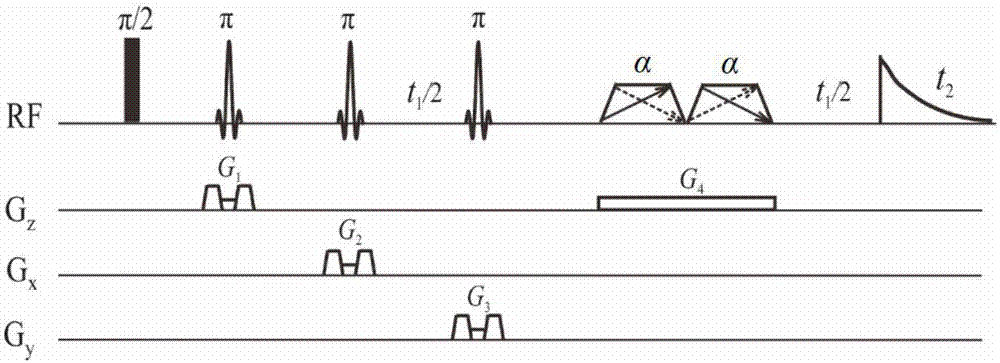
Method for obtaining single voxel one-dimensional localization spectra capable of eliminating scalar coupling modulation
InactiveCN103645453APromote localizationGood signalMagnetic measurementsPhase distortionCoupling constant
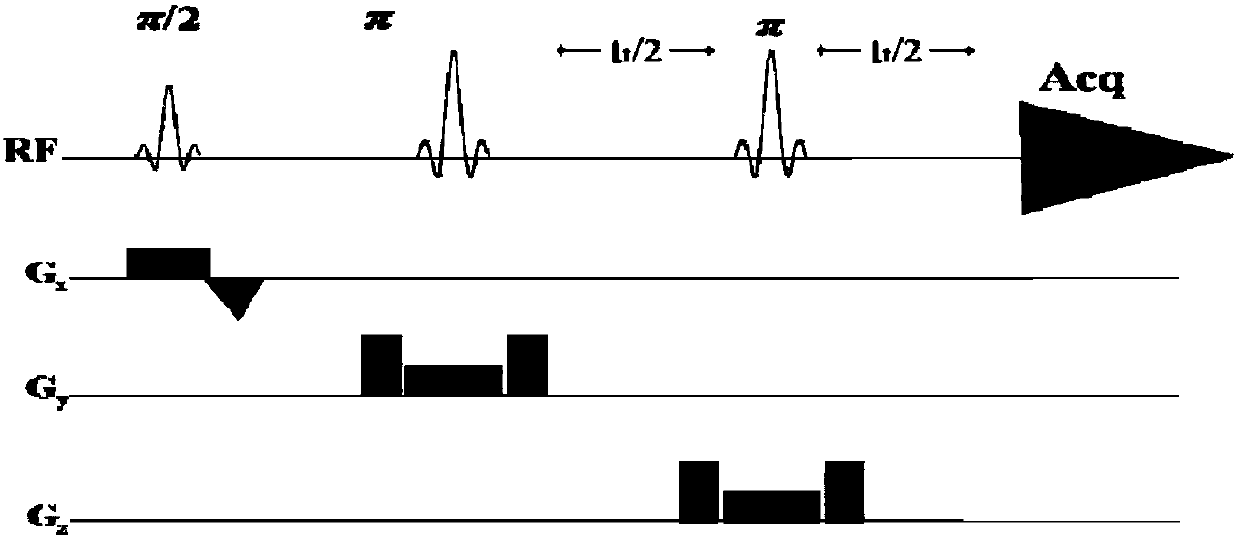
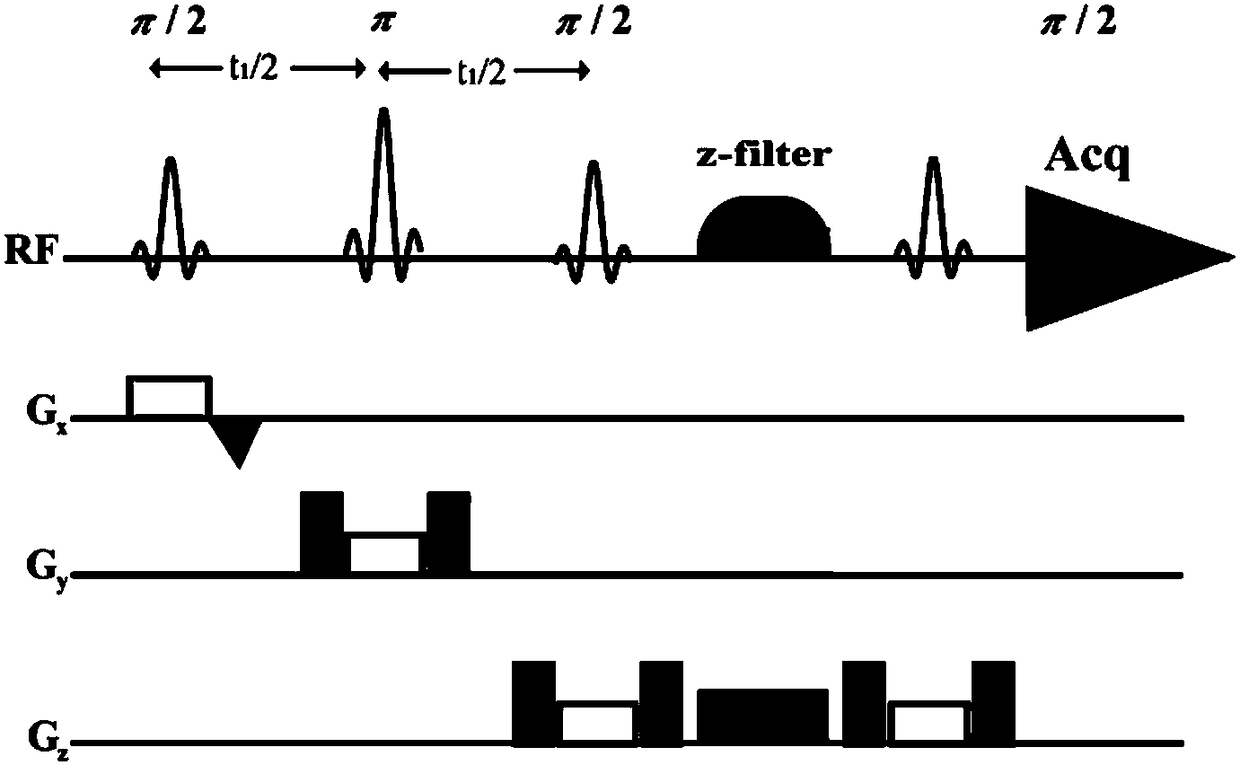
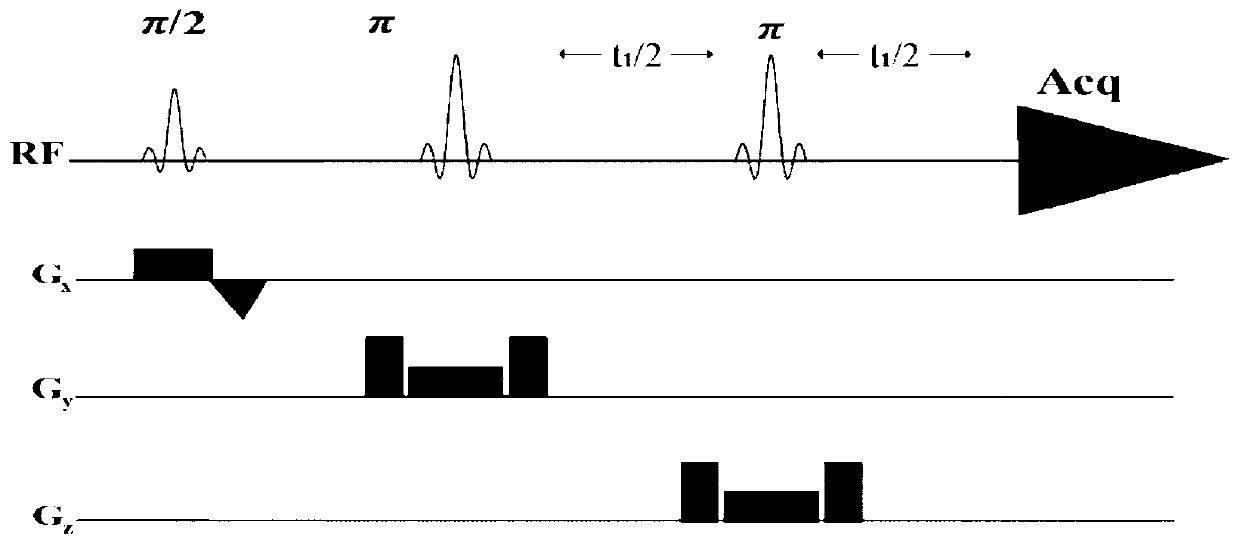
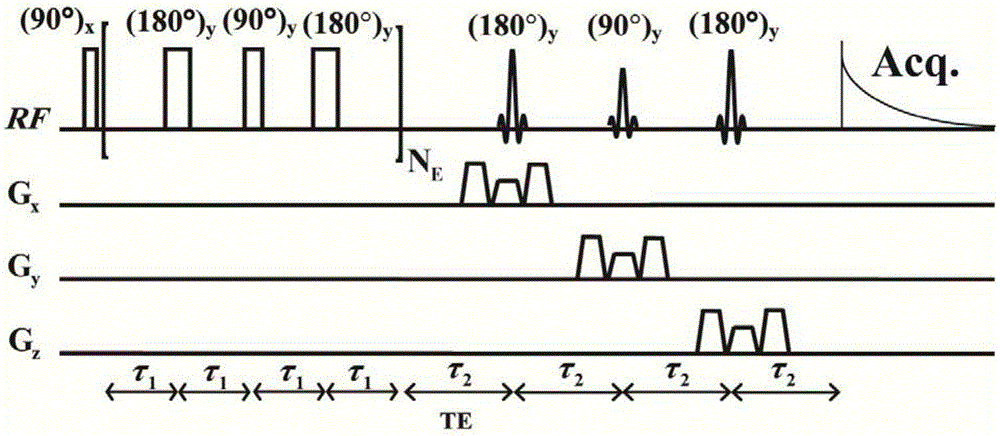
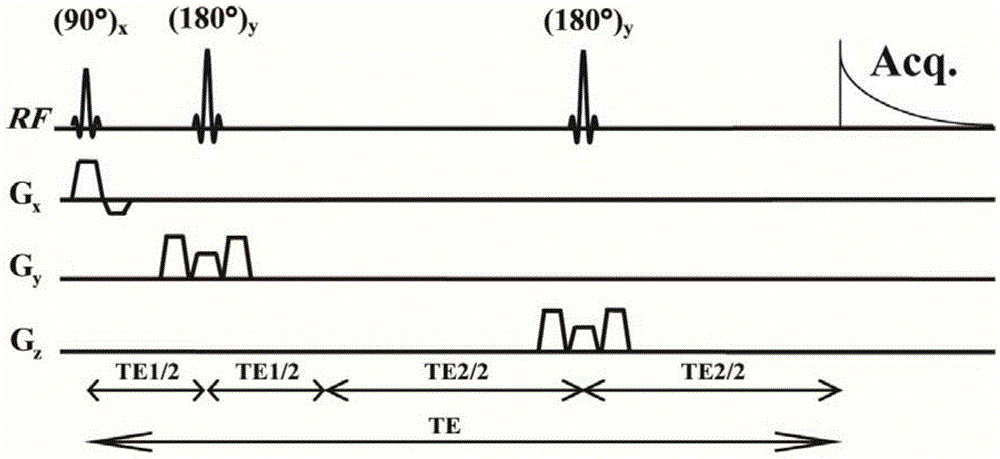
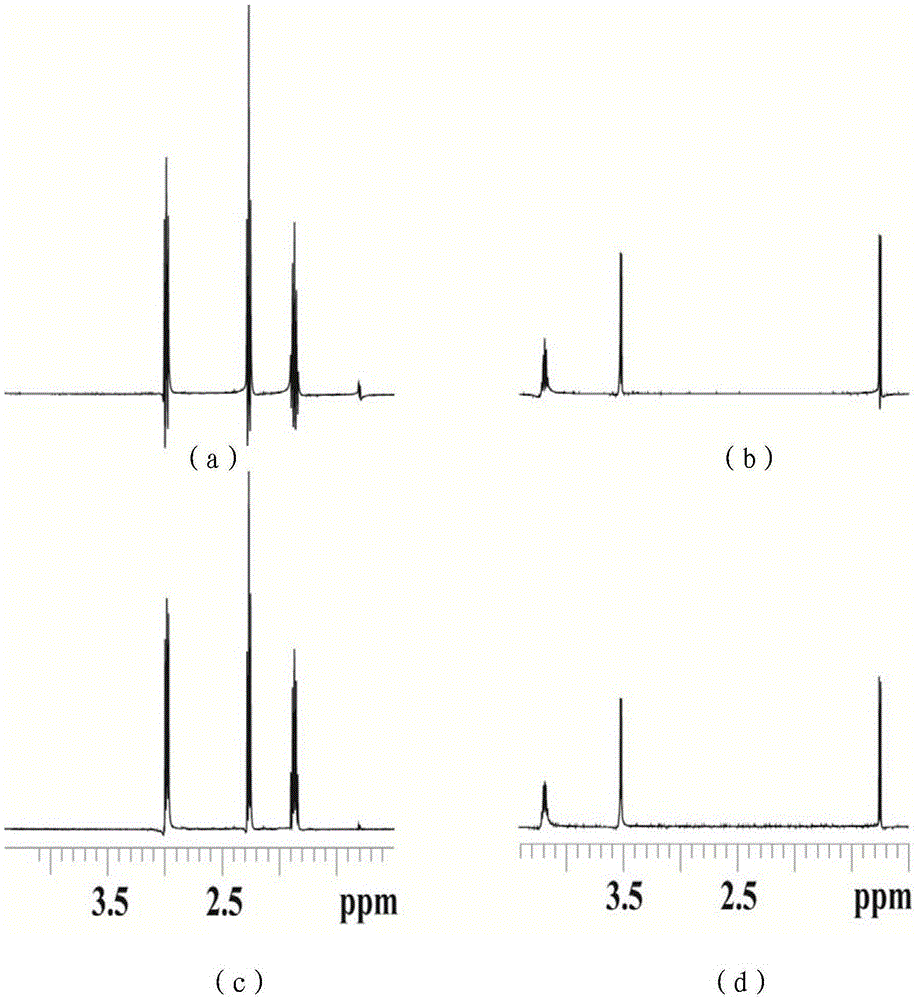
The invention provides a method for obtaining single voxel one-dimensional localization spectra capable of eliminating scalar coupling modulation. A same-direction 90-degree radio-frequency pulse is added between two spin echo modules to form a hard pulse scalar coupling re-aggregation module and a soft pulse scalar coupling re-aggregation module, the interval t between the pulses in the re-aggregation modules is set according to the coupling system condition included in a sample, and when the t and scalar coupling constant J product is much smaller than 1, the scalar coupling re-aggregation modules can eliminate scalar coupling modulation effects caused by various coupling systems. The soft pulse scalar coupling re-aggregation module completes voxel selection by combining with selecting layers and destroy gradients. According to the method, by the aid of the obtained one-dimensional localization spectra, signal amplitude and phase distortions caused by scalar coupling modulation can be prevented, perfect signals are obtained, and spectrogram attribution analysis is facilitated.
Owner:XIAMEN UNIV
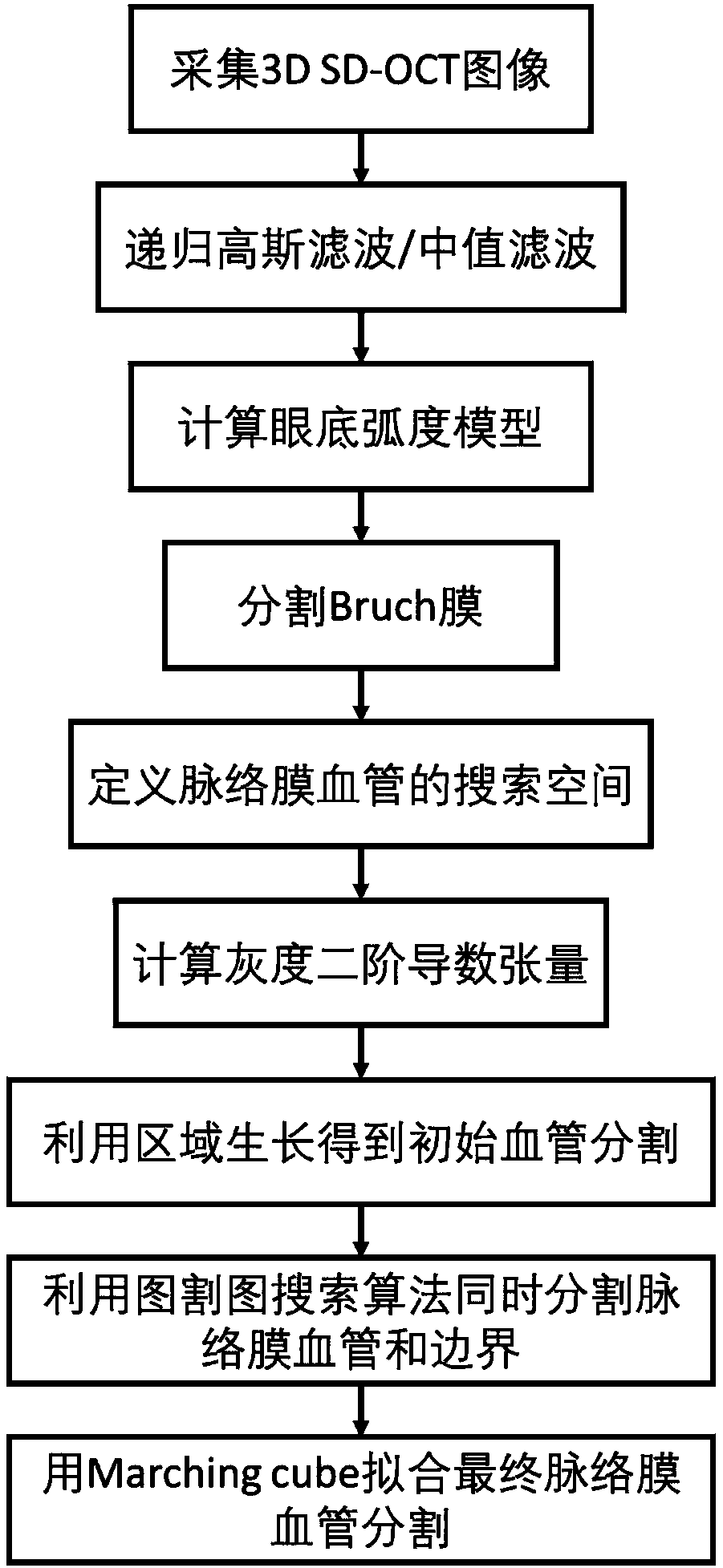
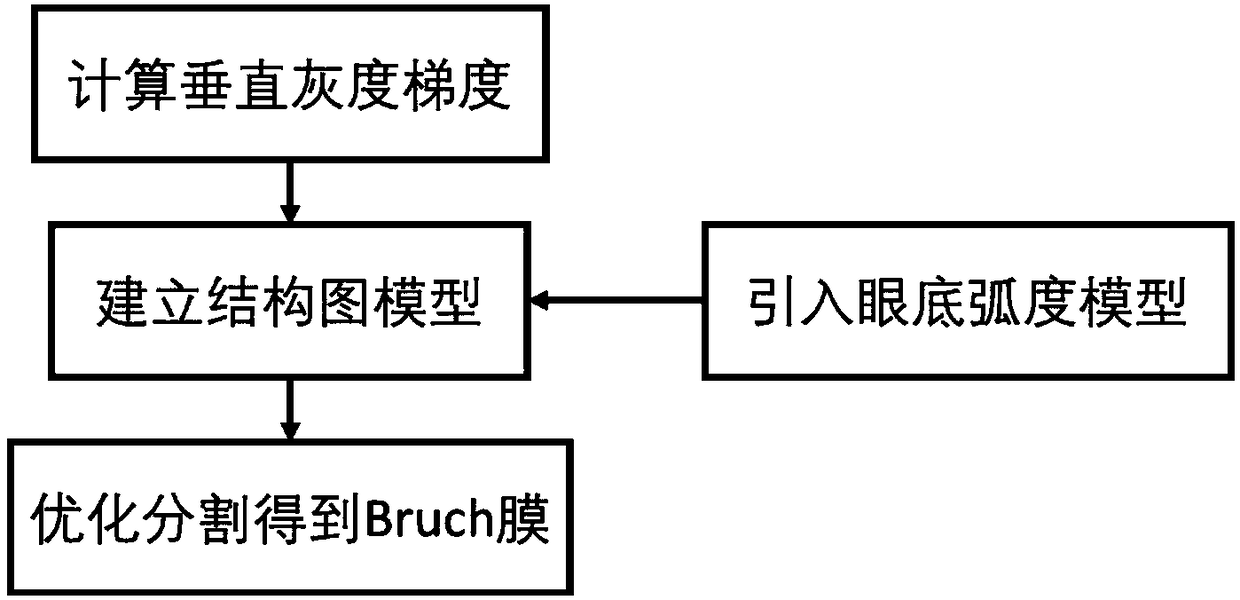
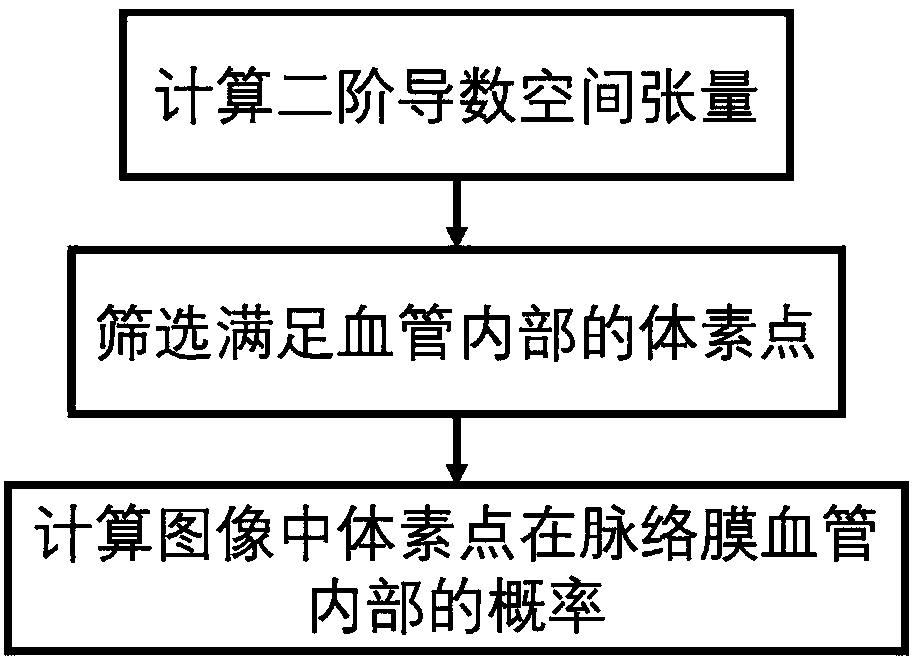
Choroid vessel segmentation method and system based on three-dimensional coherent tomographic image
ActiveCN108416793AImprove Segmentation AccuracyGood repeatabilityImage enhancementImage analysisAlgorithmTomographic image
The invention discloses a choroid vessel segmentation method and system based on a three-dimensional coherent tomographic image. The method includes the steps of collecting the three-dimensional frequency domain coherent optical tomographic image; calculating a fundus radian model according to the space size represented by a single voxel point in the three-dimensional frequency domain coherent optical tomographic image and the actual size of the image; segmenting a retinal base Bruch's membrane from the fundus radian model by using a single-layer graph searching method; calculating the space tensor of each voxel point so as to obtain the point probability of the voxel point in a choroid vessel, and screening out a pre-selected area of the choroid vessel with the point probability value larger than zero; selecting out a point probability value larger than 75% and a point probability value less than 25% from the pre-selected area of the choroid vessel to serve as the high and low thresholds of the pre-selected area growth, so as to obtain the initial segmentation of the foreground and background of the initial segmentation of the choroid vessel, and solving the optimal model of a structure diagram according to the foreground and background of the choroid vessel to obtain the accurate segmentation of the choroid vessel.
Owner:武汉华悦立远科技有限公司
Phantom for evaluating performance of magnetic resonance imaging apparatus using ultra high field
ActiveUS20160282439A1Reduce the overall heightPrevent water leakageMeasurements using NMR imaging systemsHuman bodyImaging condition
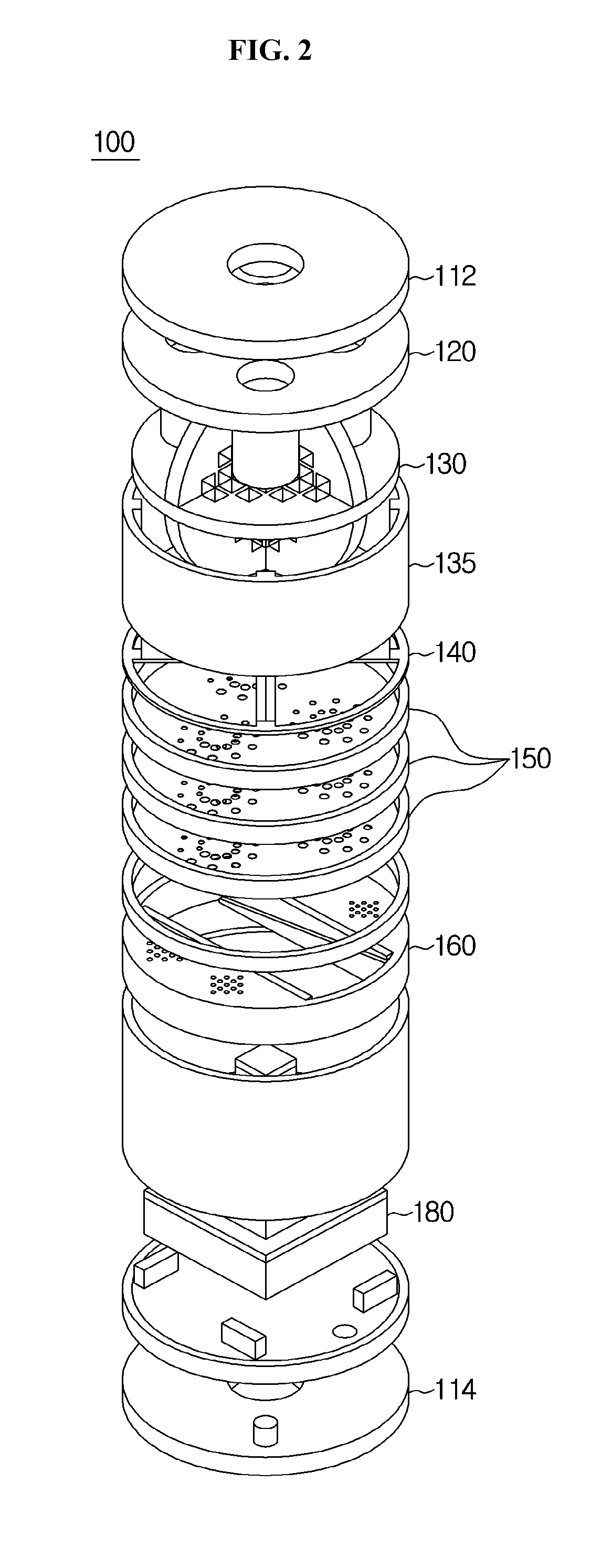
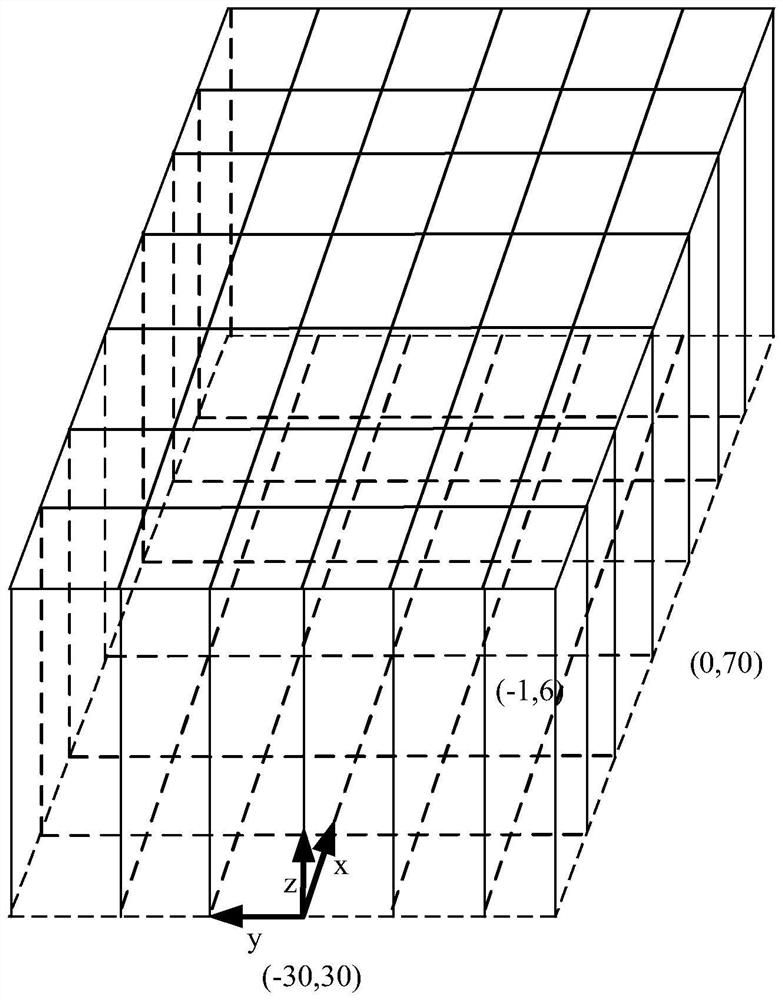
The present invention relates to a phantom for evaluating performance of an ultra high field Magnetic Resonance Imaging (MRI) apparatus. Specifically, the present invention relates to a multi-purpose phantom which can grasp a degree of diagnostic capability of an MRI apparatus using a comprehensive result of imaging conditions and variables and simultaneously analyze and evaluate performance of Magnetic Resonance Imaging (MRI), performance of Magnetic Resonance Spectroscopy (MRS) and metabolic components of a human body within a predetermined range of error and limit. The phantom for evaluating performance of an ultra high field Magnetic Resonance Imaging (MRI) apparatus may include: an outer container opened and closed using a stopper and formed with an insertion hole for injecting components of a liver metabolite and a lipid; an inner container for quantitatively evaluating the components of the liver metabolite and the lipid, acquiring an MRI image using a spin echo sequence, and acquiring a relaxation time through spectroscopy using a single voxel technique of the MRI apparatus; a geometric accuracy evaluation apparatus installed at an upper end portion of the outer container in a shape of three-dimensional lattice type frame; slice position evaluation apparatuses of different shapes, capable of measuring a position of a slice in a middle of the outer container; a contrast resolution evaluation apparatus configured of a plurality of holes in the middle of the outer container to perform evaluation at regular intervals; a spatial resolution evaluation apparatus installed inside the outer container, in which a plurality of hole bundles configured of space evaluation holes is formed to have a same diameter and arranged in an evaluation frame in parallel at regular intervals to have holes of different diameters in each of the hole bundles; a slice thickness evaluation apparatus installed at a height a same as that of the spatial resolution evaluation apparatus and attached to the outer container to be formed in a shape of a stair; and a brain metabolite evaluation apparatus configured of a plurality of layers at a lower portion of the outer container.
Owner:THE CATHOLIC UNIV OF KOREA IND ACADEMIC COOPERATION FOUND
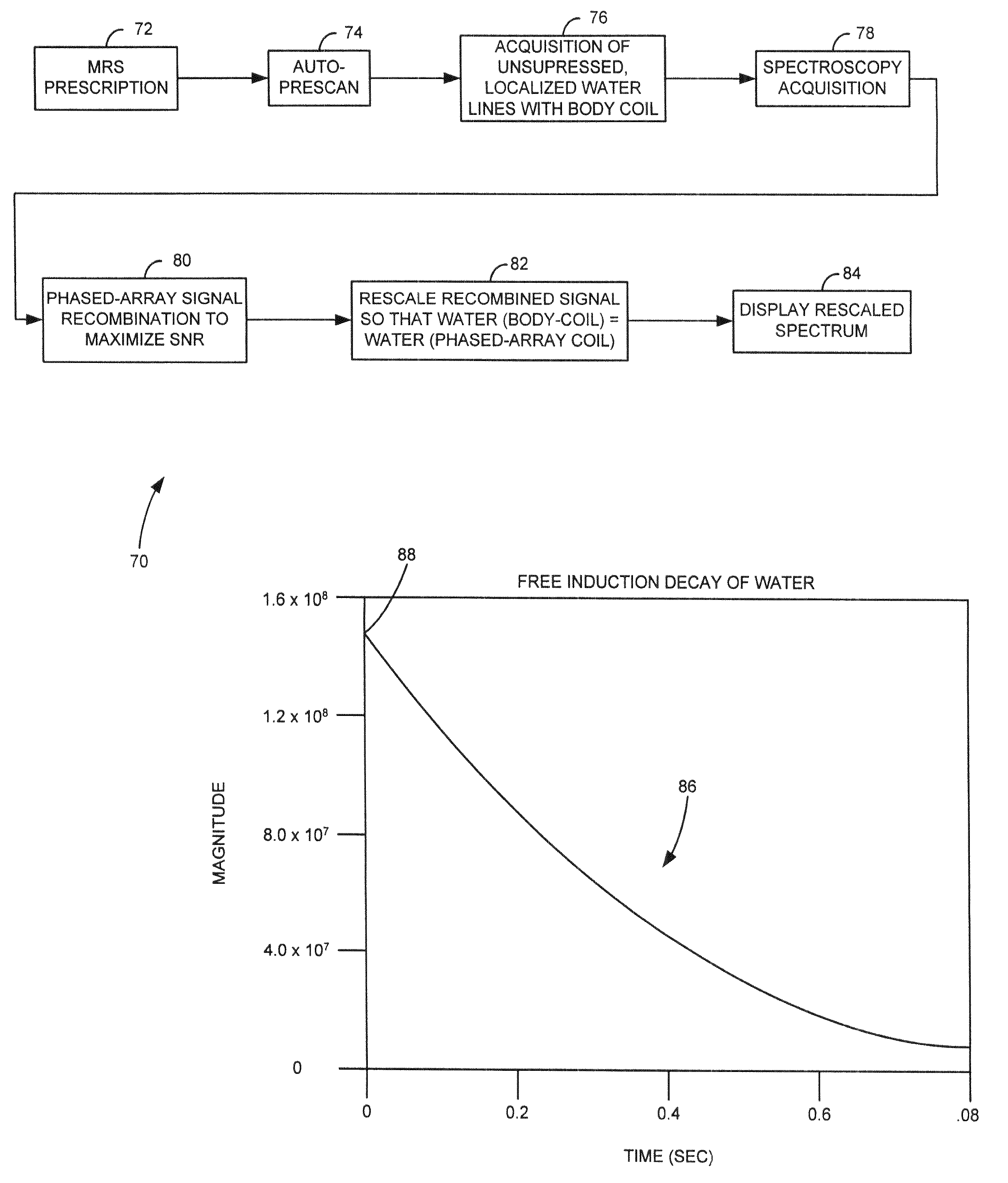
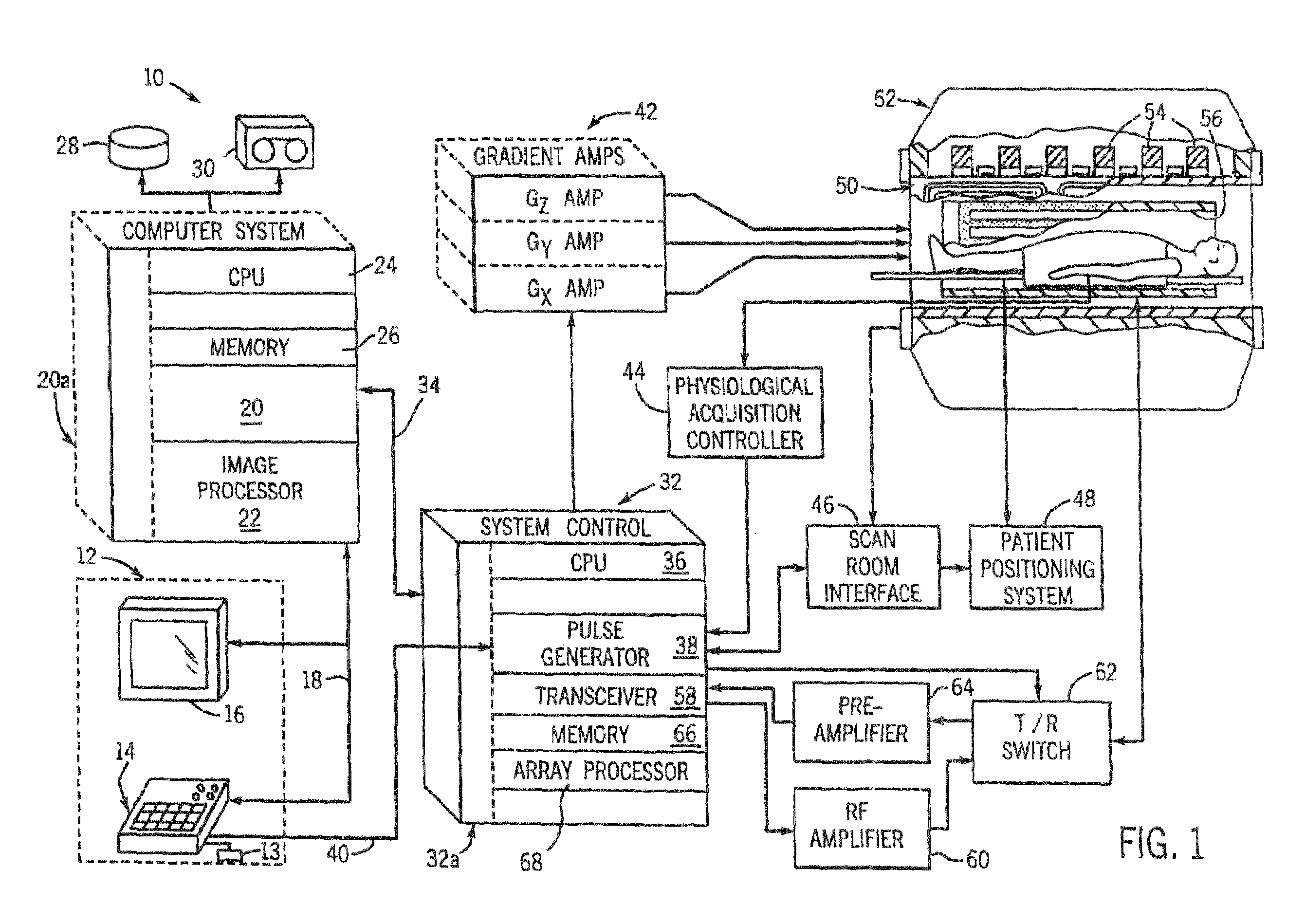
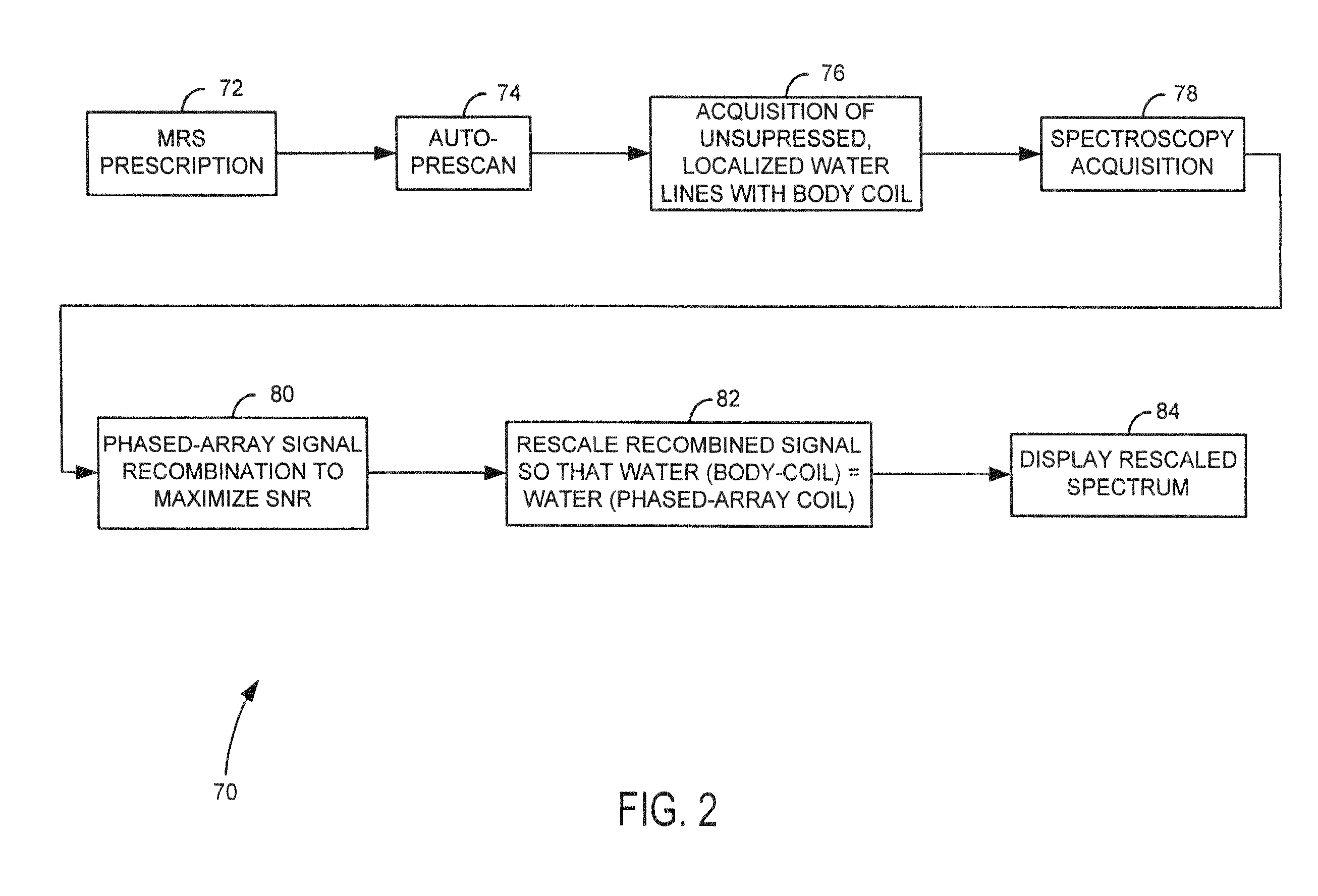
Method and system of scaling MR spectroscopic data acquired with phased-array coils
The present invention is directed to a system and method of resealing spectroscopic data acquired with phased-array or surface coils to absolute “local” units and, hence, preserves the industry-desirable quantitative approach to single voxel MRS. An MRS signal is acquired from water using a body coil and is used as a reference signal to scale MR spectroscopic data acquired with a plurality of RF receive coils, i.e. phased-array or surface coils. In the resealing process, the amplitude of water in the MRS data will be set to match the amplitude of the water reference signal.
Owner:GENERAL ELECTRIC CO
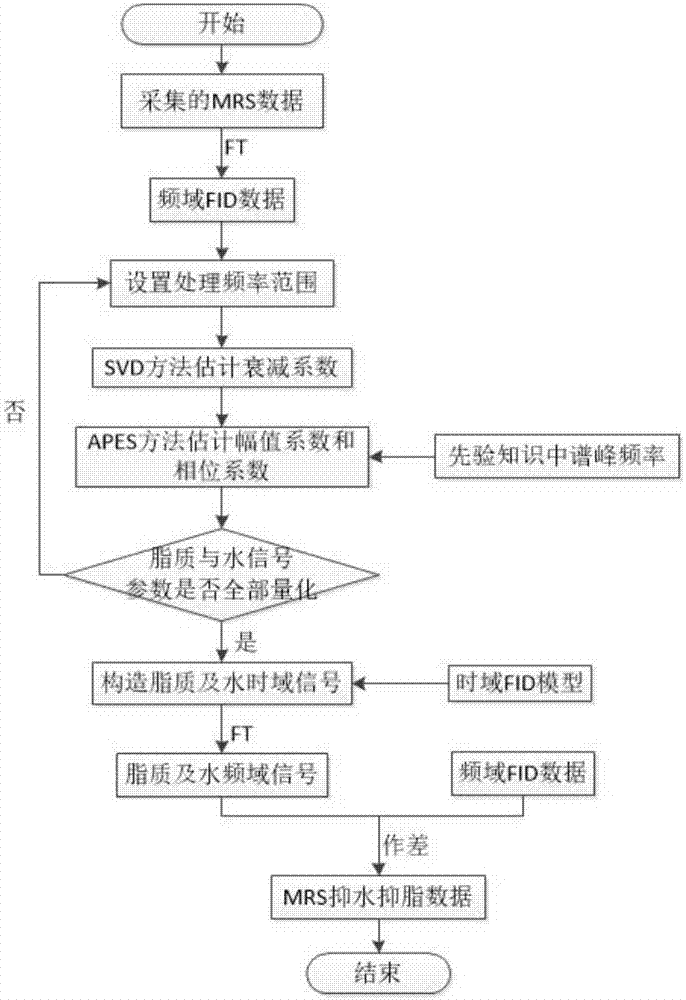
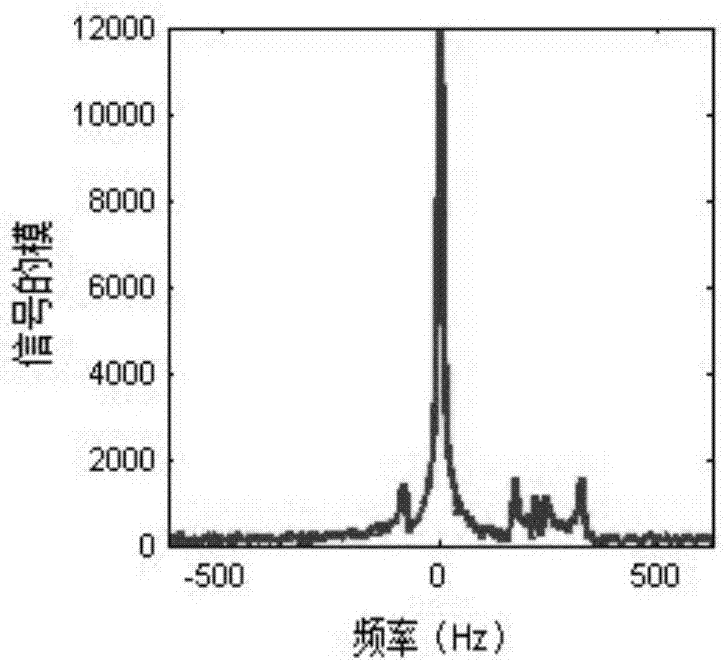
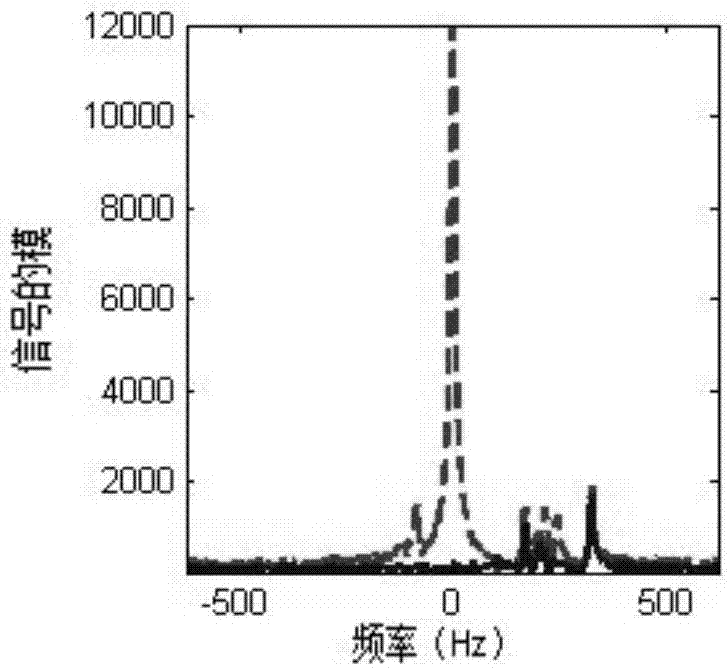
Frequency-domain-based method for lipid and water suppression treatment of magnetic resonance spectrum imaging data
The invention belonging to the field of medical imaging provides a frequency-domain-based method for lipid and water suppression treatment of magnetic resonance spectrum imaging data. The method comprises: one-dimensional Fourier transform is carried out on collected single voxel magnetic resonance spectrum time-domain data to obtain a frequency-domain FID signal; according to prior knowledge, four lipid peaks and one water signal frequency range are determined; a nonlinear parameter attenuation coefficient in a spectrum time-domain model by using a singular value decomposition method; on the basis of a spectrum peak frequency in the prior knowledge and the attenuation coefficient obtained by quantification, the amplitude and phase of a linear parameter are estimated by using an amplitude-phase adaptive FIR filtering method; the frequency range is changed and the previous two steps are executed circularly until quantification of the four group of lipid and water signal parameters is completed; the time-domain FID model and the lipid and water signal quantification parameters form lipid and water time-domain signals; and one-dimensional Fourier transform is carried out on the time-domain signals, difference processing is carried out on the frequency domain and an original spectral signal, thereby obtaining magnetic resonance spectrum imaging data after lipid and water suppression treatment.
Owner:杭州全景医学影像诊断有限公司
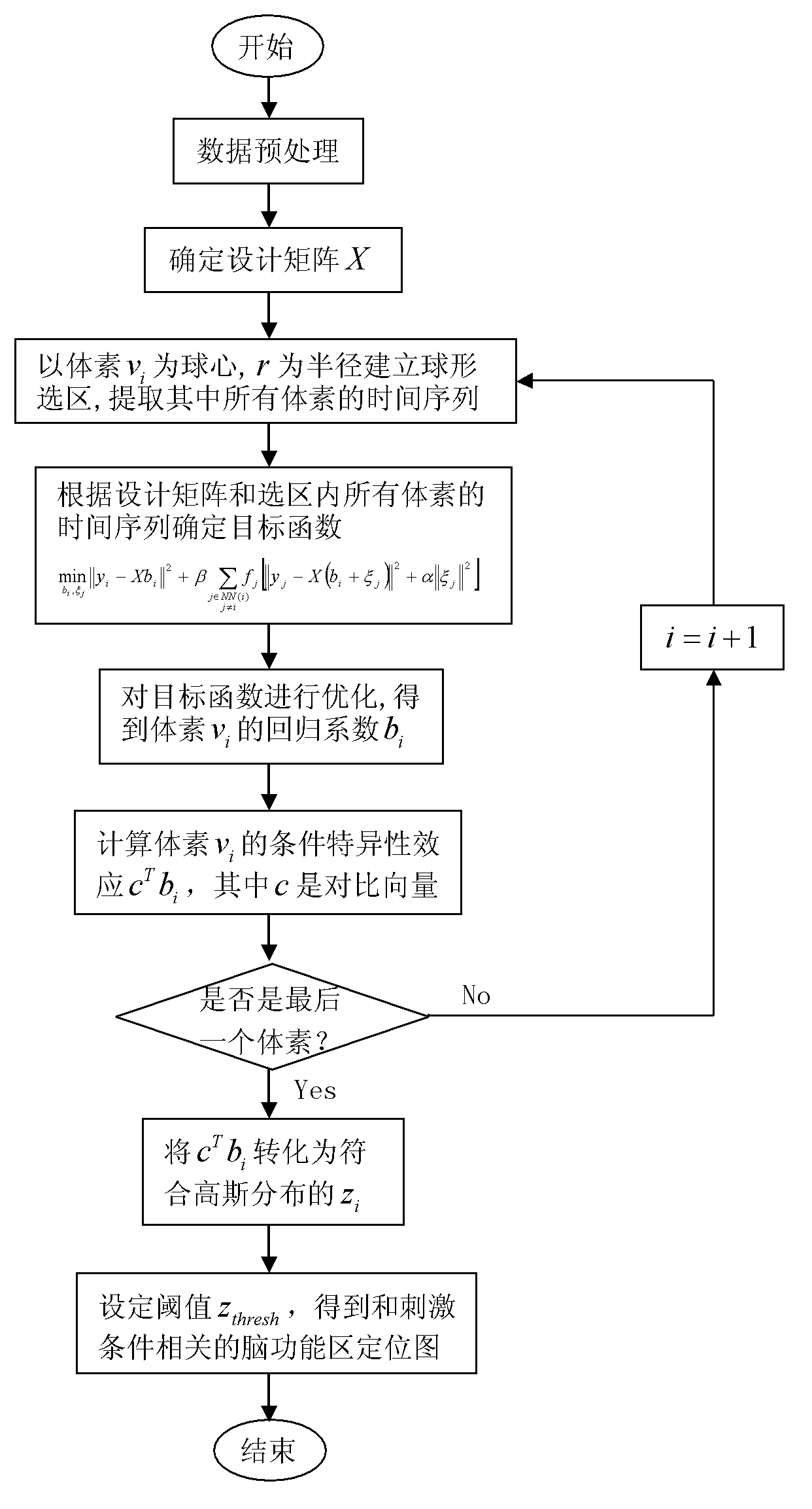
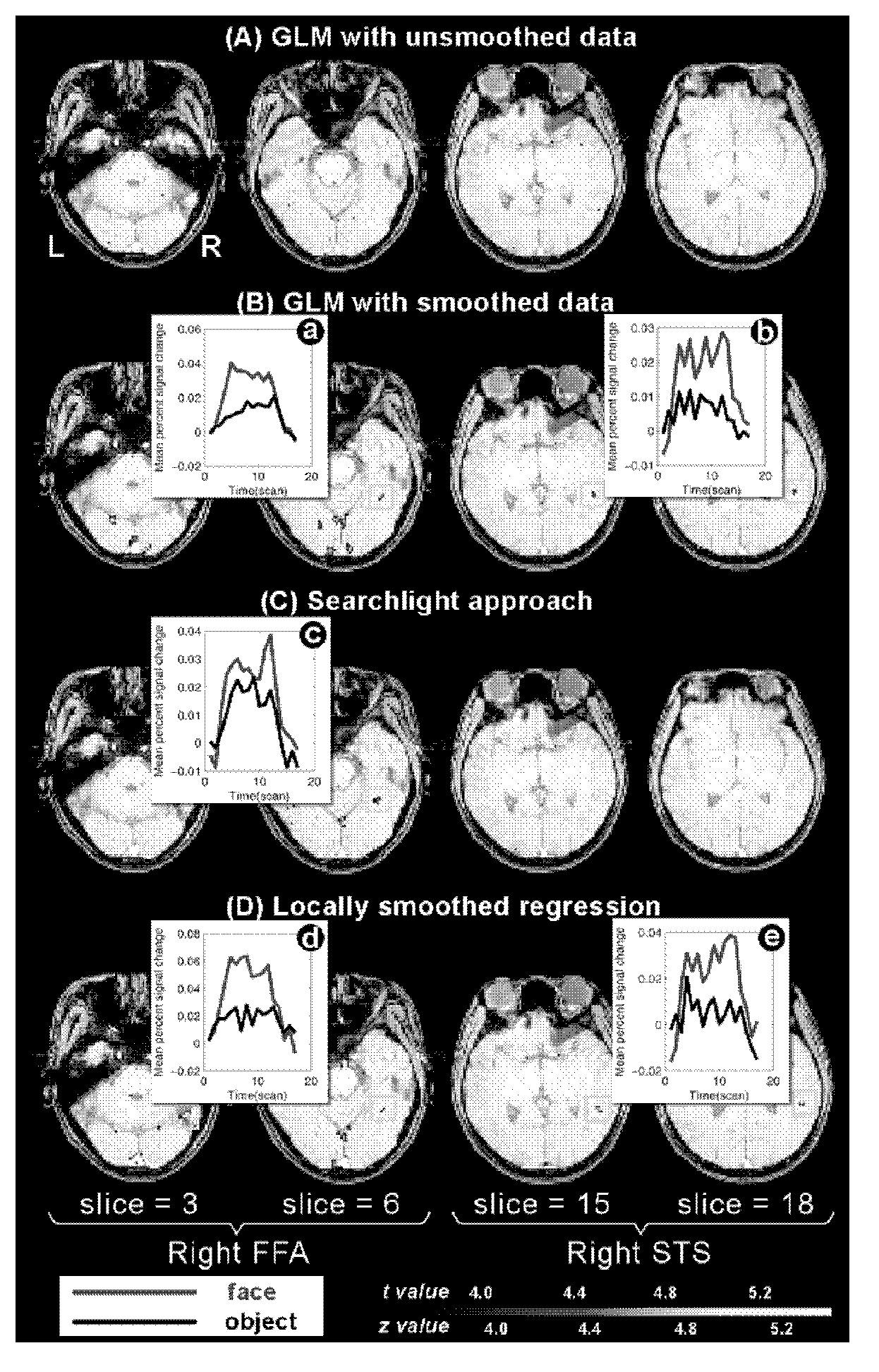
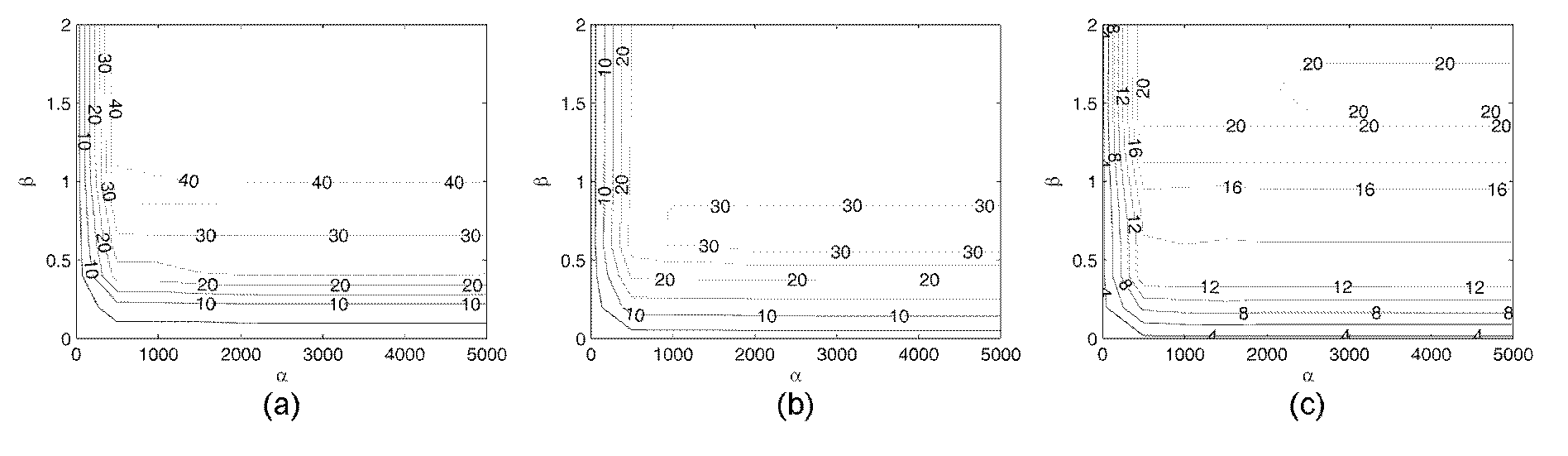
Brain functional region positioning method based on local smoothing regressions
The invention provides a brain functional region positioning method based on local smoothing regressions. The brain functional region positioning method based on the local smoothing regressions comprises the following steps of pretreating data and deciding a design matrix X; taking a voxel vi as the center of a sphere and r as a semidiameter for the establishment of a spherical selected region and extracting the time sequence of all the voxels in the spherical selected region; according to the time sequence of all the voxels in the spherical selected region and the design matrix, forming an objective function and optimizing the objective function; calculating a condition specificity effect of the voxel vi; turning to the next voxel vi+1 and repeating steps from S2 to S4 till the execution of the steps on each voxel of a whole brain; and setting a threshold value for a whole brain perception mapping so as to obtain a brain functional region positioning map relevant with stimulus conditions. All the generalized linear models based on the regressions of the single voxels and based on Gaussian smoothing filtering can be regarded as special cases of the invention; the brain functional region positioning method based on the local smoothing regressions can be integrated into a framework used for a searchlight method; after the obtainment of regression coefficients, mahalanobis distances between various predictor coefficients are calculated; and through the adjustment on hyper-parameters alpha and beta, smoothing effects of various degrees are obtained.
Owner:INST OF AUTOMATION CHINESE ACAD OF SCI
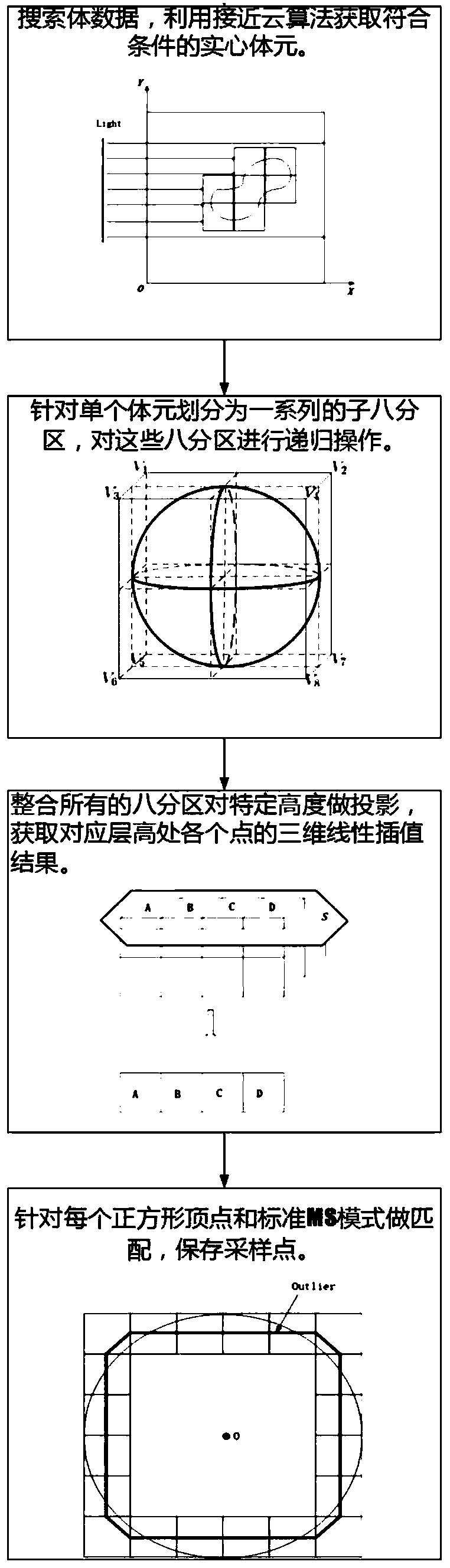
3D printing slicing method for implicit expression medical model
ActiveCN110533770AEasy to shapeImprove efficiencyAdditive manufacturing apparatusMedical automated diagnosisAlgorithmConsumables
The invention discloses a 3D printing slicing method for an implicit expression medical model, and relates to computer graphics and 3D printing. The method comprises the following steps of searching the volume data to obtain solid volume elements meeting conditions; dividing a single voxel into a series of sub eight partitions, and performing recursive operation; integrating all the eight partitions to project the specific height, and obtaining three-dimensional linear interpolation of each point at the corresponding layer height; matching each square vertex with a standard stepping square mode, and storing sampling points; acquiring a plurality of adjacent sampling points and retracting, and calculating a retracting distance; obtaining all sampling points and adjacent points of the sampling points, and constructing a complete inner layer contour; checking whether the current sampling point has an error; removing known self-intersection points, and optimizing the shape of an inner-layer contour; retracting inwards to increase a virtual layer; constructing a parallel scanning line field, and calculating intersection point coordinates of the scanning line field and the virtual layer;adopting labyrinth filling for alternate wiring to achieve internal filling . The method reduces consumables, time and space expenses..
Owner:XIAMEN UNIV
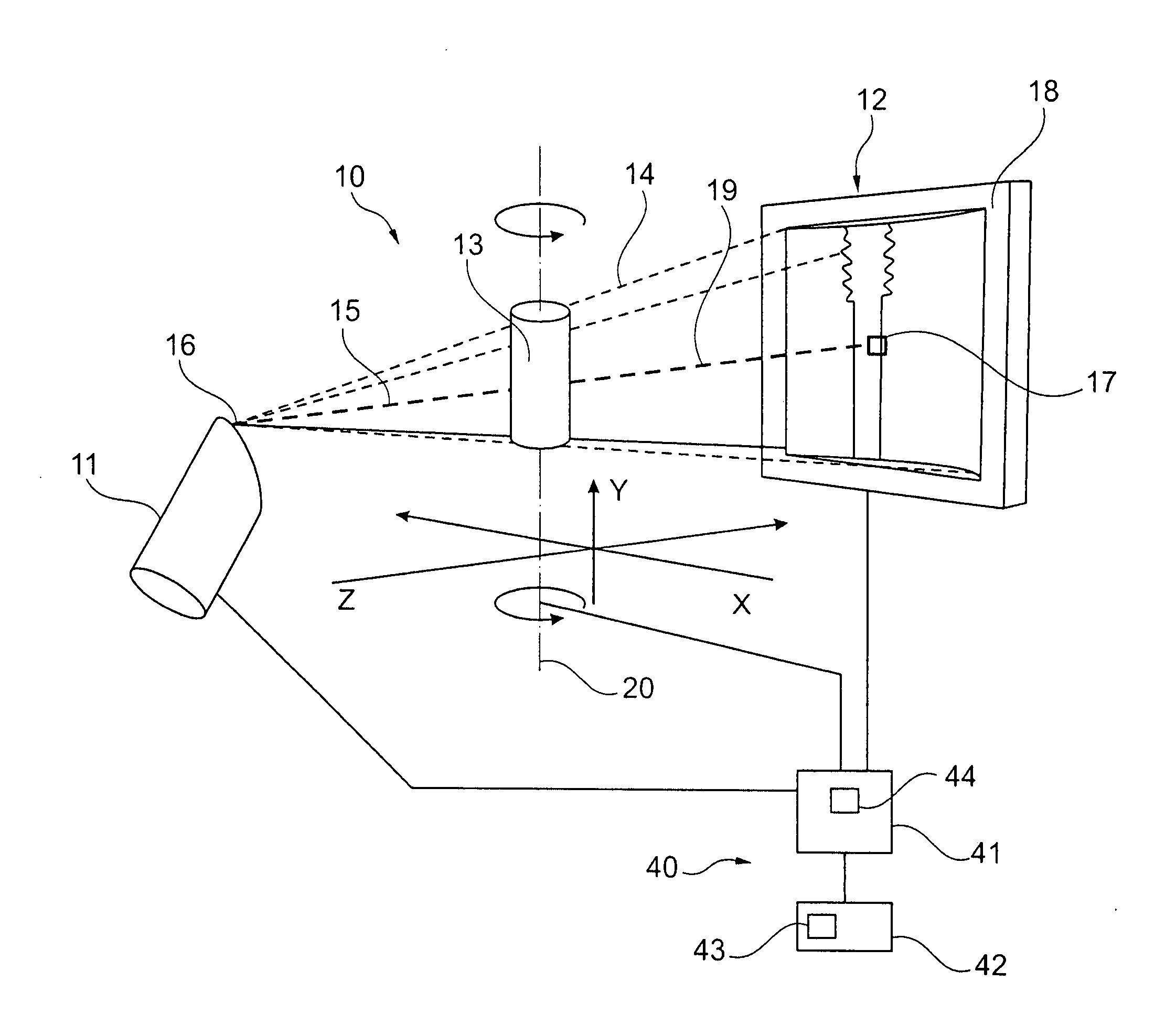
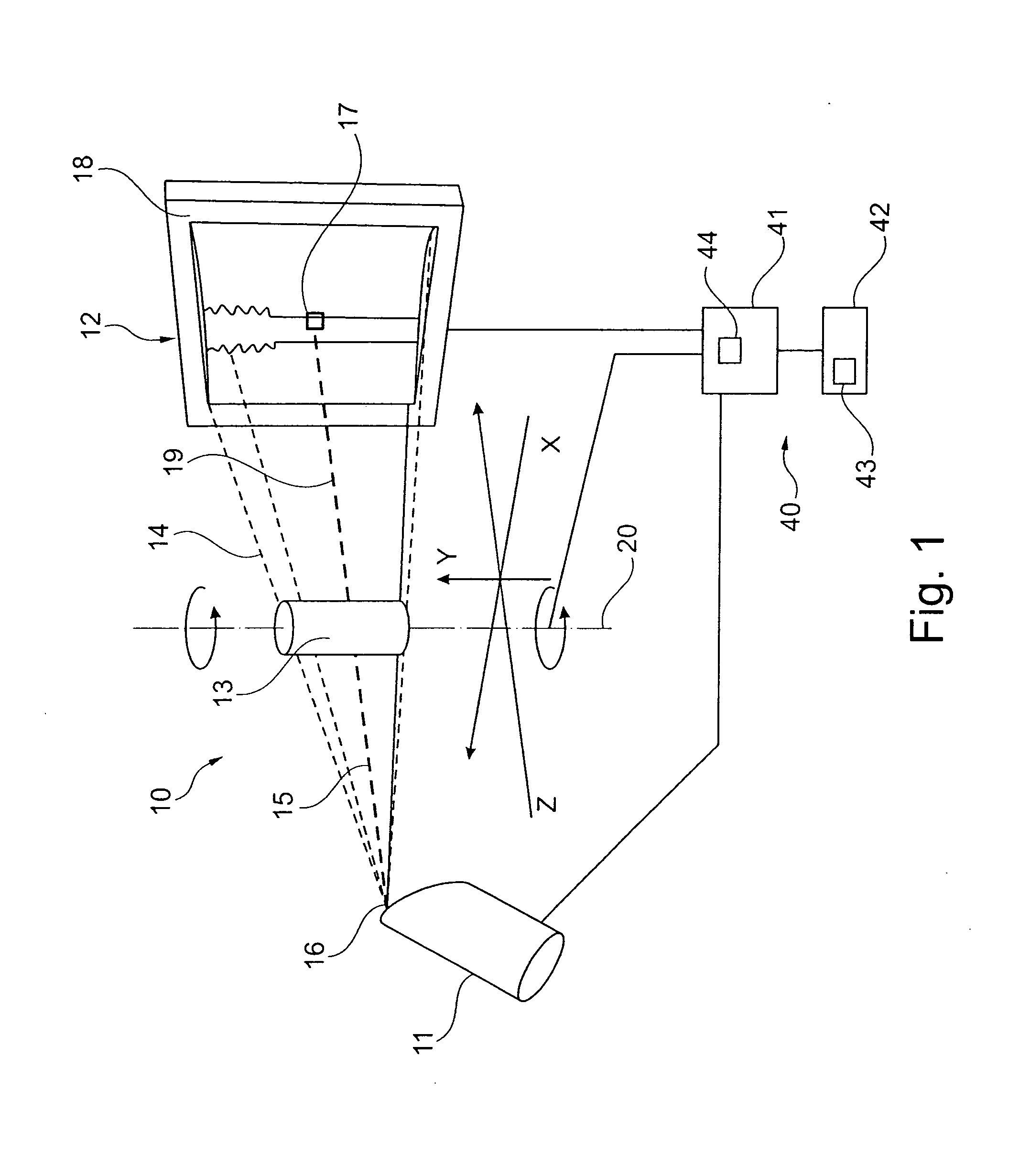
Computed tomography method, computer software, computing device and computed tomography system for determining a volumetric representation of a sample
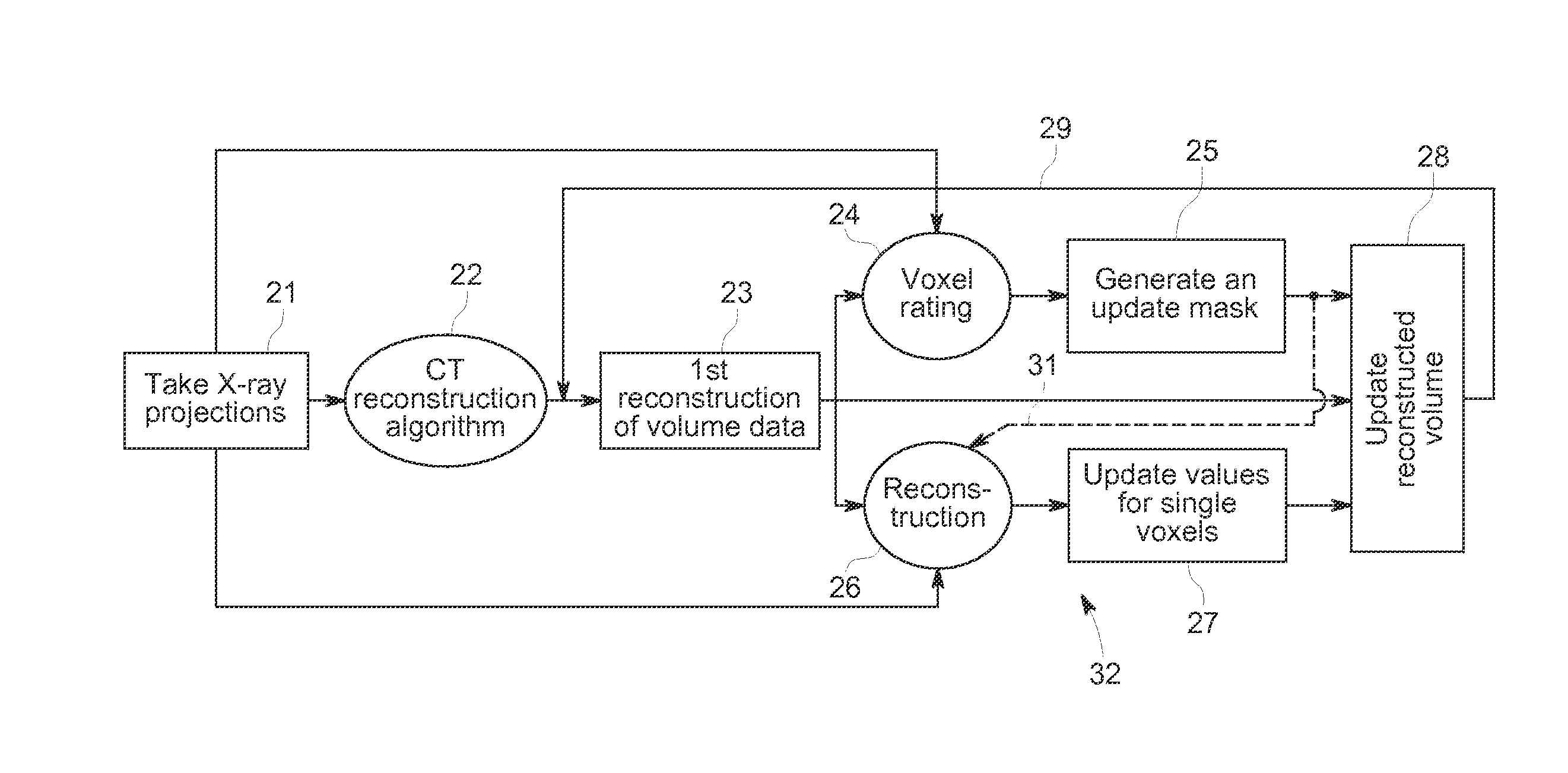
ActiveUS20110129055A1Improve accuracyImprove developmentReconstruction from projectionMaterial analysis using wave/particle radiationVolumetric dataX-ray
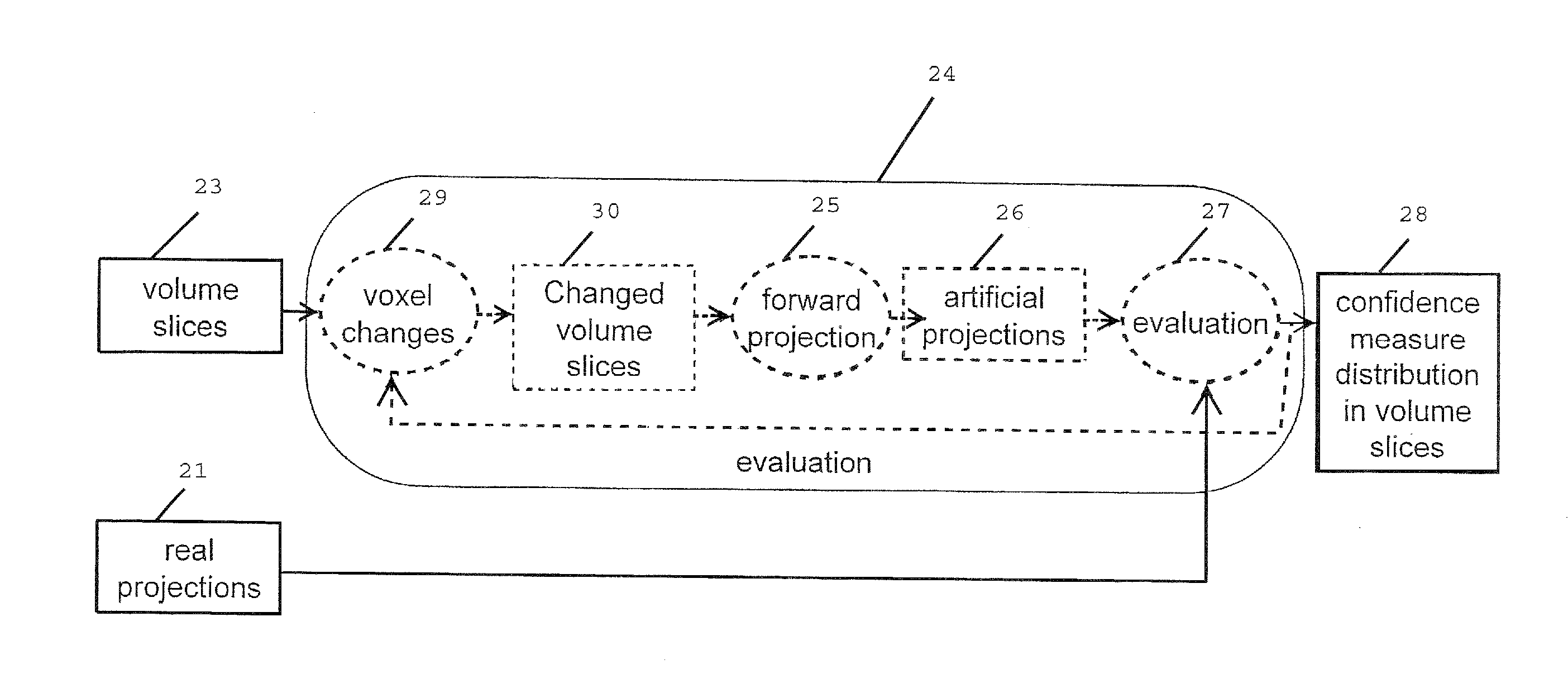
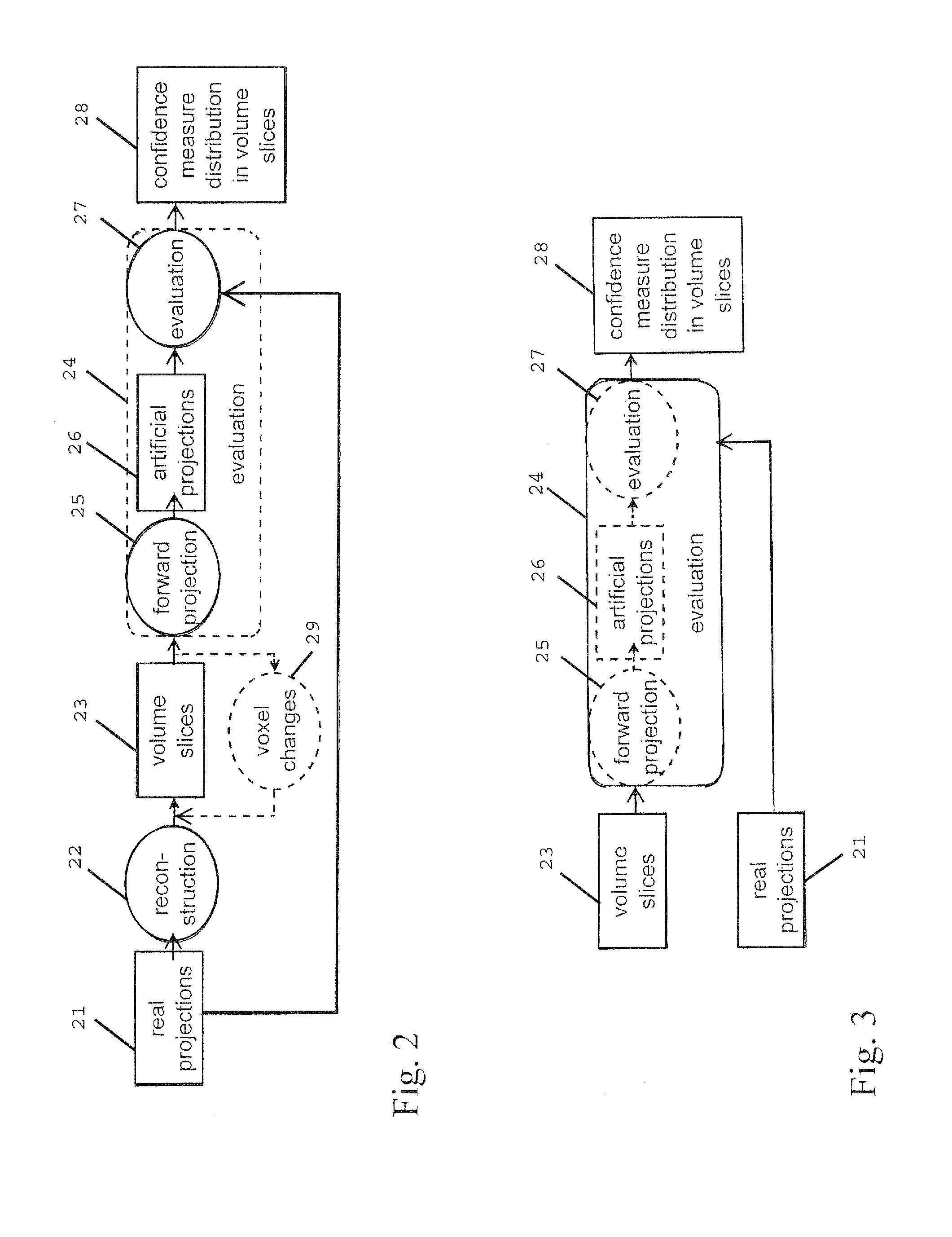
A computed tomography method for determining a volumetric representation of a sample comprises using reconstructed volume data of the sample from x-ray projections of the sample taken by an x-ray system, computing a set of artificial projections of said sample by a forward projection from said reconstructed volume data , and determining, essentially from process data of said reconstruction including said reconstructed volume data and / or said x-ray projections, individual confidence measures for single voxels of said volume data based on calculating, for each of said measured x-ray projections, the difference between the contribution of this measured x-ray projection to the voxel under inspection and the contribution from a corresponding artificial projection to the voxel under inspection.
Owner:GE SENSING & INSPECTION TECH GMBH
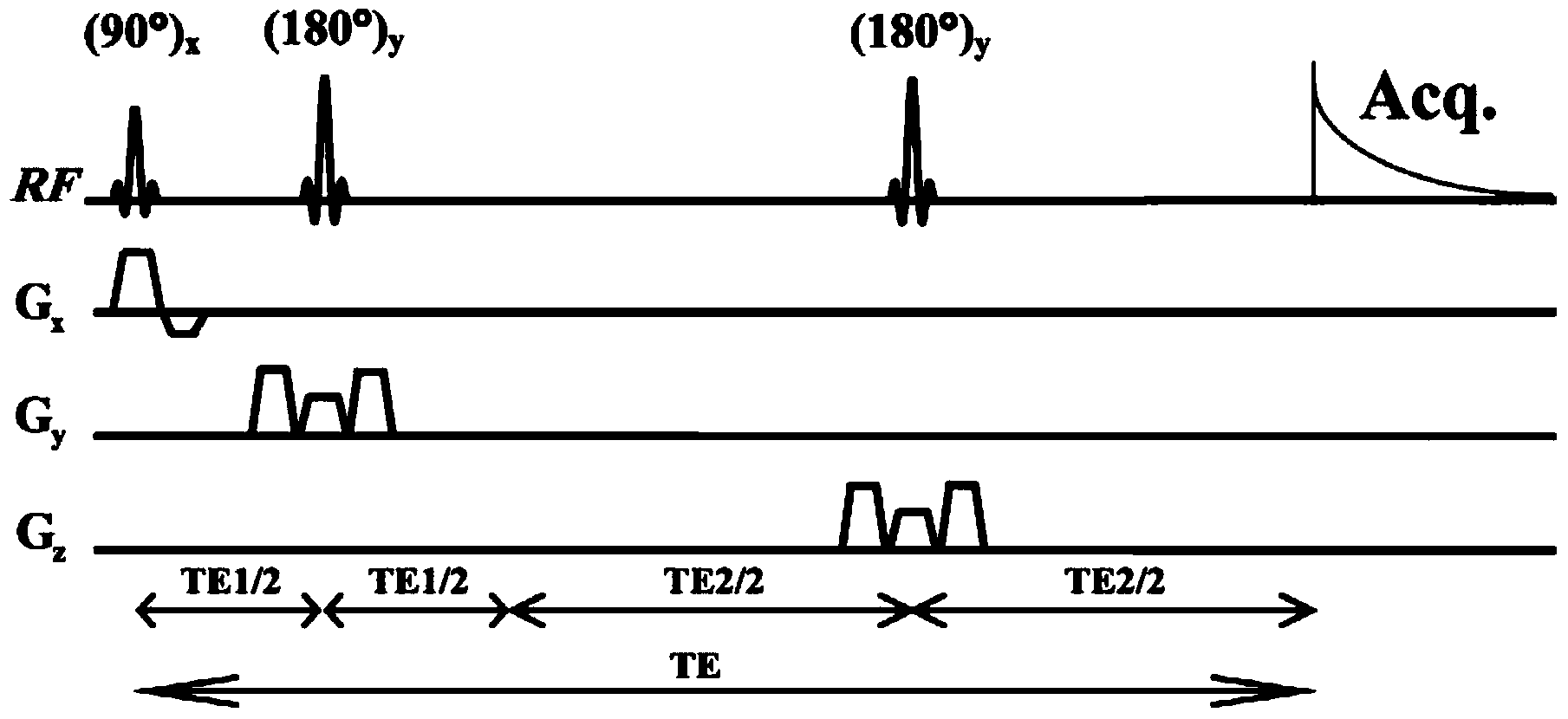
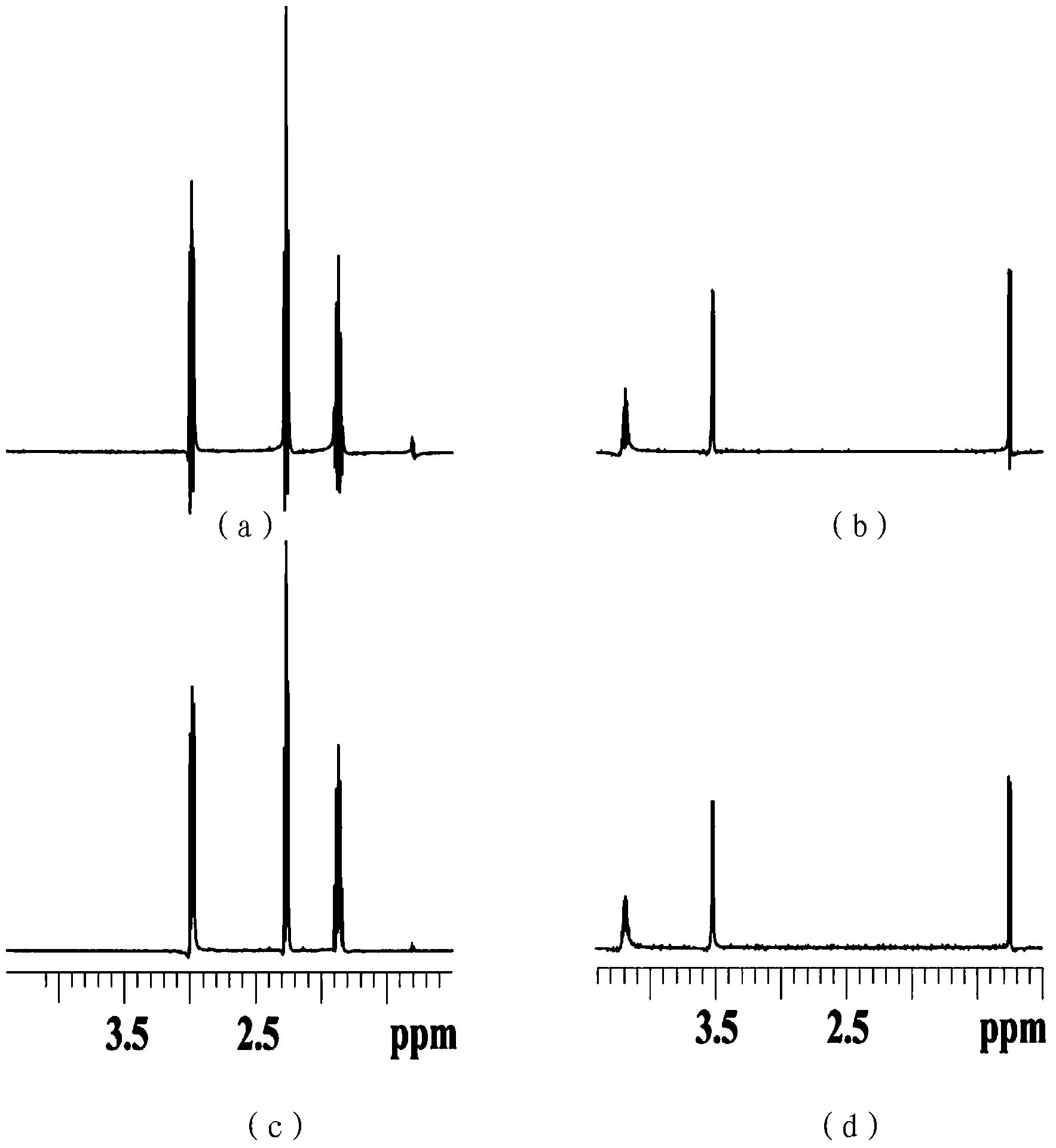
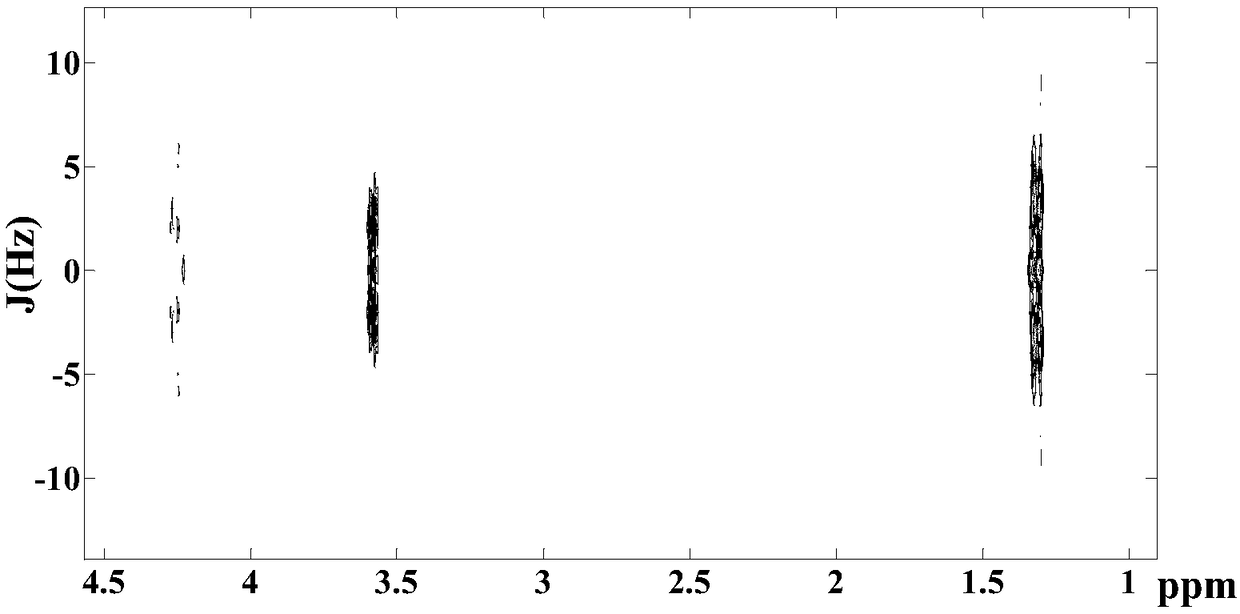
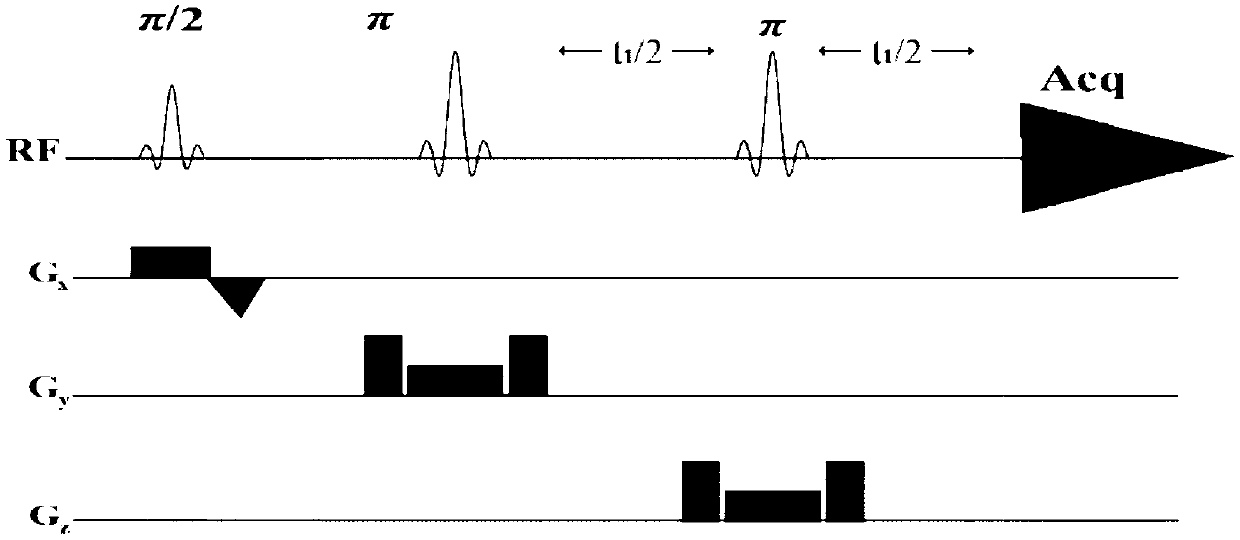
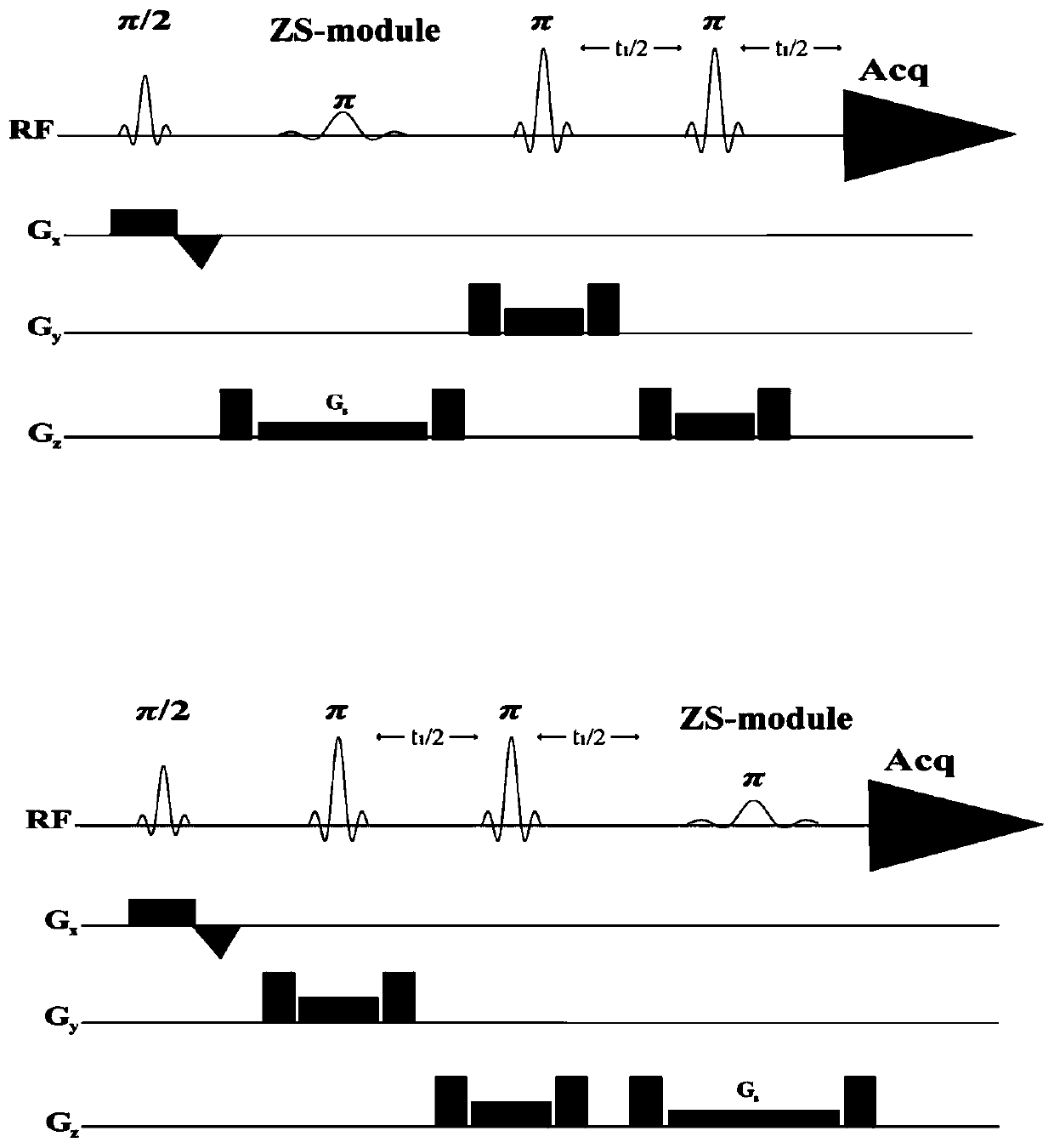
Method for realizing pure absorption line type two-dimensional magnetic resonance single-voxel localized J decomposition spectrum
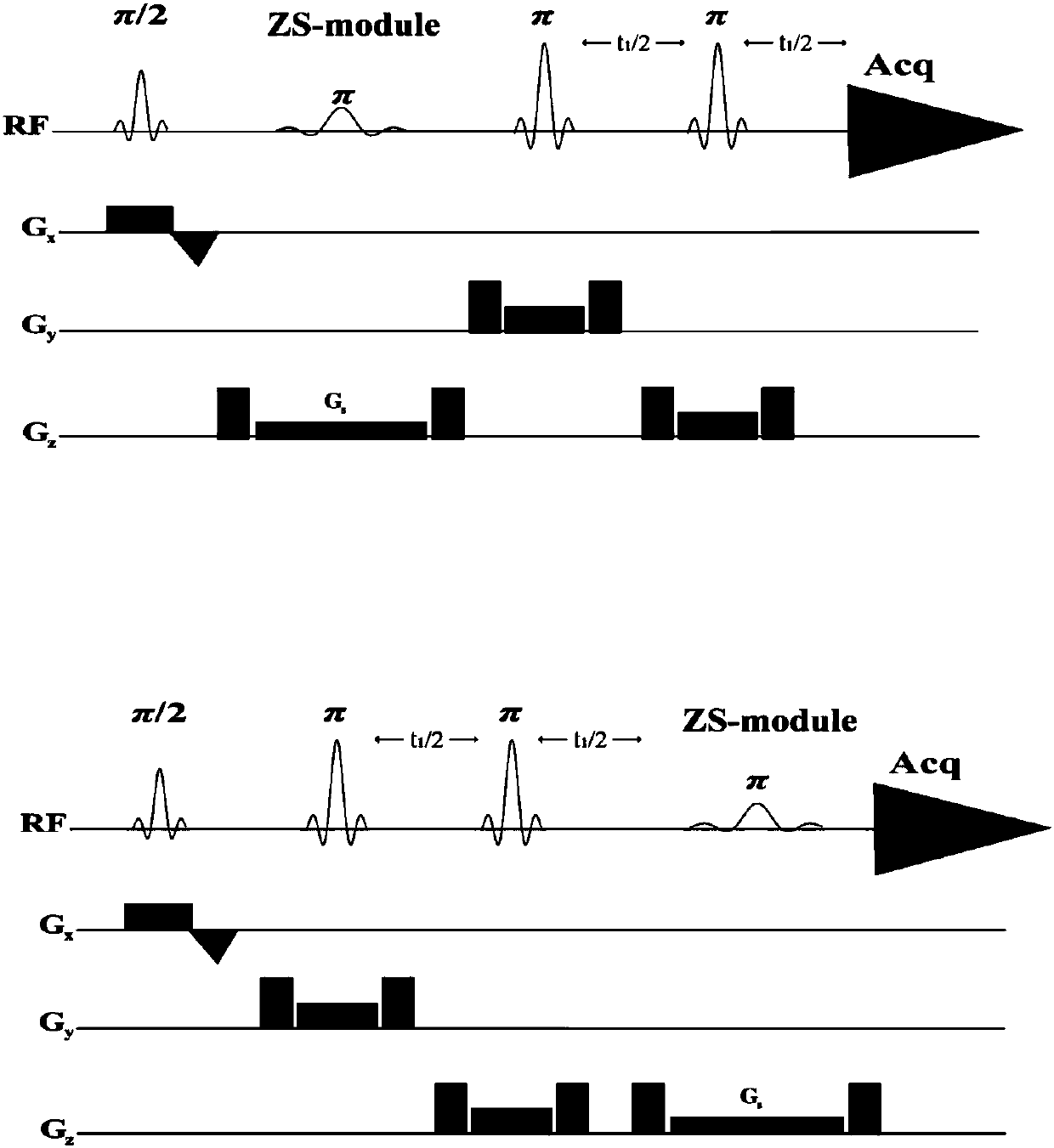
The invention provides a method for realizing a pure absorption line type two-dimensional magnetic resonance single-voxel localized J decomposition spectrum. ZS modules are added at different positions to form an N sequence and an R sequence, the experimental result of the R sequence symmetrically flipped along an indirect dimension F1 = 0 Hz, and superimposing the symmetrically flipped experimental result of the R sequence and the result of the N sequence to obtain the pure absorption line type single-voxel two-dimensional localized J decomposition spectrum. The N sequence is formed through adding the ZS module after the first 90 DEG localized pulse, the R sequence is formed through adding the ZS module after the last 180 DEG localized pulse, the delay times at both sides of the last 180DEG of each of the sequences are uniform and are t1 / 2, and indirect dimensional F1 evolution is performed. The ZS module consists of a selective 180 DEG soft pulse, a Z-directional gradient magnetic field and a symmetric spoiled gradient, so the selective 180 DEG soft pulse realizes the evolution refocusing of different nuclei at different spatial positions, and the coherence order of the resonance nuclei transfers; and the ZS modules of the N sequence and the R sequence have different positions, so final two-dimensional signal composition modes are different.
Owner:XIAMEN UNIV
Laser radar 3D point cloud ground detection method
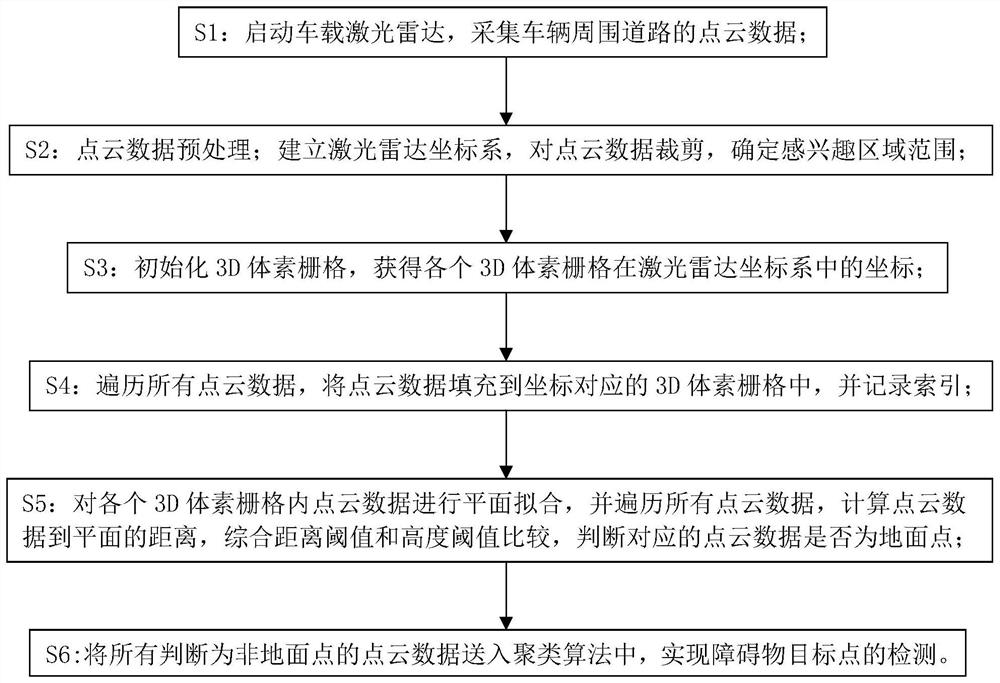
PendingCN114488190AFit the real situationAchieve ground filtering effectCharacter and pattern recognitionElectromagnetic wave reradiationCluster algorithmRadar
The invention discloses a laser radar 3D point cloud ground detection method. The problem that an expected ground filtering effect is difficult to achieve in the prior art is solved. The method comprises the following steps: collecting point cloud data of roads around a vehicle; cutting the point cloud data, and determining a region-of-interest range; initializing 3D voxel grids, and obtaining coordinates of each 3D voxel grid in a laser radar coordinate system; traversing all the point cloud data, filling 3D voxel grids corresponding to the coordinates with the point cloud data, and recording indexes; carrying out plane fitting on the point cloud data in each 3D voxel grid, traversing all the point cloud data, calculating the distance from the point cloud data to a plane, comparing a distance threshold value with a height threshold value, and judging whether the corresponding point cloud data is a ground point or not; and sending all point cloud data which are judged to be non-ground points into a clustering algorithm to realize detection of obstacle target points. And for a scene of a single voxel, ground points are identified and distinguished, and an expected ground filtering effect is achieved.
Owner:ZHEJIANG LEAPMOTOR TECH CO LTD
Single-voxel localized 1-D high-resolution homonuclear decoupling spectrum method
InactiveCN108107391AEliminate phase distortionWide applicabilityMeasurements using NMR spectroscopyMagnetic variable regulationSpectroscopyChirp
The invention provides a single-voxel localized 1-D high-resolution homonuclear decoupling spectrum method, and relates to a magnetic resonance apparatus. On the basis of a traditional 2D localized correlated spectroscopy, the method adds linear adiabatic sweep (chirp) pulses and gradients applied at the same time to form a z-filter module that can effectively suppress a zero-quantum coherent pathand other paths so as to obtain a pure Lorentz absorption linetype nuclear magnetic resonance spectrum. The selection of voxels is accomplished together by selective pulses combined with selection layers and destruction gradients. The post-processing of the experimentally acquired data can yield a 1-D spectrum with only chemical shift information. The 1-D localized spectrum obtained by the invention can eliminate the phase distortion caused by the zero quantum coherence, thereby having the advantages of high spectral resolution and the like. This method can be used for the study of the biological tissue and living body localized spectrum to obtain more concise and perfect spectral information, so that the localized spectrum has a wider range of applications.
Owner:XIAMEN UNIV
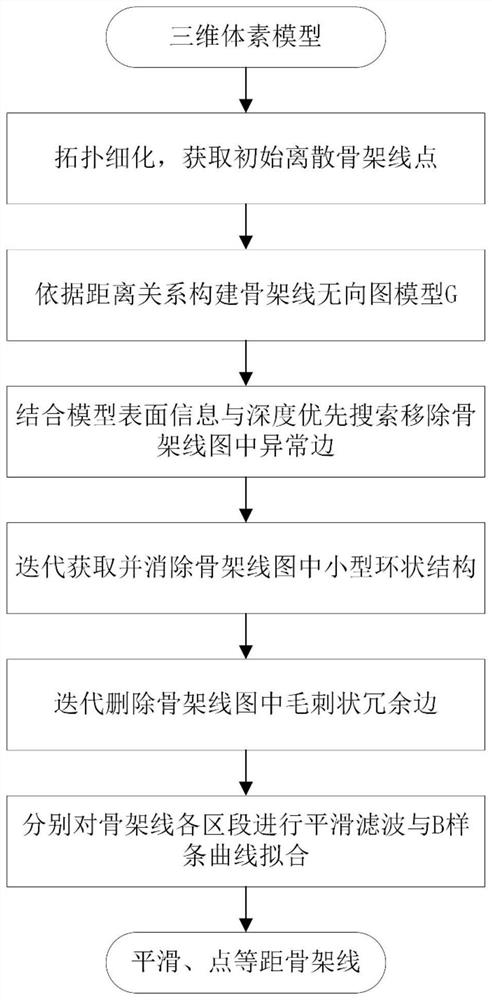
Skeleton line extraction method suitable for complex multi-cavity three-dimensional model
PendingCN114119620AThe result is accurateImprove robustnessImage enhancementImage analysisMoving averageUndirected graph
The invention relates to a skeleton line extraction method suitable for a complex multi-cavity three-dimensional model, and the method comprises the steps: taking a voxel model as input, firstly, based on a topological refinement algorithm, gradually deleting simple voxel points which do not affect a local connection relation through parallel iteration refinement, and obtaining a basic skeleton line with a single pixel width; secondly, an undirected graph model is constructed for the basic skeleton line according to the distance relation, and edges, located outside the model or at outlier positions, in a graph are removed through pretreatment; and then, iteratively detecting a small basic ring structure in the combined image, and removing local redundant edges and burr-shaped noise edges in skeleton lines in combination with the degree and length information of vertexes. And finally, fitting each section on the skeleton line by adopting a moving average algorithm and a B-spline curve section to obtain a smooth skeleton line conforming to a model topological structure. The method is accurate in result and high in robustness, and can play a very important role in application aspects of three-dimensional model deformation, topology, form acquisition and the like.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY
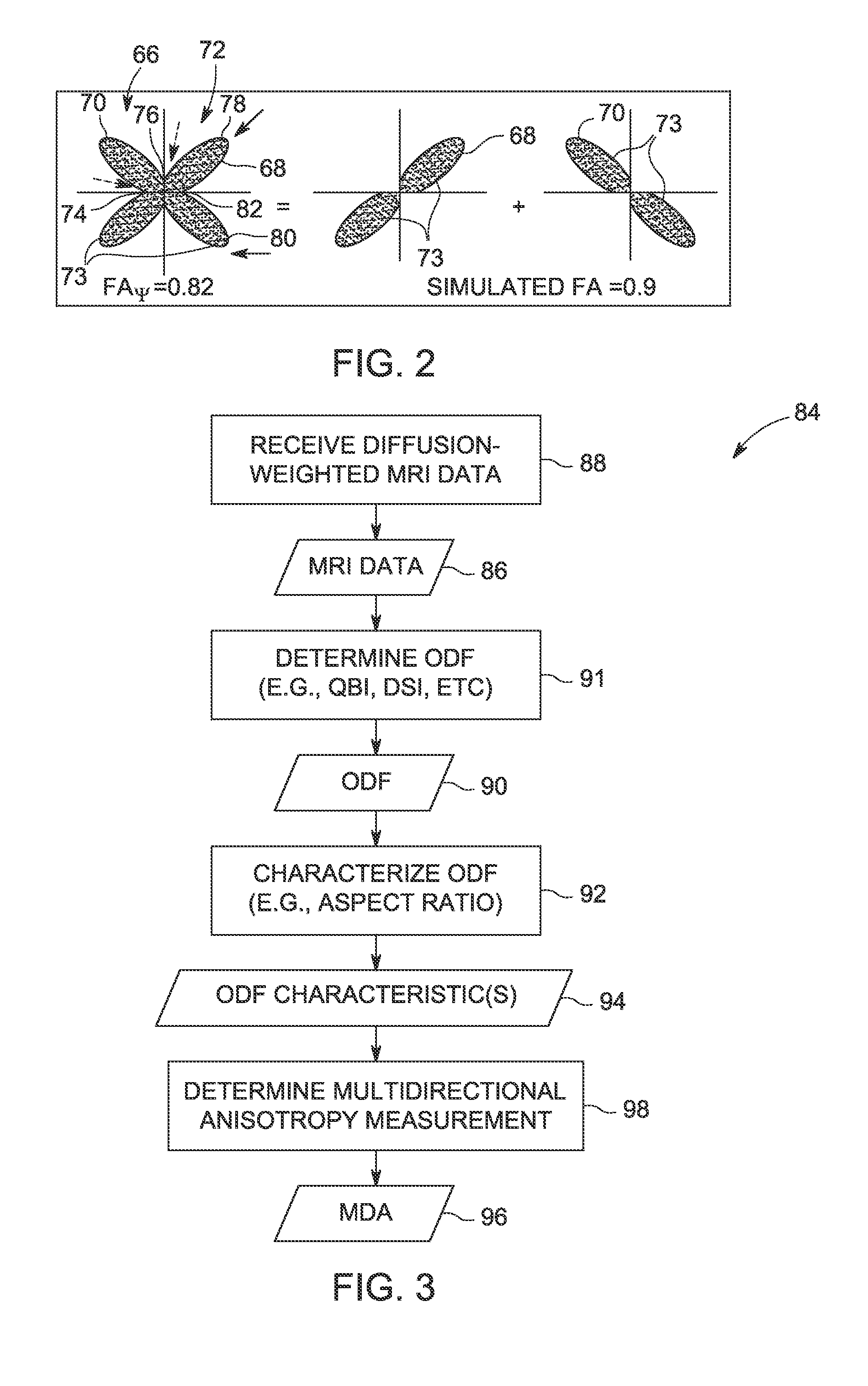
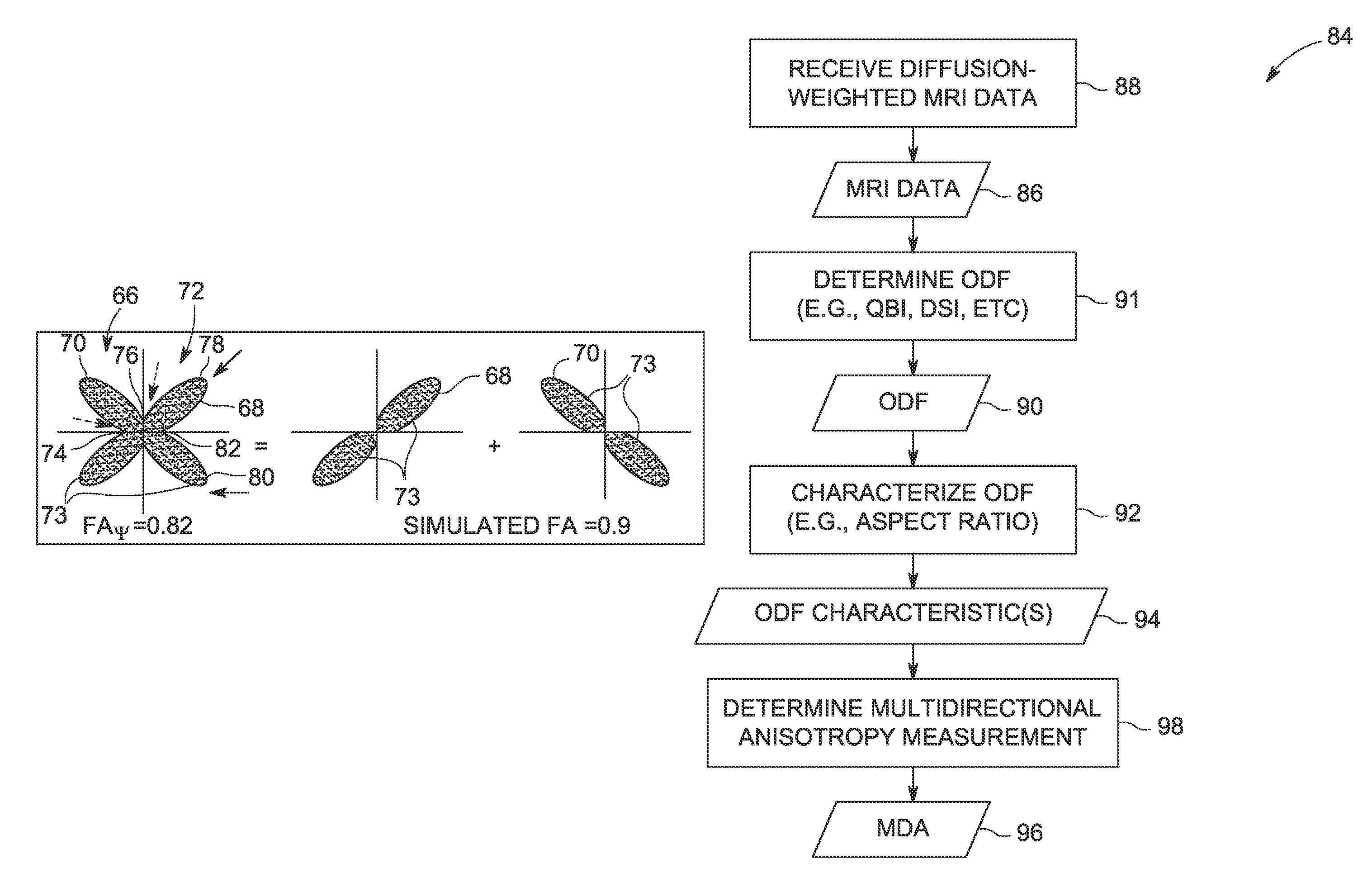
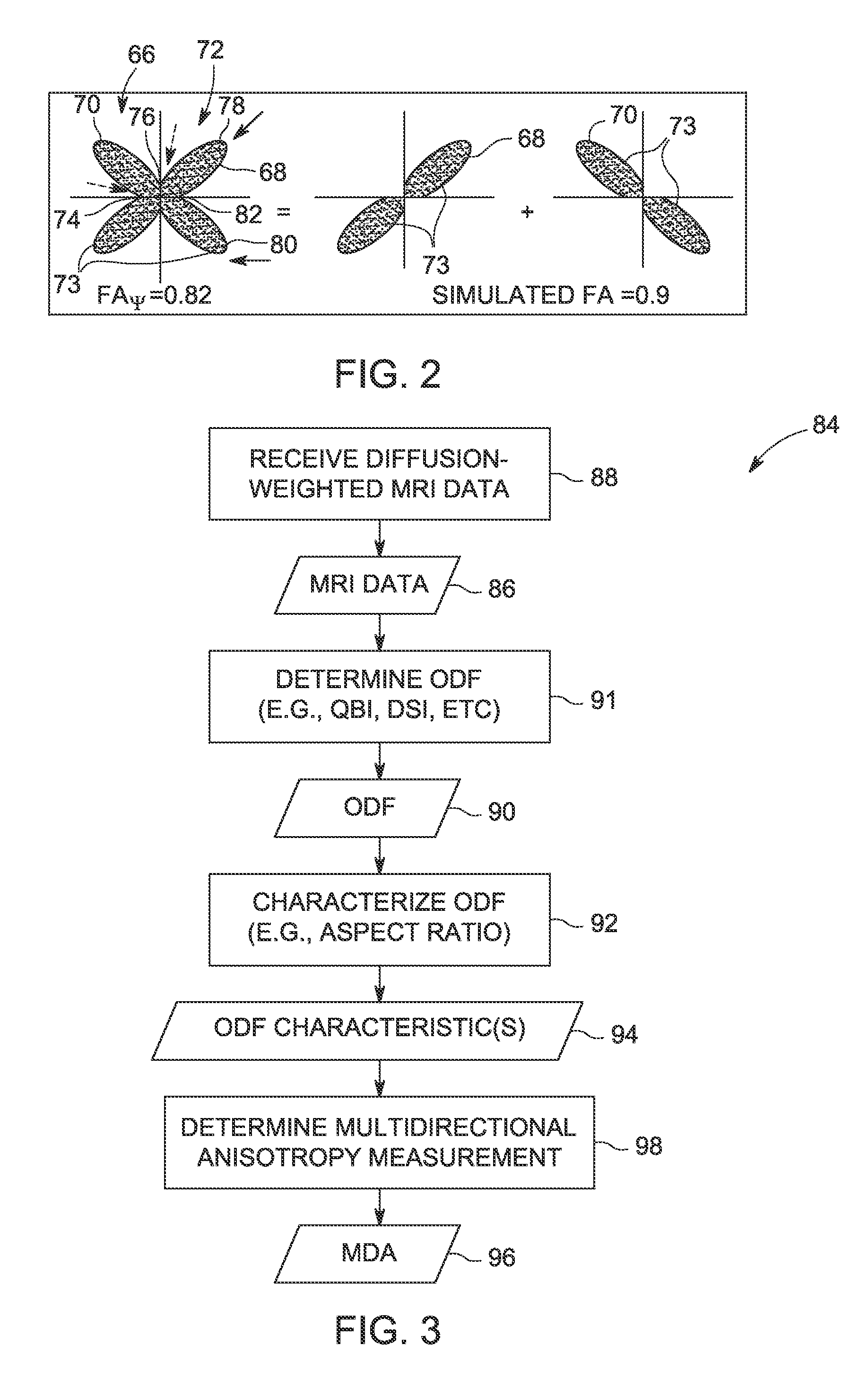
Methods for measuring diffusional anisotropy
ActiveUS20130271130A1Measurements using NMR imaging systemsElectric/magnetic detectionFiberDiffusion
A method for measuring diffusional anisotropy in diffusion-weighted magnetic resonance imaging. The method includes determining an orientation diffusion function (ODF) for one or more fibers within a single voxel, wherein the ODF includes lobes representative of a probability of diffusion in a given direction for the one or more fibers. The method also includes characterizing an aspect ratio of the lobes. The method further includes determining a multi-directional anisotropy metric for the one or more fibers based on the aspect ratio of the lobes.
Owner:GENERAL ELECTRIC CO
Computed tomography method, and system
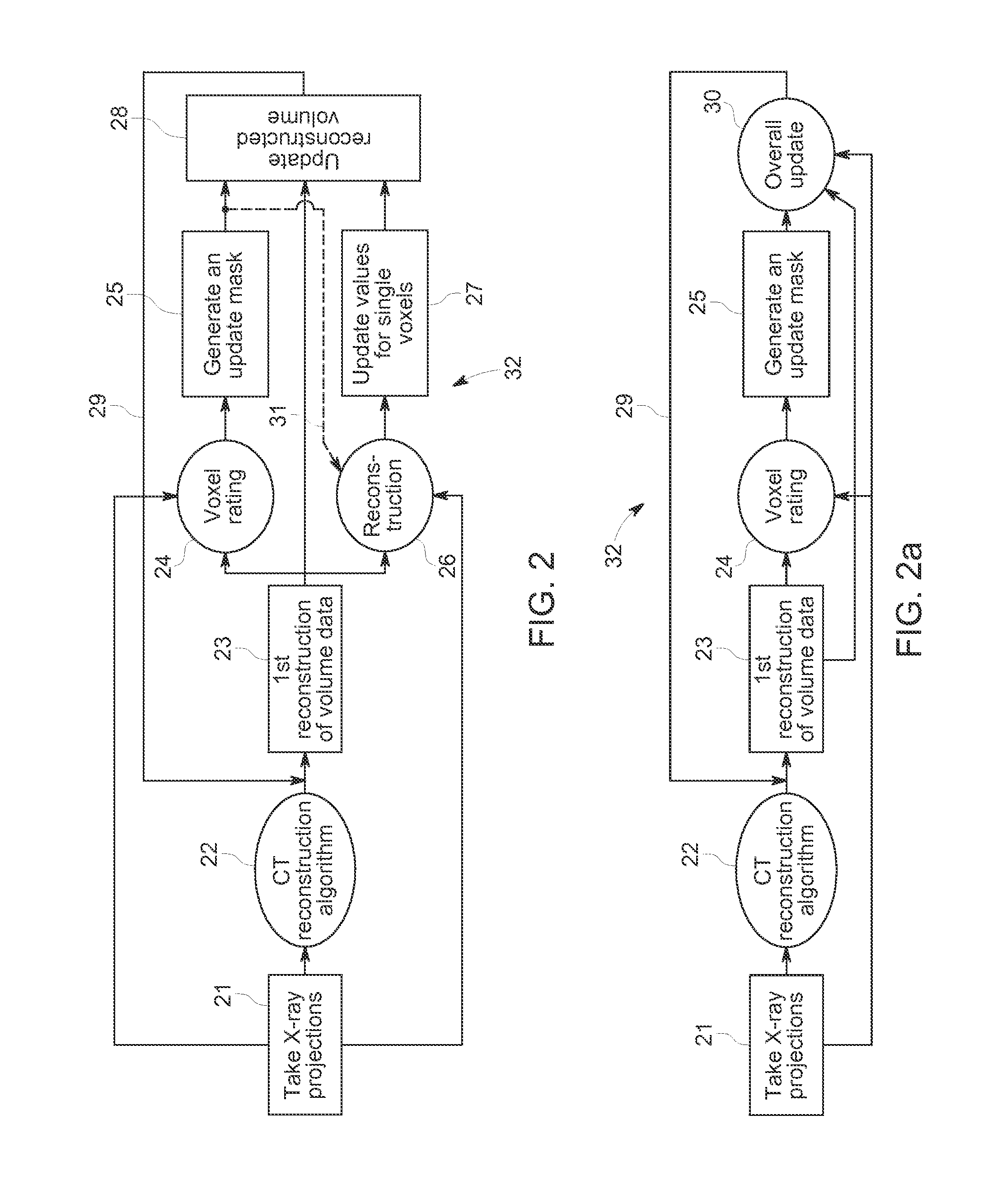
ActiveUS8977022B2Reduce rebuild timeImprove data qualityImage analysisCharacter and pattern recognitionComputed tomographyVolumetric data
A computed tomography method for determining a volumetric representation of a sample comprising reconstruction initial volume data of the sample from x-ray projections of the sample taken by an x-ray system, determining a part of the reconstructed initial volume data to be updated, and executing an iterative update process configured to generate, using an iterative reconstruction method, updated volume data only for the part of the volume data determined to be updated. Determining the part of the sample volume to be updated comprises individually evaluating every single voxel in the reconstructed initial volume data, based on available quality information for the reconstructed initial volume data, whether or not the voxel fulfils a predetermined condition indicating that an update is required for the voxel, and the iterative update process generates the updated volume data only for the voxels which have been determined that an update is required.
Owner:GE SENSING & INSPECTION TECH GMBH
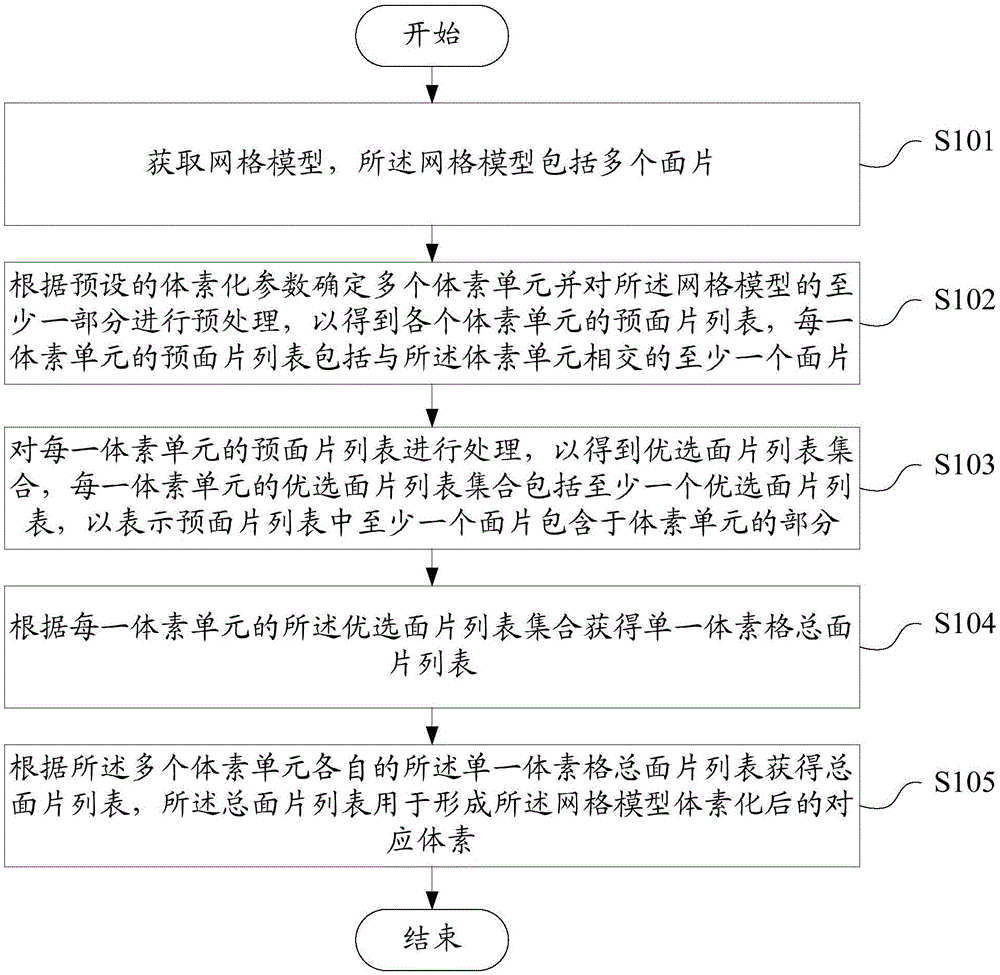
Grid model voxelization method and apparatus
InactiveCN106600671AVoxelization is convenientVoxelization shortcut3D-image renderingComputer scienceSingle voxel
Provided is a grid model voxelization method and apparatus. The method includes the following steps of acquiring a grid model; determining a plurality of voxel units and preprocessing at a part of the grid model according to the preset voxelization parameters to obtain the pre-facet list of each voxel unit; processing the pre-facet list of each voxel unit to obtain a preferable facet list set, the preferable facet list set of each voxel unit includes at least one preferable facet list to express that at least one facet in the pre-facet list is included in the voxel unit; acquiring the total facet list of a single voxel grid according to the preferable facet list set of each voxel unit; acquiring the total facet list according to the total facet list of their respective single voxel grids of the plurality of voxel units, the total facet list is used for forming the corresponding voxels of the grid model after the voxelization. The grid model voxelization method and apparatus can completely guarantee all of the details of the grid model in the voxelization result in an accurate, efficient and robustness manner.
Owner:海宁思核云计算技术有限公司
Computed tomography method, computer program, computing device and computed tomography system
ActiveUS20130163842A1Reduce rebuild timeImprove data qualityImage analysisCharacter and pattern recognitionX-rayComputing tomography
A computed tomography method for determining a volumetric representation of a sample comprising reconstruction initial volume data of the sample from x-ray projections of the sample taken by an x-ray system, determining a part of the reconstructed initial volume data to be updated, and executing an iterative update process for generating configured to generate, using an iterative reconstruction method, updated volume data only for the part of the volume data determined to be updated. Determining the part of the sample volume to be updated comprises individually evaluating every single voxel in the reconstructed initial volume data, based on available quality information for the reconstructed initial volume data, whether or not the voxel fulfils a predetermined condition indicating that an update is required for the voxel, and the iterative update process generates the updated volume data only for the voxels which have been determined that an update is required.
Owner:GE SENSING & INSPECTION TECH GMBH
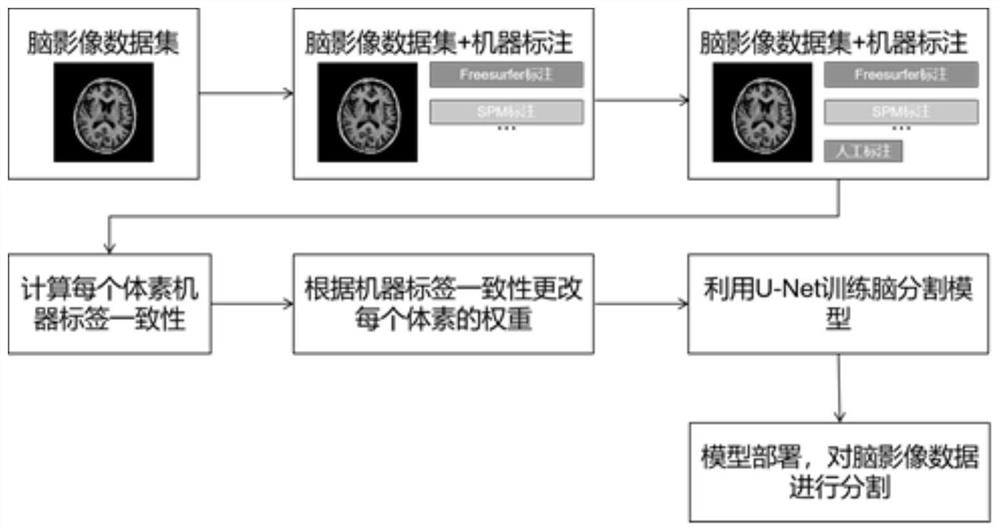
Method for training brain image segmentation model and brain image segmentation method
PendingCN113379757AReduce in quantityLow costImage enhancementImage analysisPattern recognitionData set
The invention relates to a method for training a brain image segmentation model. The method comprises the following steps: collecting a brain image data set; segmenting the brain image by using a plurality of analysis software to obtain a plurality of groups of machine labels; performing manual labeling on a part of the brain image to obtain an artificial label; and based on the brain image data set and the associated label, iteratively training the segmentation model to be trained by circularly executing the following steps to obtain a target segmentation model: inputting the brain image into the segmentation model to be trained to obtain a predicted segmentation result; according to the consistency between the machine labels of the single voxels, determining the weights of the labels; calculating a loss function value according to the difference between the predicted segmentation result and each label and the weight; adjusting network parameters of the to-be-trained segmentation model based on loss function value minimization to obtain a current iteration segmentation model; wherein the weight of the machine label of the single voxel is negatively correlated with the consistency. The invention further relates to a brain image segmentation method and a brain image segmentation device.
Owner:XUANWU HOSPITAL OF CAPITAL UNIV OF MEDICAL SCI +1
Computed tomography method, computer software, computing device and computed tomography system for determining a volumetric representation of a sample
ActiveUS8526570B2Improve accuracyQuality improvementReconstruction from projectionMaterial analysis using wave/particle radiationSoft x rayX-ray
A computed tomography method for determining a volumetric representation of a sample comprises using reconstructed volume data of the sample from x-ray projections of the sample taken by an x-ray system, computing a set of artificial projections of said sample by a forward projection from said reconstructed volume data, and determining, essentially from process data of said reconstruction including said reconstructed volume data and / or said x-ray projections, individual confidence measures for single voxels of said volume data based on calculating, for each of said measured x-ray projections, the difference between the contribution of this measured x-ray projection to the voxel under inspection and the contribution from a corresponding artificial projection to the voxel under inspection.
Owner:GE SENSING & INSPECTION TECH GMBH
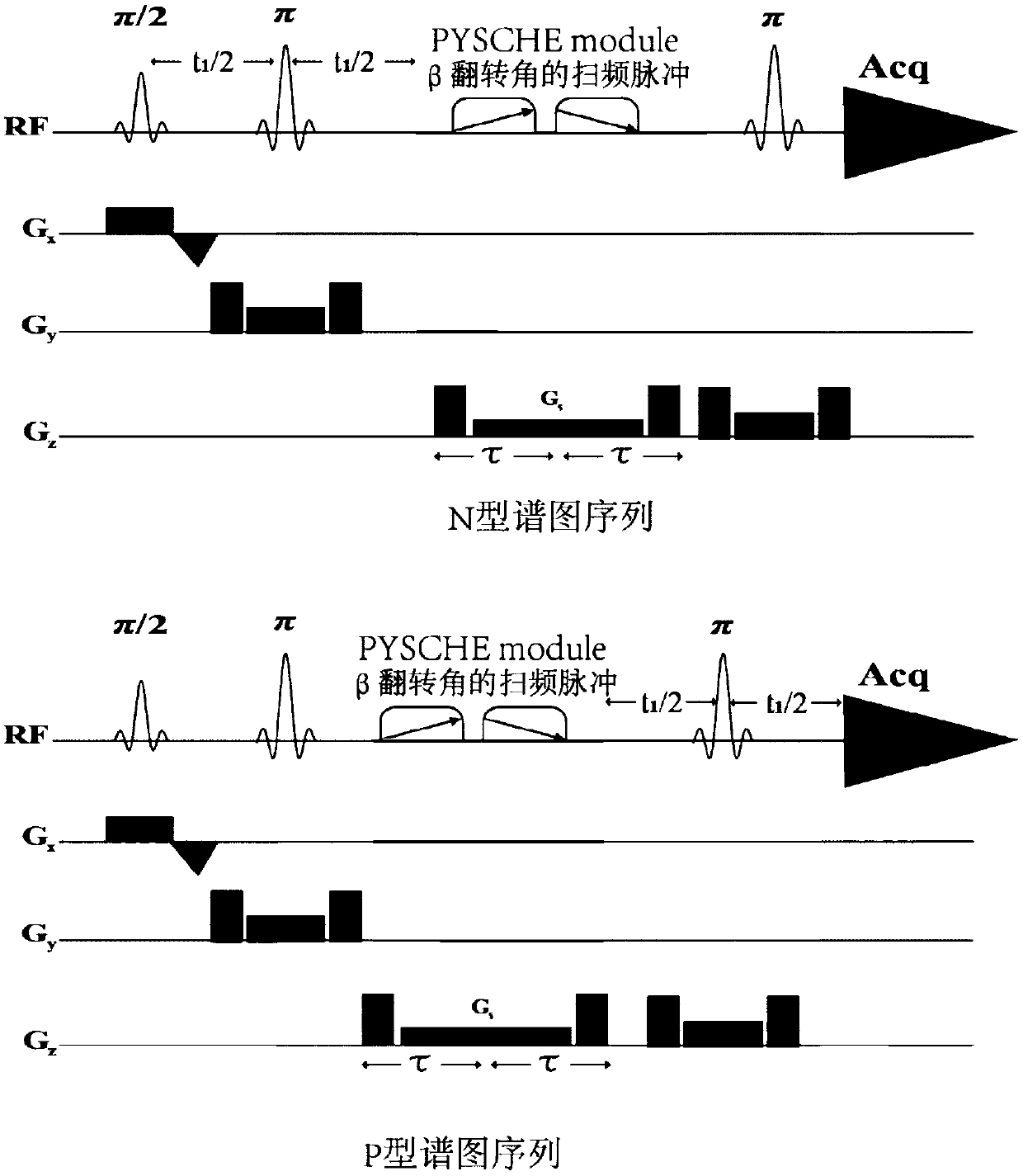
Method for realizing single-voxel localized two-dimensional phase-sensitive J-decomposition spectrum
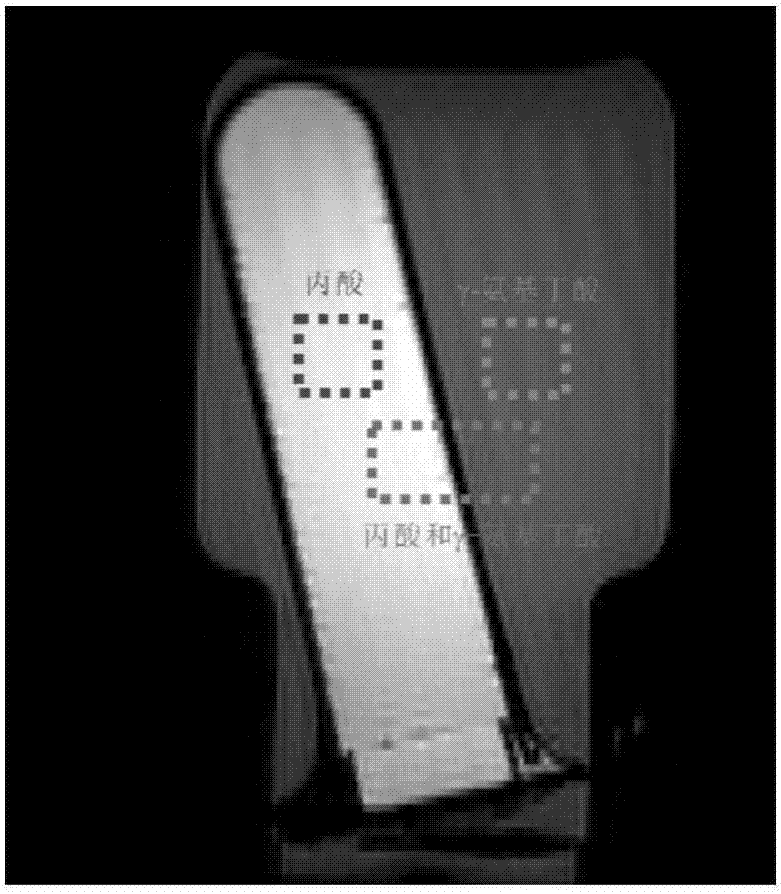
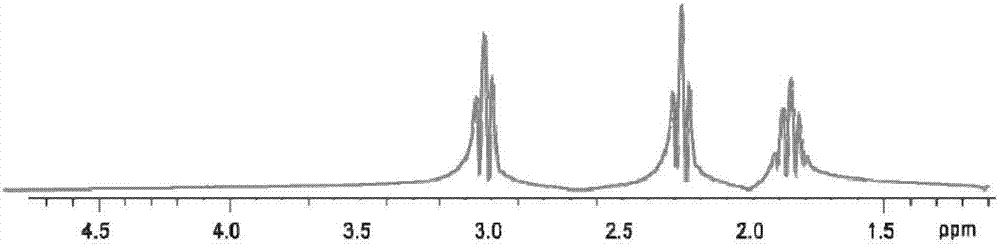
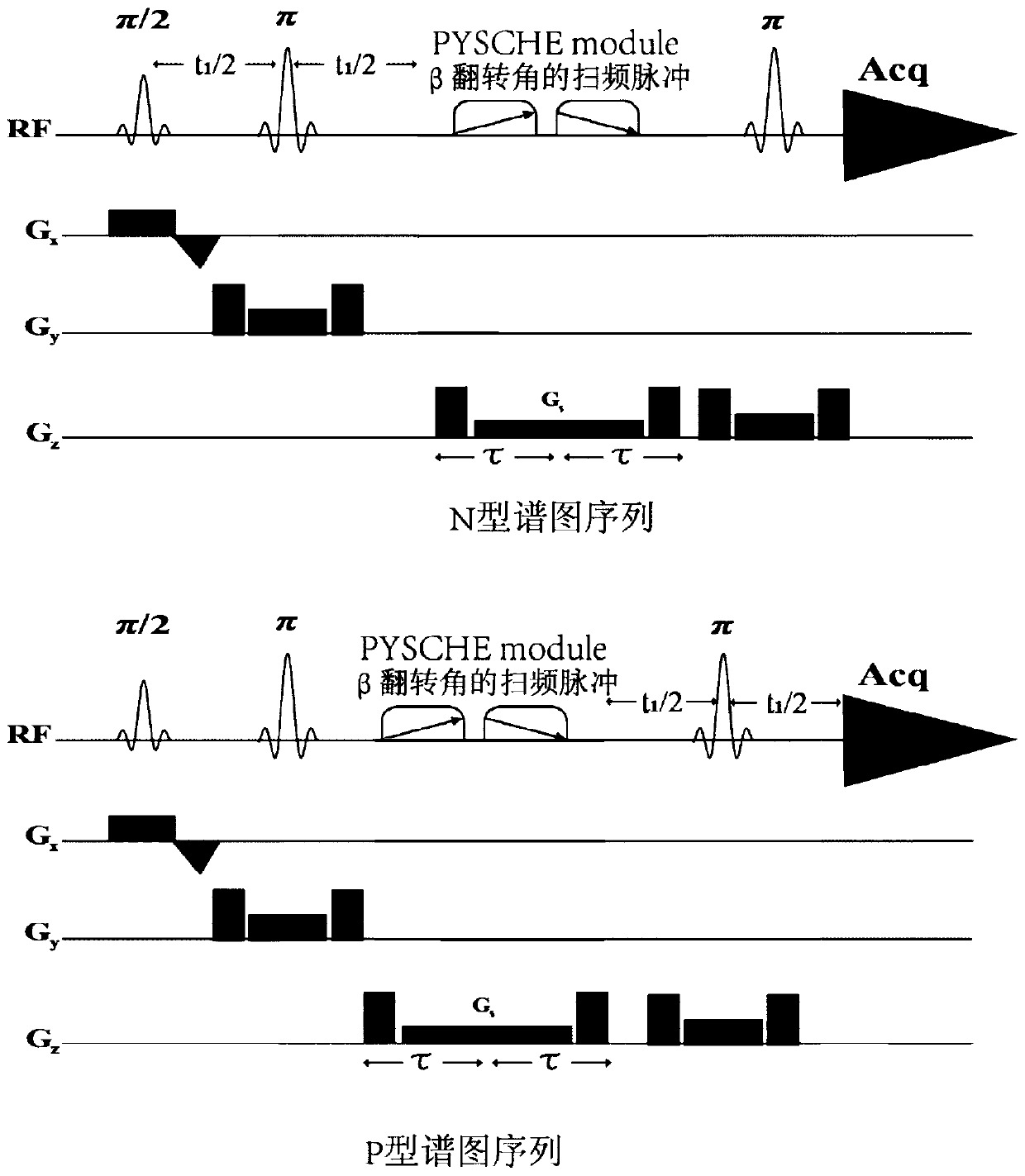
InactiveCN108680884AHigh resolutionMeasurements using NMR spectroscopyDiagnostic recording/measuringDecompositionPhase sensitive
The invention provides a method for realizing a single-voxel localized two-dimensional phase-sensitive J-decomposition spectrum. On the basis of a JPRESS sequence, a new sequence formed by a PYSCHE decoupling module formed by one pair of small-angle inverted sweep pulses and corresponding weak gradient fields and phased gradient fields is inserted before two 180-degree localized pulses, wherein the sequence is formed by N sequences formed at two sides of the 180-degree localized pulses before the PYSCHE decoupling module and P sequences formed at the two sides of the 180-degree localized pulses after the PYSCHE decoupling module during indirect dimensional evolution time; and the two-dimensional spectrum obtained by an N sequence experiment is overturned symmetrically along the indirect dimension F1=10 Hz and then the overturned two-dimensional spectrum is overlapped with a two-dimensional spectrum obtained by a P sequence experiment to obtain single-voxel localized two-dimensional phase-sensitive J-decomposition spectrum.
Owner:XIAMEN UNIV
Two-dimensional Magnetic Resonance Single-Voxel Localized J-Decomposition Spectroscopy Method for Realizing Pure Absorption Lineshape
InactiveCN108169273BHigh resolutionAnalysis using nuclear magnetic resonanceParticle physicsMaterials science
The invention provides a method for realizing a pure absorption line type two-dimensional magnetic resonance single-voxel localized J decomposition spectrum. ZS modules are added at different positions to form an N sequence and an R sequence, the experimental result of the R sequence symmetrically flipped along an indirect dimension F1 = 0 Hz, and superimposing the symmetrically flipped experimental result of the R sequence and the result of the N sequence to obtain the pure absorption line type single-voxel two-dimensional localized J decomposition spectrum. The N sequence is formed through adding the ZS module after the first 90 DEG localized pulse, the R sequence is formed through adding the ZS module after the last 180 DEG localized pulse, the delay times at both sides of the last 180DEG of each of the sequences are uniform and are t1 / 2, and indirect dimensional F1 evolution is performed. The ZS module consists of a selective 180 DEG soft pulse, a Z-directional gradient magnetic field and a symmetric spoiled gradient, so the selective 180 DEG soft pulse realizes the evolution refocusing of different nuclei at different spatial positions, and the coherence order of the resonance nuclei transfers; and the ZS modules of the N sequence and the R sequence have different positions, so final two-dimensional signal composition modes are different.
Owner:XIAMEN UNIV
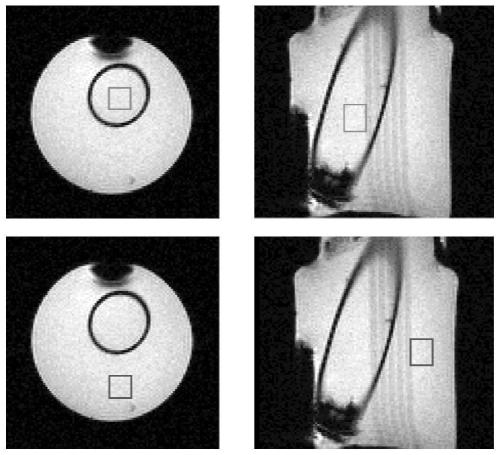
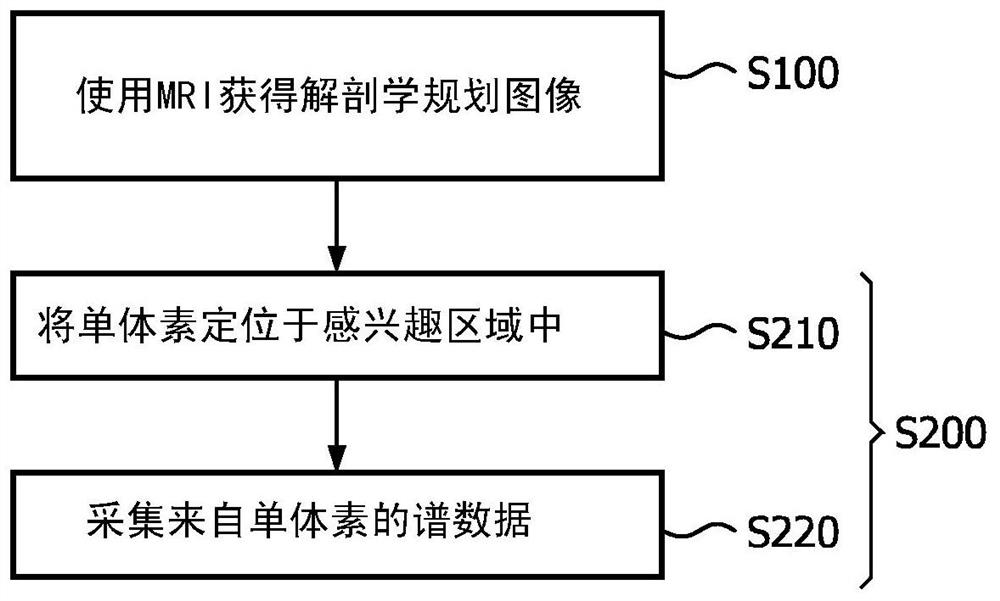
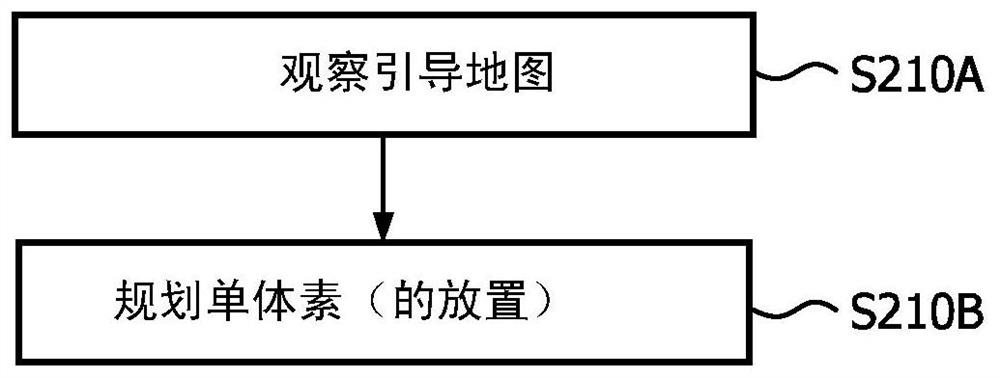
Imaging System for Single Voxel Spectroscopy
A guide map is created for placing a spectroscopic voxel in a region of interest in single voxel magnetic resonance spectroscopy. An anatomically planned image of the region of interest is obtained by MRI. stepping spectroscopic voxels across the region of interest, measuring a characteristic of the magnetic field used in the MRI at each location of the imaging voxel, and using the measurements to derive an indication of a magnetic field over the region of interest A guided FWHM map of the magnetic field uniformity / inhomogeneity. The guide map is created by superimposing the guide FWHM map on the anatomical planning image. A spectroscopic voxel having a size corresponding to the spectroscopic voxel is placed within the region of interest according to the guide map. Thereafter, spectral data is acquired from the region of interest confined to the single voxel.
Owner:KONINKLJIJKE PHILIPS NV
Measuring diffusional anisotropy of ODF lobes having maxima peaks and minima troughs with diffusion weighted MRI
A method for measuring diffusional anisotropy in diffusion-weighted magnetic resonance imaging. The method includes determining an orientation diffusion function (ODF) for one or more fibers within a single voxel, wherein the ODF includes lobes representative of a probability of diffusion in a given direction for the one or more fibers. The method also includes characterizing an aspect ratio of the lobes. The method further includes determining a multi-directional anisotropy metric for the one or more fibers based on the aspect ratio of the lobes.
Owner:GENERAL ELECTRIC CO
A Single-Voxel Localized One-Dimensional Pure Chemical Shift NMR Spectroscopy Method
ActiveCN106645255BExtended NDT ApplicationsSimplify spectral informationAnalysis using nuclear magnetic resonanceSignal classificationMultiple signal classification
An NMR (nuclear magnetic resonance) single voxel localization and one-dimensional pure chemical shift spectroscopy method comprises steps as follows: a to-be-detected sample is put in a detection cavity of a magnetic resonance imaging instrument; the position of the sample in the detection cavity is adjusted to ensure that an interested region is located at the center of the detection cavity for tuning, shimming and power and frequency correction; pai / 2 non-selective radio-frequency pulse width of an excitation sample is measured, and measured pai / 2 non-selective radio-frequency pulse can turn the magnetization vector from the Z-axis longitudinal direction to the XY transverse plane; a single voxel localization and one-dimensional pure chemical shift spectroscopy pulse sequence is imported to the imaging instrument, and a signal excitation module, a single voxel localization module and a pure chemical shift evolution module of the single voxel localization and one-dimensional pure chemical shift spectroscopy pulse sequence are opened; sequence parameters are set, and data sampling is executed; after data are sampled, sampled data are subjected to post-processing, post-processing includes two-dimensional Fourier transform, spectrogram indirect dimension projection and DMUSIC (decay multiple signal classification), and single voxel localization and one-dimensional pure chemical shift spectroscopy with high resolution and high signal-to-noise ratio is obtained.
Owner:XIAMEN UNIV
A Method for Realizing Two-Dimensional Phase-Sensitive J-Decomposition Spectroscopy of One-Voxel Localization
InactiveCN108680884BHigh resolutionMeasurements using NMR spectroscopyDiagnostic recording/measuringDecompositionPhase sensitive
The invention provides a method for realizing a single-voxel localized two-dimensional phase-sensitive J-decomposition spectrum. On the basis of a JPRESS sequence, a new sequence formed by a PYSCHE decoupling module formed by one pair of small-angle inverted sweep pulses and corresponding weak gradient fields and phased gradient fields is inserted before two 180-degree localized pulses, wherein the sequence is formed by N sequences formed at two sides of the 180-degree localized pulses before the PYSCHE decoupling module and P sequences formed at the two sides of the 180-degree localized pulses after the PYSCHE decoupling module during indirect dimensional evolution time; and the two-dimensional spectrum obtained by an N sequence experiment is overturned symmetrically along the indirect dimension F1=10 Hz and then the overturned two-dimensional spectrum is overlapped with a two-dimensional spectrum obtained by a P sequence experiment to obtain single-voxel localized two-dimensional phase-sensitive J-decomposition spectrum.
Owner:XIAMEN UNIV
A method for metabolite signal quantitation for magnetic resonance spectroscopy data
ActiveUS20210190891A1Reduce dependenceIncrease capacityMeasurements using NMR spectroscopyMeasurements using NMR imaging systemsMagnetic resonance spectroscopicMetabolite
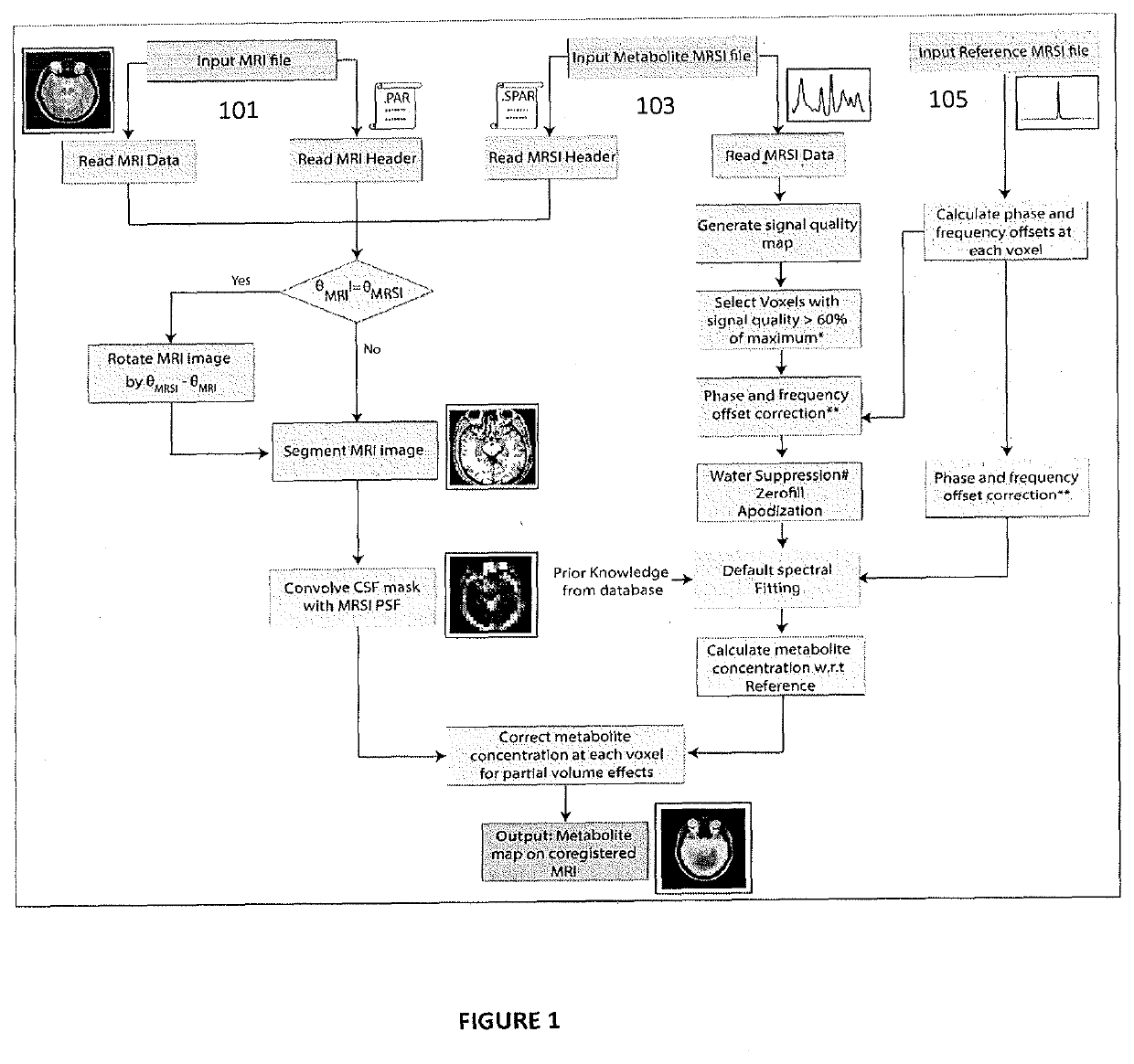
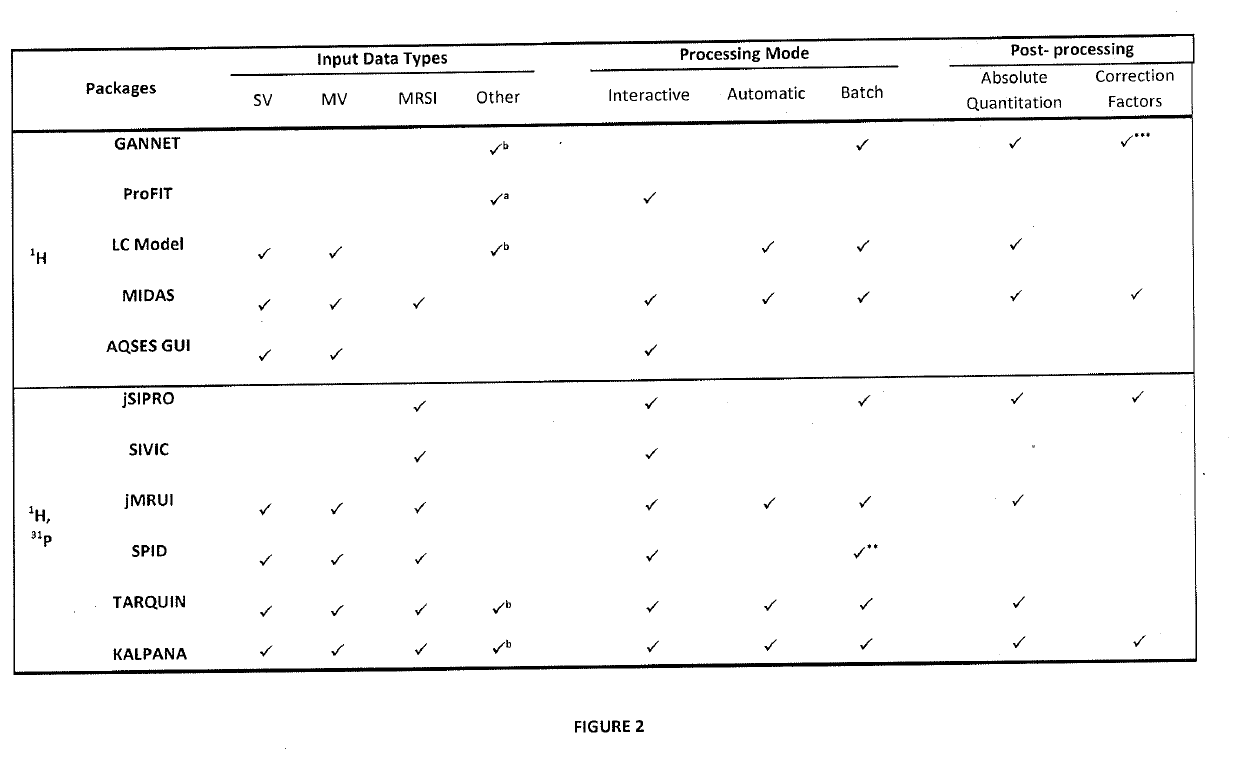
It is an integrative platform for visualization, preprocessing and quantitation of MRS data acquired using single voxel, multi voxel magnetic resonance spectroscopy imaging (MRSI) and MEshcher-GArwood Point-RESolved Spectroscopy (MEGA-PRESS) acquisition methods. The method integrates both time- and frequency-domain signal processing methods on a single platform. The method is optimized for proton (1H) and phosphorous (31P) MRS data. It employs the use of iterative baseline estimation and fitting procedure to provide improved quantitation accuracy. The method can be used in both interactive and automatic mode to cater to the needs of researchers and clinicians.
Owner:SEC +1
A method for obtaining single-voxel one-dimensional localized spectra with scalar coupling modulation eliminated
InactiveCN103645453BPromote localizationGood signalDiagnostic recording/measuringSensorsPhase distortionCoupling constant
The invention provides a method for obtaining single voxel one-dimensional localization spectra capable of eliminating scalar coupling modulation. A same-direction 90-degree radio-frequency pulse is added between two spin echo modules to form a hard pulse scalar coupling re-aggregation module and a soft pulse scalar coupling re-aggregation module, the interval t between the pulses in the re-aggregation modules is set according to the coupling system condition included in a sample, and when the t and scalar coupling constant J product is much smaller than 1, the scalar coupling re-aggregation modules can eliminate scalar coupling modulation effects caused by various coupling systems. The soft pulse scalar coupling re-aggregation module completes voxel selection by combining with selecting layers and destroy gradients. According to the method, by the aid of the obtained one-dimensional localization spectra, signal amplitude and phase distortions caused by scalar coupling modulation can be prevented, perfect signals are obtained, and spectrogram attribution analysis is facilitated.
Owner:XIAMEN UNIV
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com