Patents
Literature
89 results about "Interactor" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
An interactor is also a person who interacts with the members of the audience.
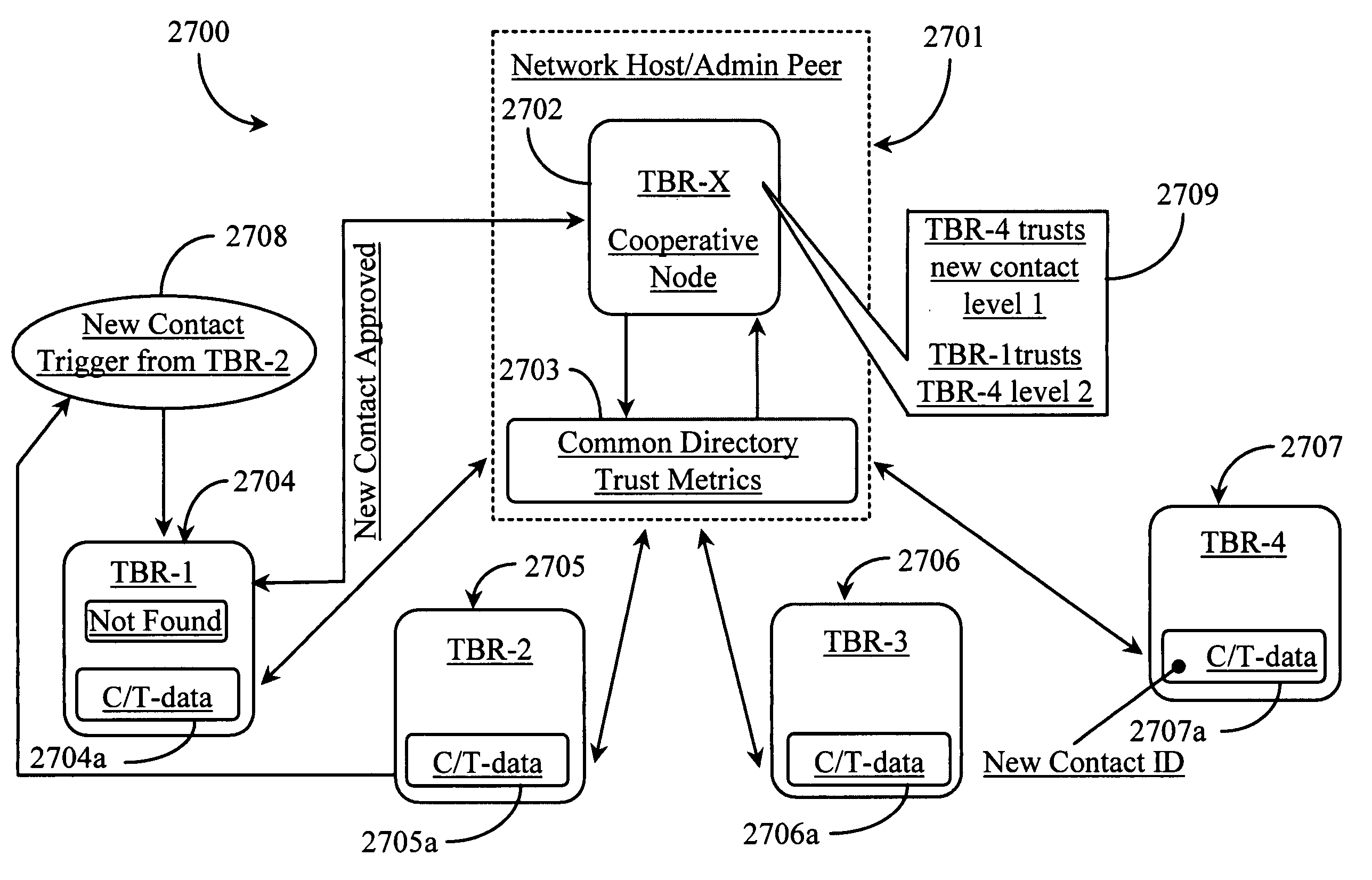
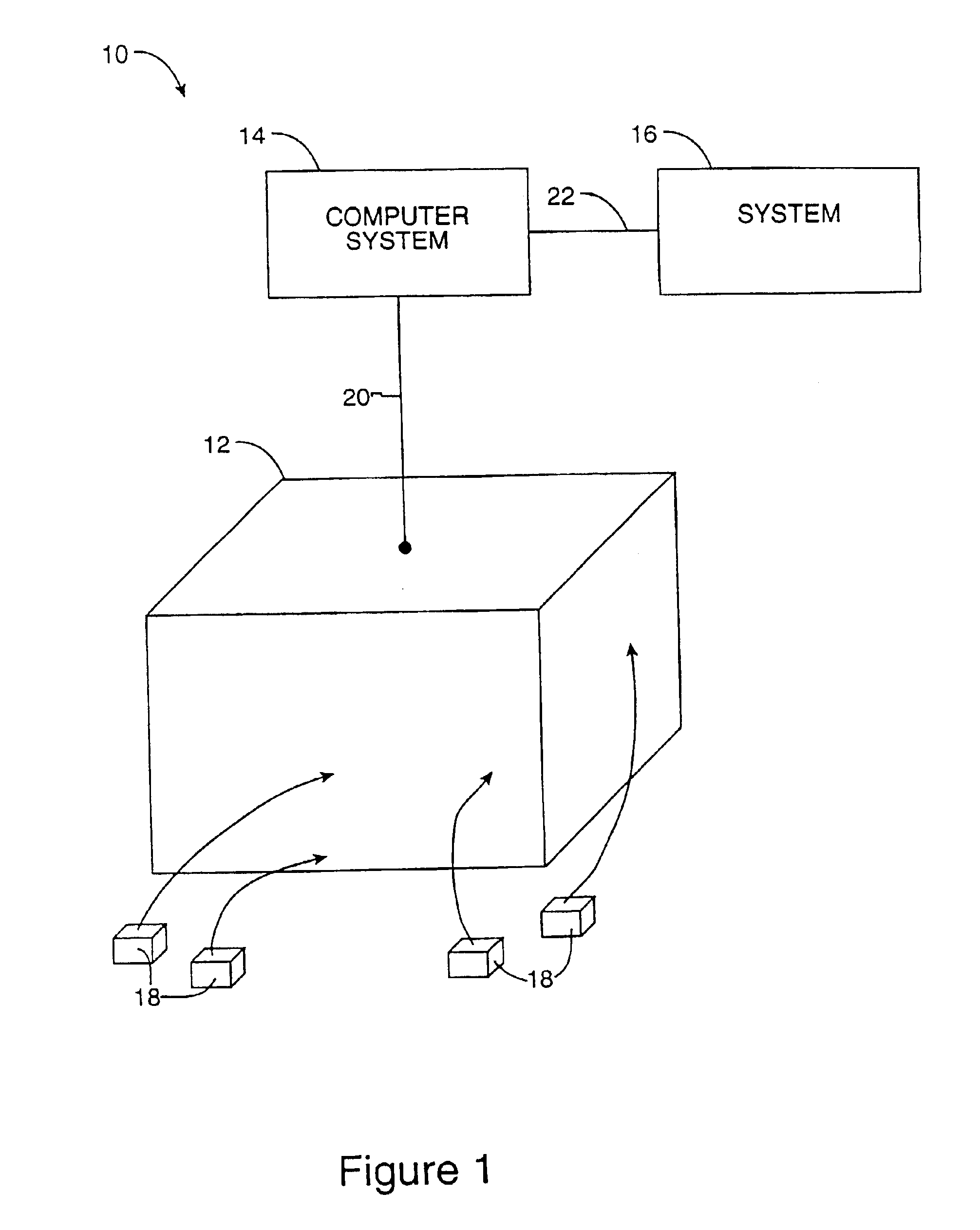
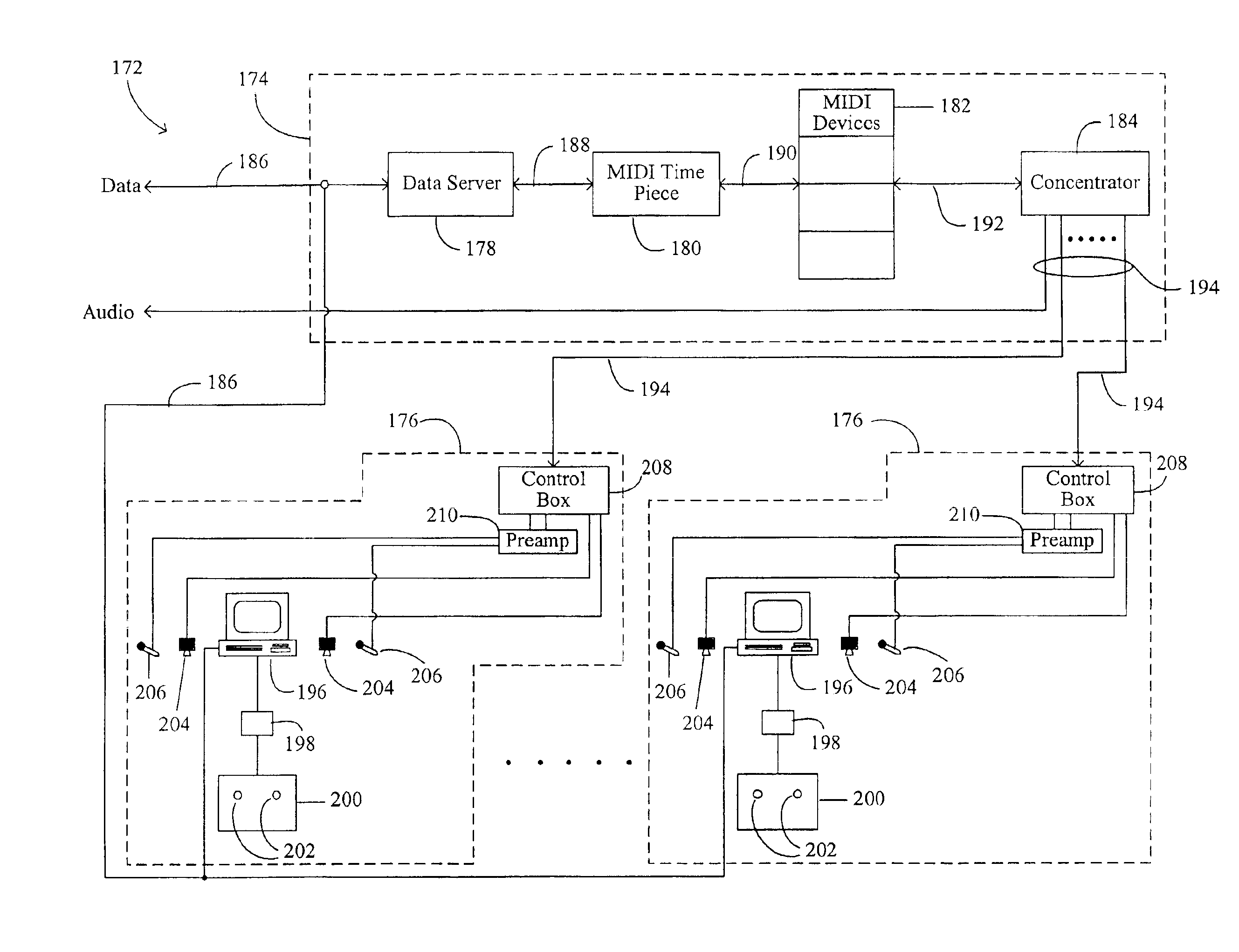
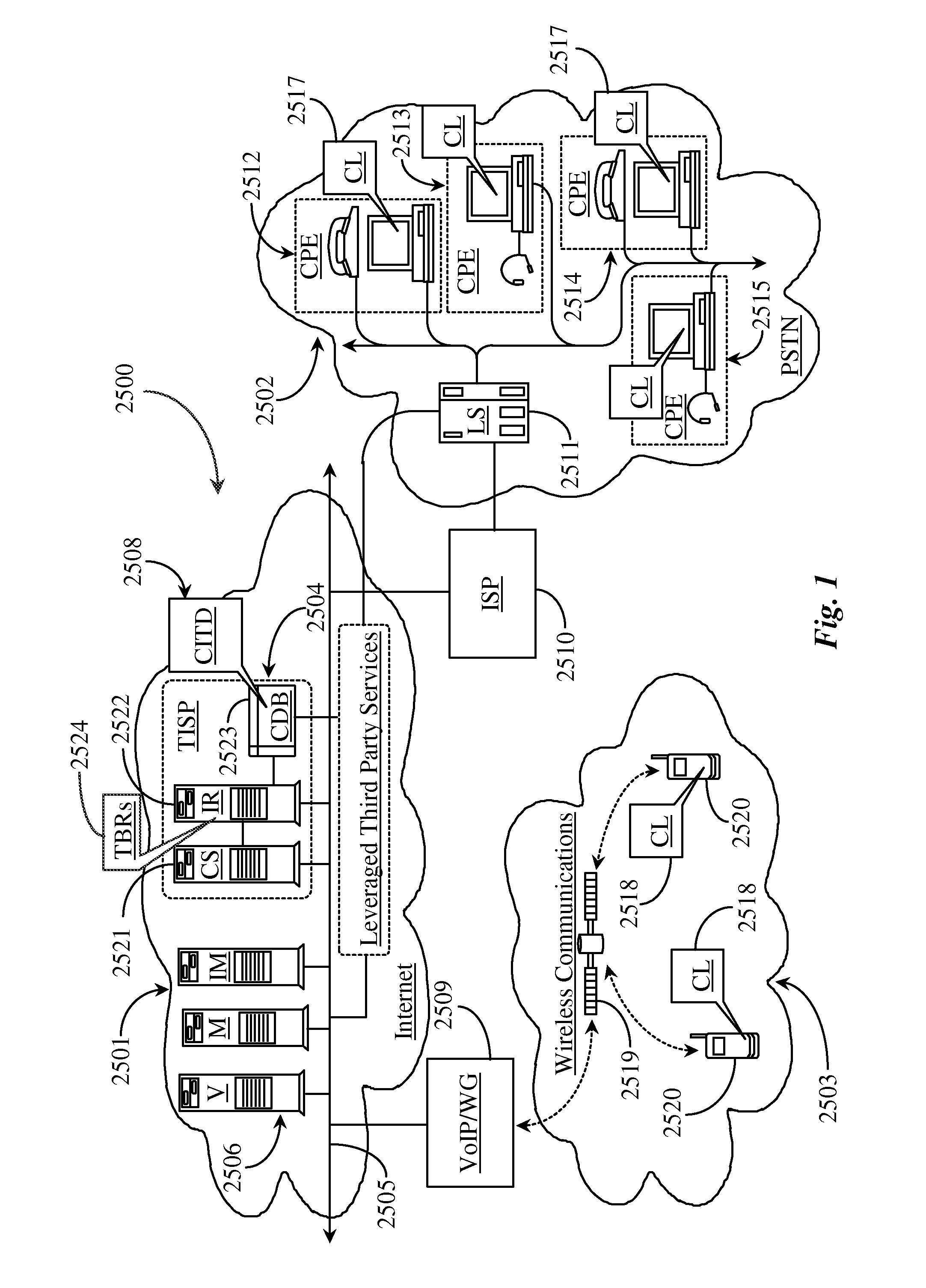
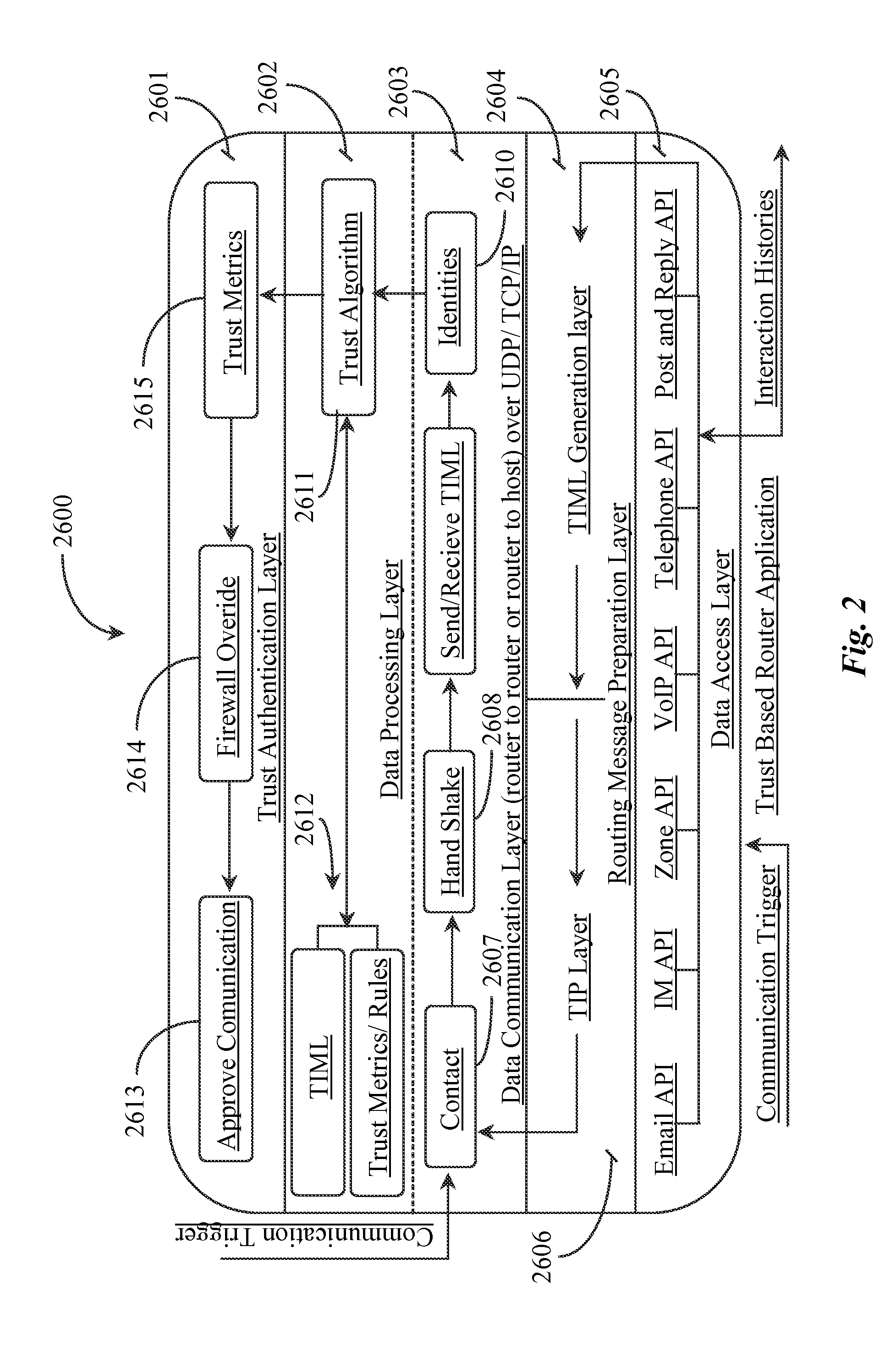
Methods and apparatus for enabling a dynamic network of interactors according to personal trust levels between interactors
A system for causing routing of a communication event includes a sending platform for initiating and sending the communication event, a communications network for carrying the communication event, a receiving platform for receiving the communication event for final routing, and a router resident at least in the receiving platform for preparing and executing or forwarding for execution a routing instruction for handling the incoming communication event or notification thereof, the routing instruction thus executed, overriding a default routing instruction, the overriding routing instruction initiated upon discovery by the router of some level of trust metric between the sender and intended recipient of the event.
Owner:FORTE INTERNET SOFTWARE
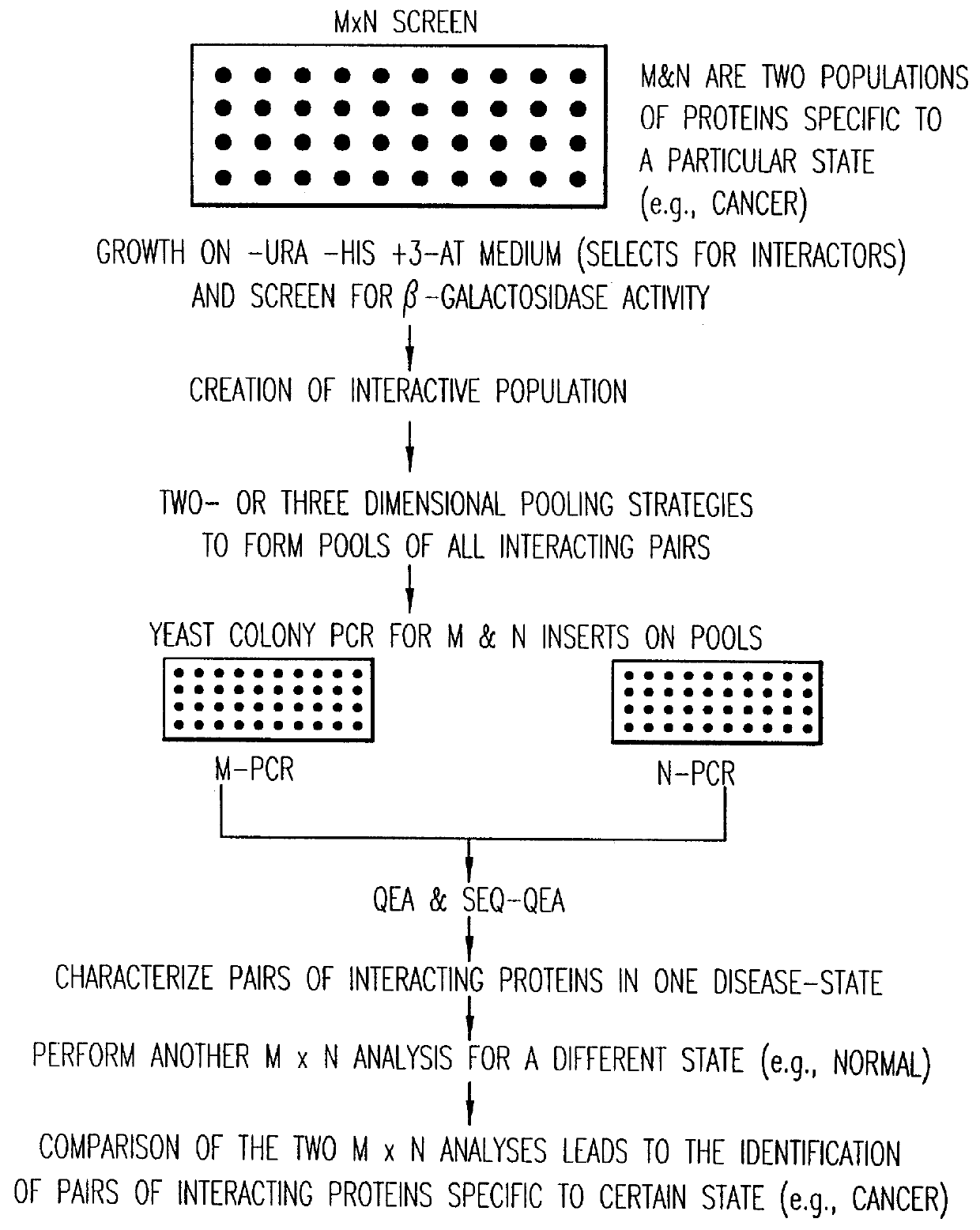
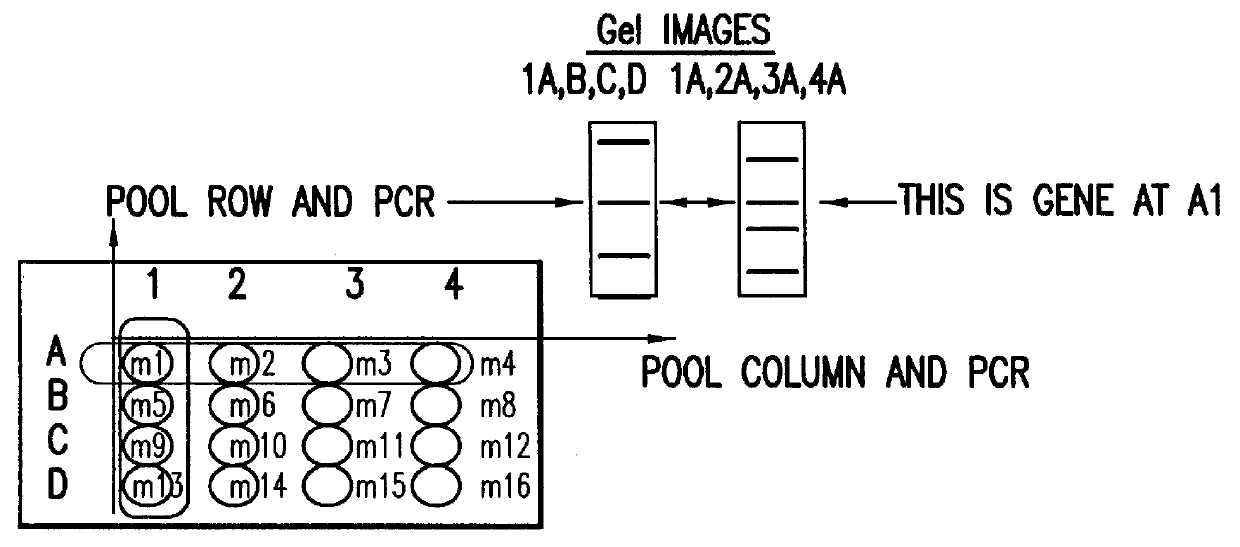
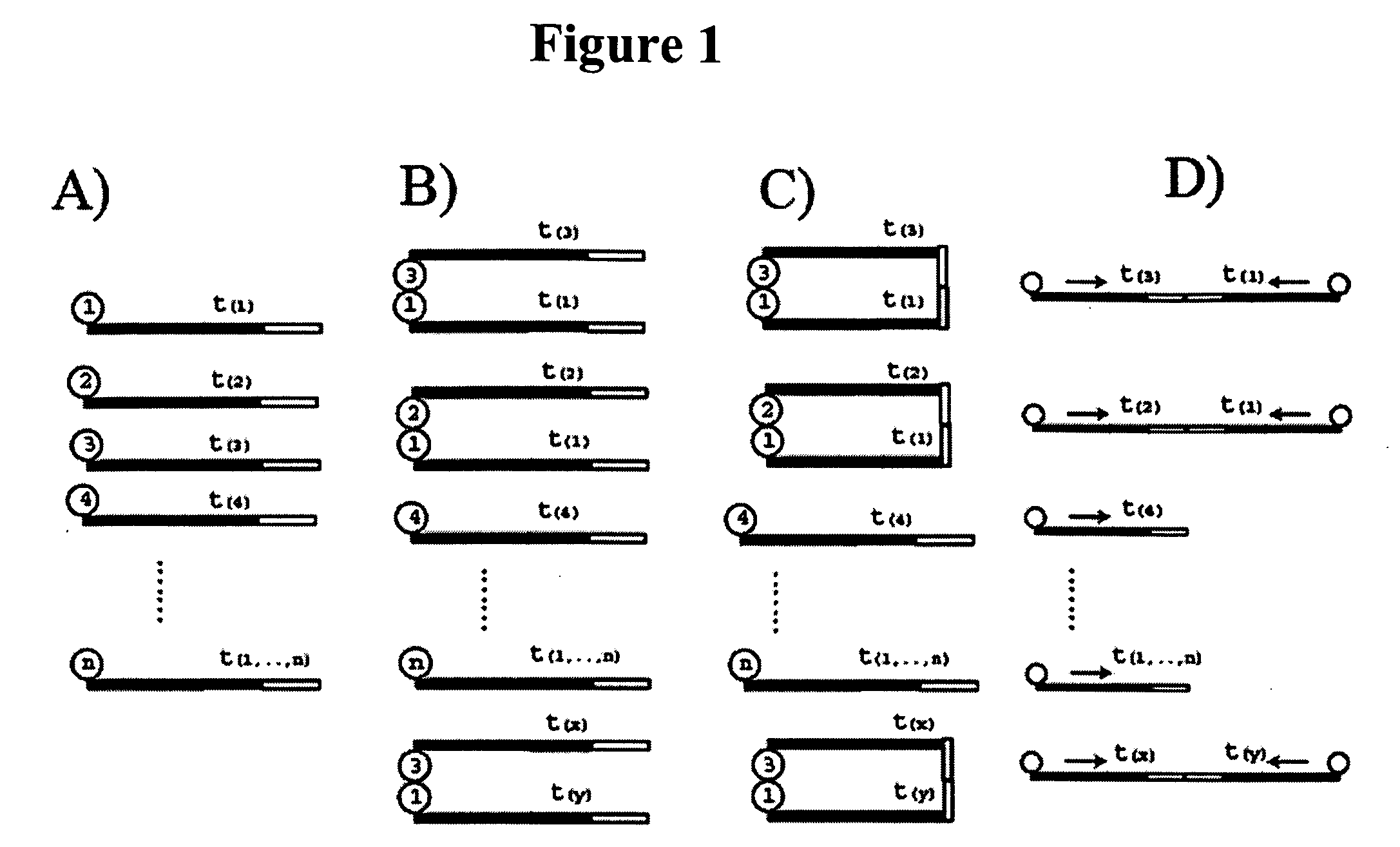
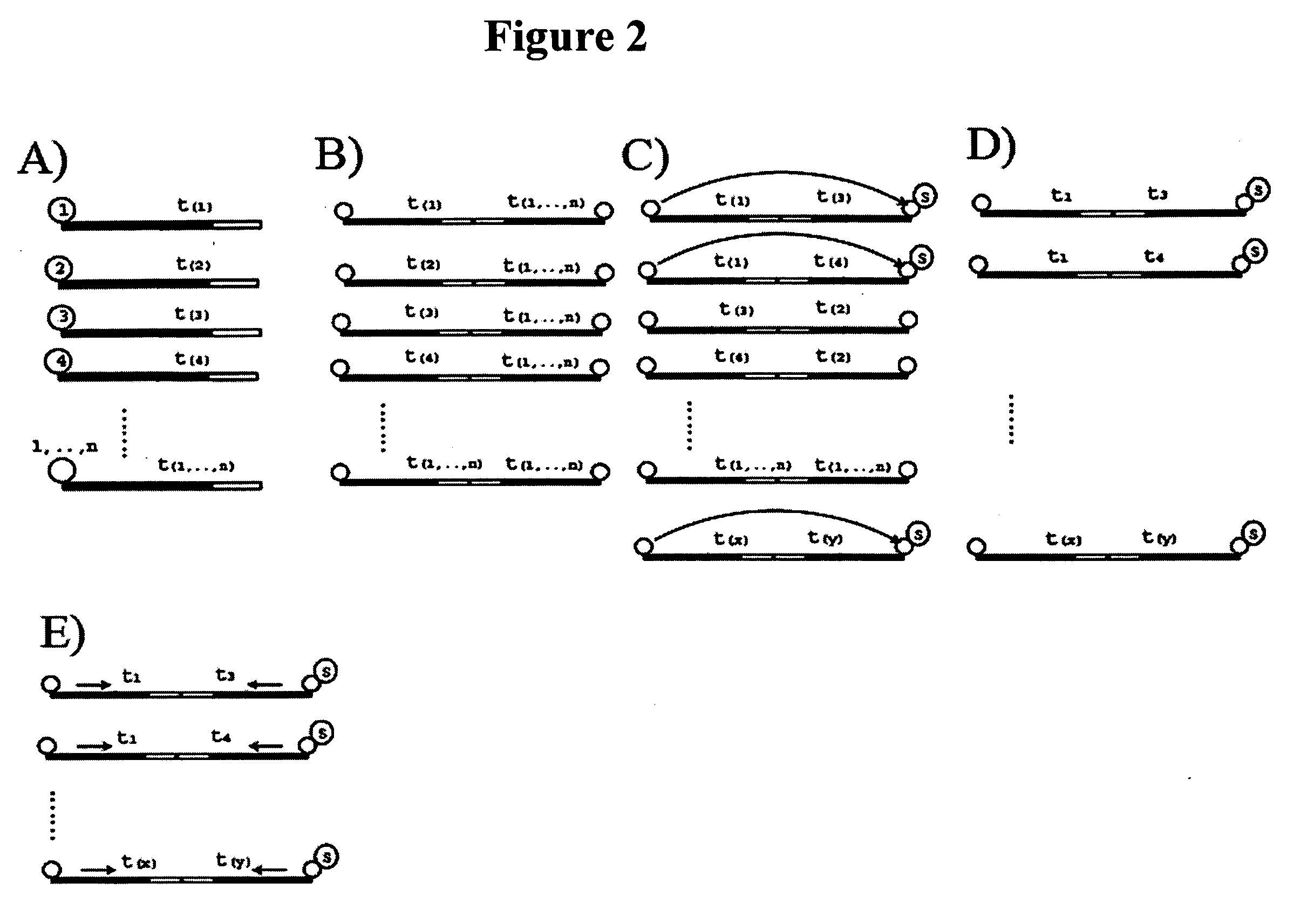
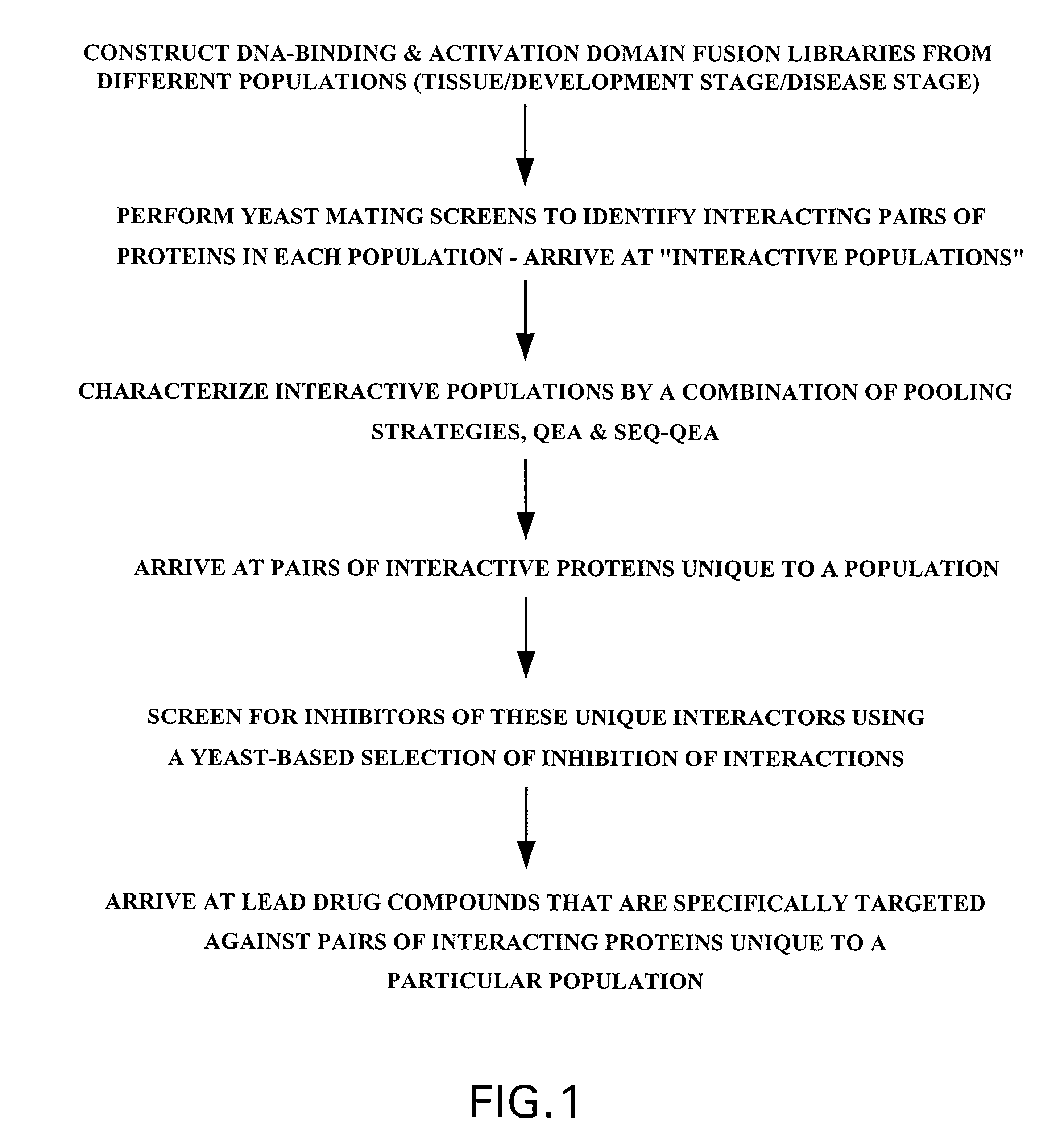
Identification and comparison of protein-protein interactions that occur in populations and identification of inhibitors of these interactors
InactiveUS6057101AEfficient screeningLess experimentally significant and specific indicationMaterial nanotechnologyFungiDiseaseBinding site
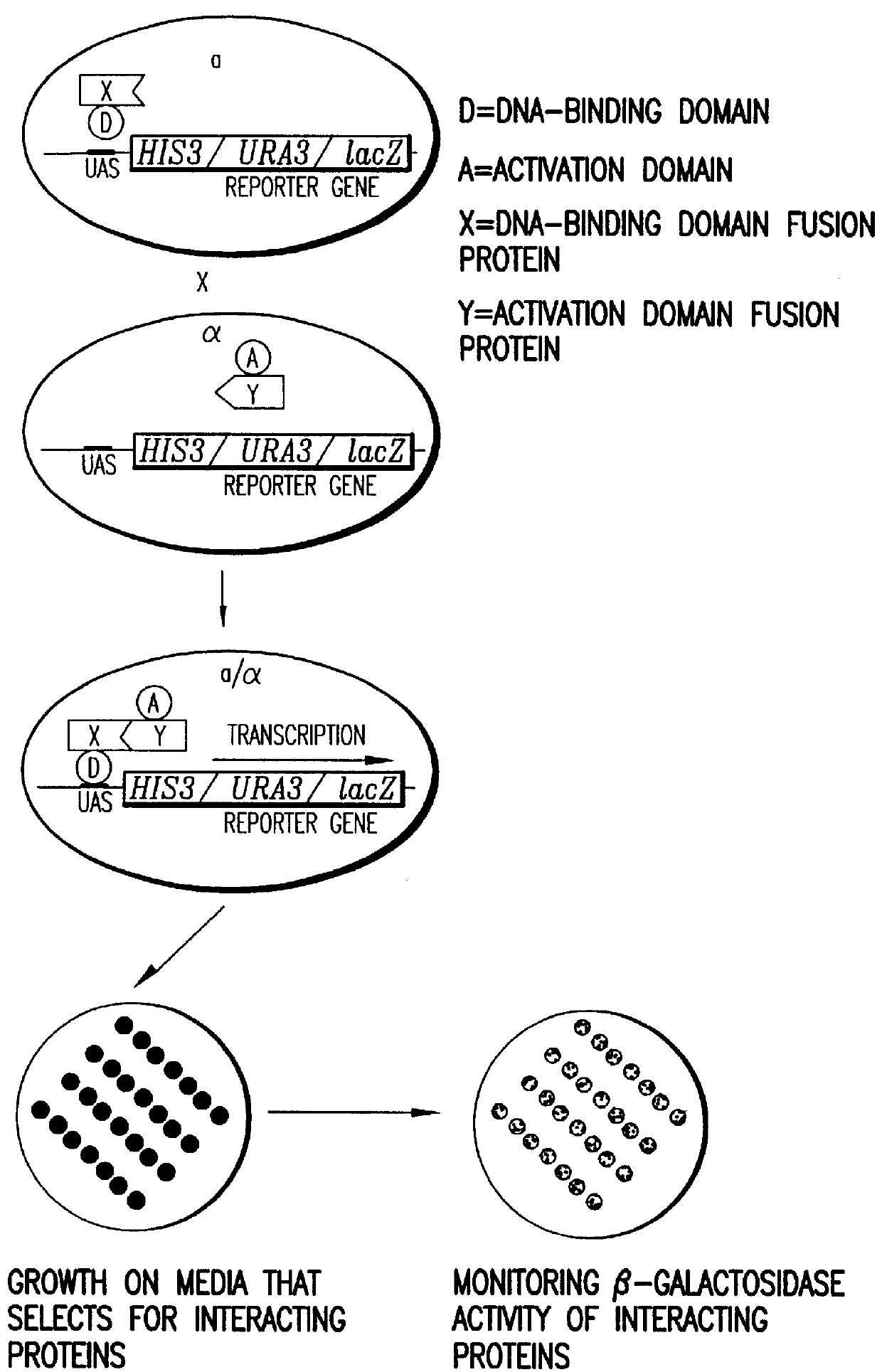
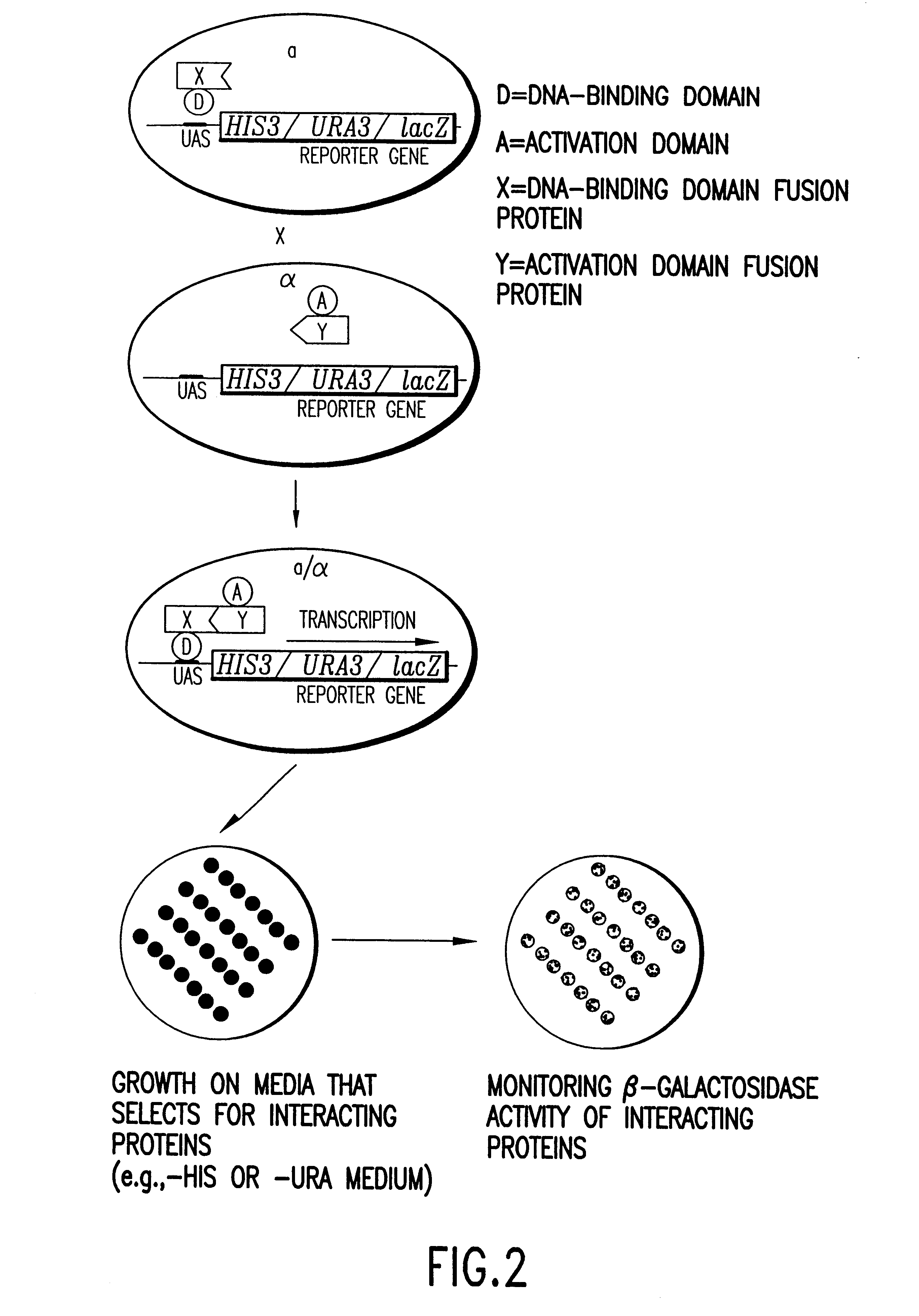
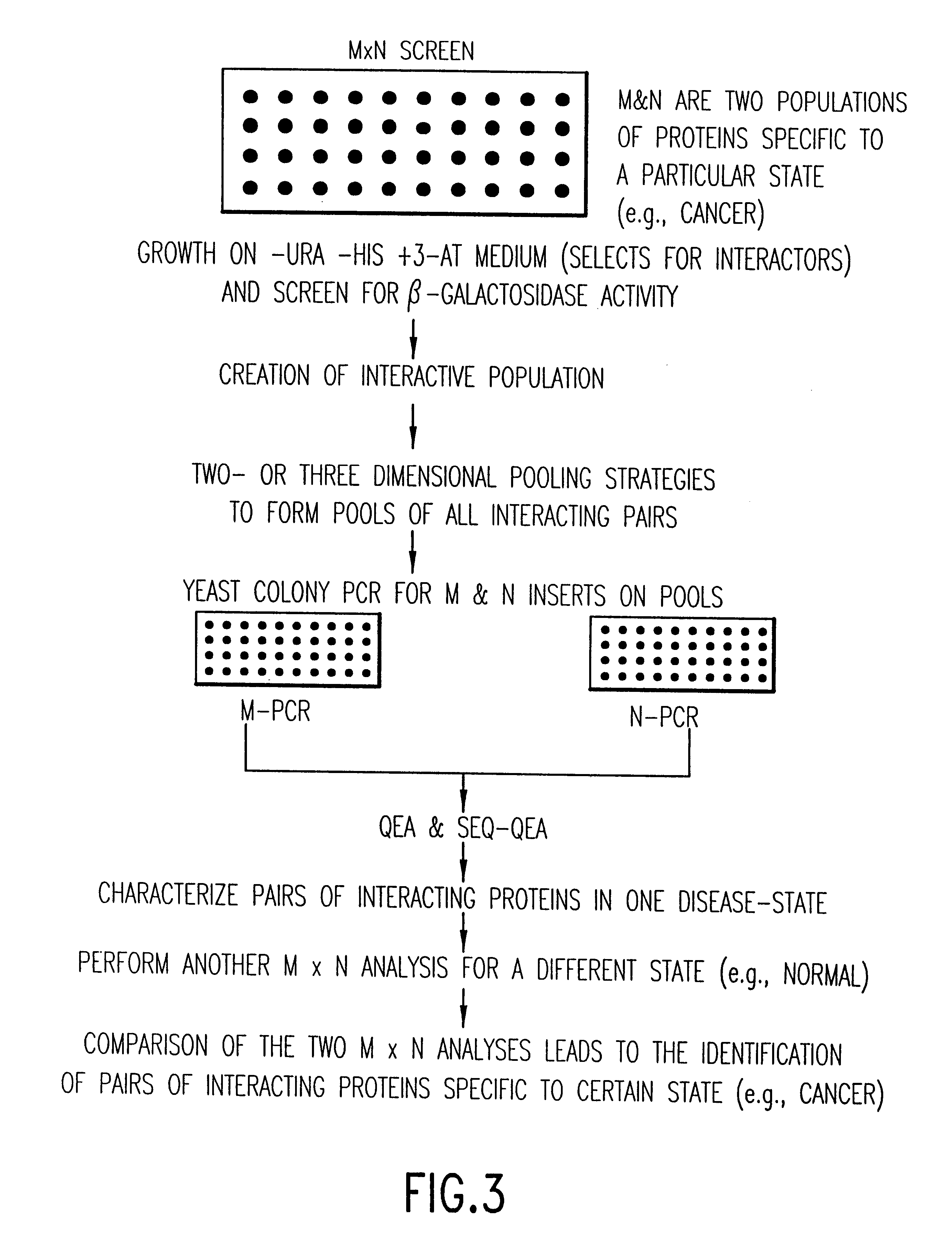
Methods are described for detecting protein-protein interactions, among two populations of proteins, each having a complexity of at least 1,000. For example, proteins are fused either to the DNA-binding domain of a transcriptional activator or to the activation domain of a transcriptional activator. Two yeast strains, of the opposite mating type and carrying one type each of the fusion proteins are mated together. Productive interactions between the two halves due to protein-protein interactions lead to the reconstitution of the transcriptional activator, which in turn leads to the activation of a reporter gene containing a binding site for the DNA-binding domain. This analysis can be carried out for two or more populations of proteins. The differences in the genes encoding the proteins involved in the protein-protein interactions are characterized, thus leading to the identification of specific protein-protein interactions, and the genes encoding the interacting proteins, relevant to a particular tissue, stage or disease. Furthermore, inhibitors that interfere with these protein-protein interactions are identified by their ability to inactivate a reporter gene. The screening for such inhibitors can be in a multiplexed format where a set of inhibitors will be screened against a library of interactors. Further, information-processing methods and systems are described. These methods and systems provide for identification of the genes coding for detected interacting proteins, for assembling a unified database of protein-protein interaction data, and for processing this unified database to obtain protein interaction domain and protein pathway information.
Owner:CURAGEN CORP
Regulation analysis by cis reactivity, RACR
ActiveUS20070020669A1Facilitates high throughput analysisHigh throughput analysisSugar derivativesMicrobiological testing/measurementPresent methodInteractor
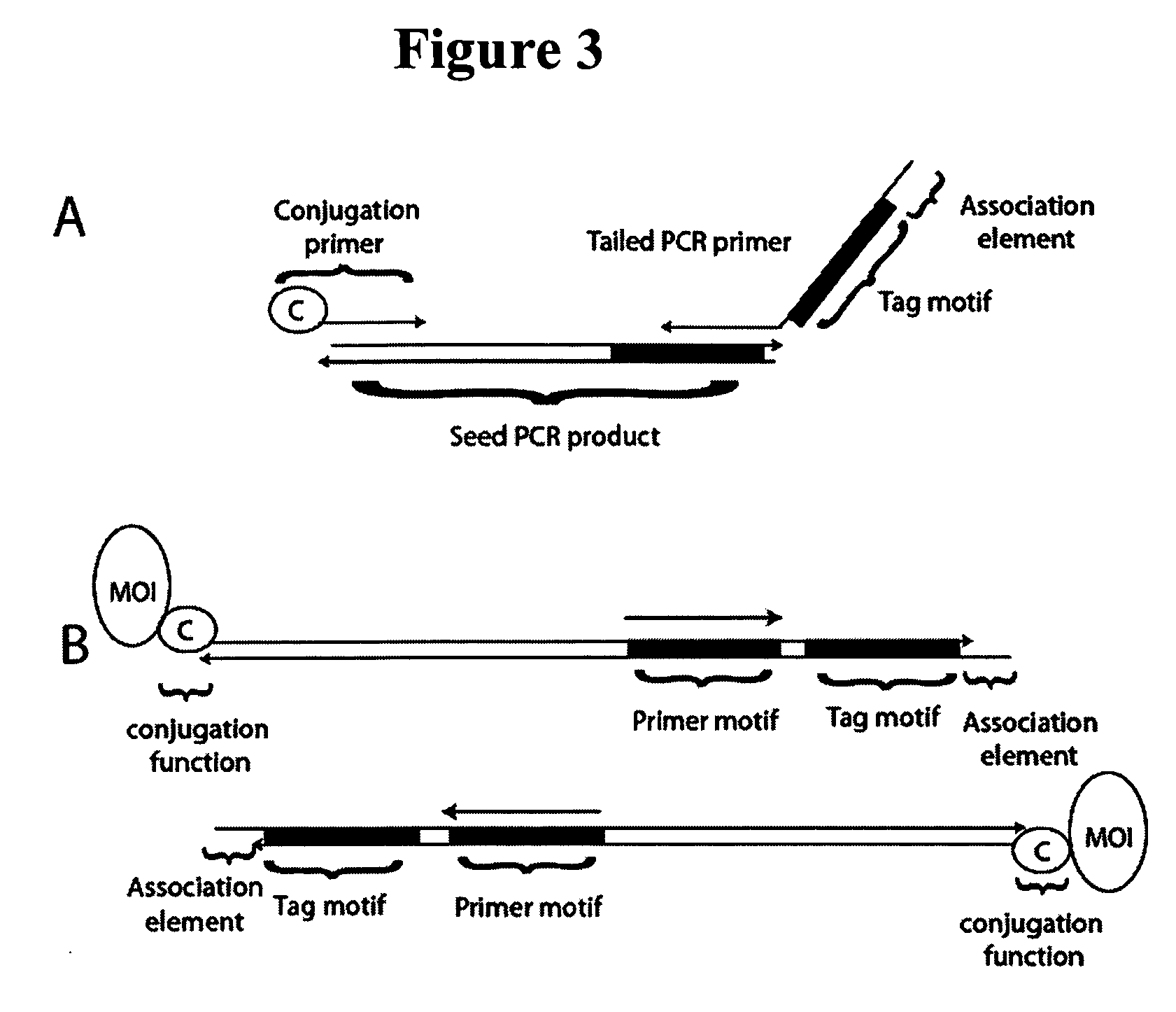
Methods of detecting affinity interactions between at least two molecules of interest are provided. The method comprises: a. forming a plurality of interactors by coupling each molecule of interest with at least one nucleic acid moiety comprising an identification sequence element and at an association element; b. promoting an association between at least two nucleic acid moieties from different interactors to form a plurality of unique associated oligonucleotides, wherein each nucleic acid moiety may form more than one unique associated oligonucleotide, and wherein each unique associated oligonucleotide comprises at least two identification sequence elements derived from the at least two nucleic acid moieties; c. selecting the plurality of unique associated oligonucleotides; and d. subjecting the selected associated oligonucleotides to an analysis that permits detection of the at least two identification sequence elements. Similar methods directed to detecting functional interactions, libraries of interactors employable in the present methods, and kits comprising those libraries are also provided.
Owner:OLINK PROTEOMICS AB
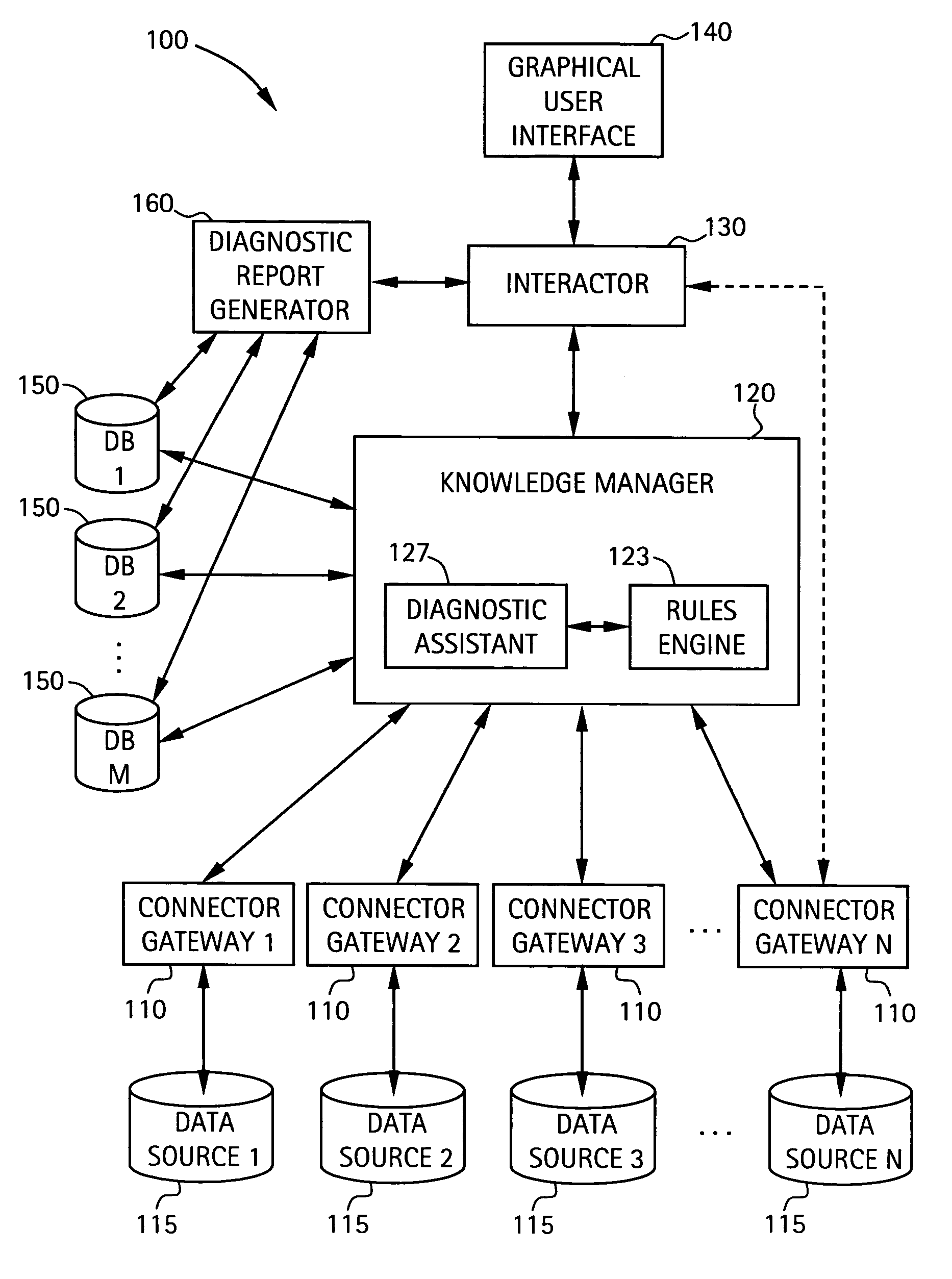
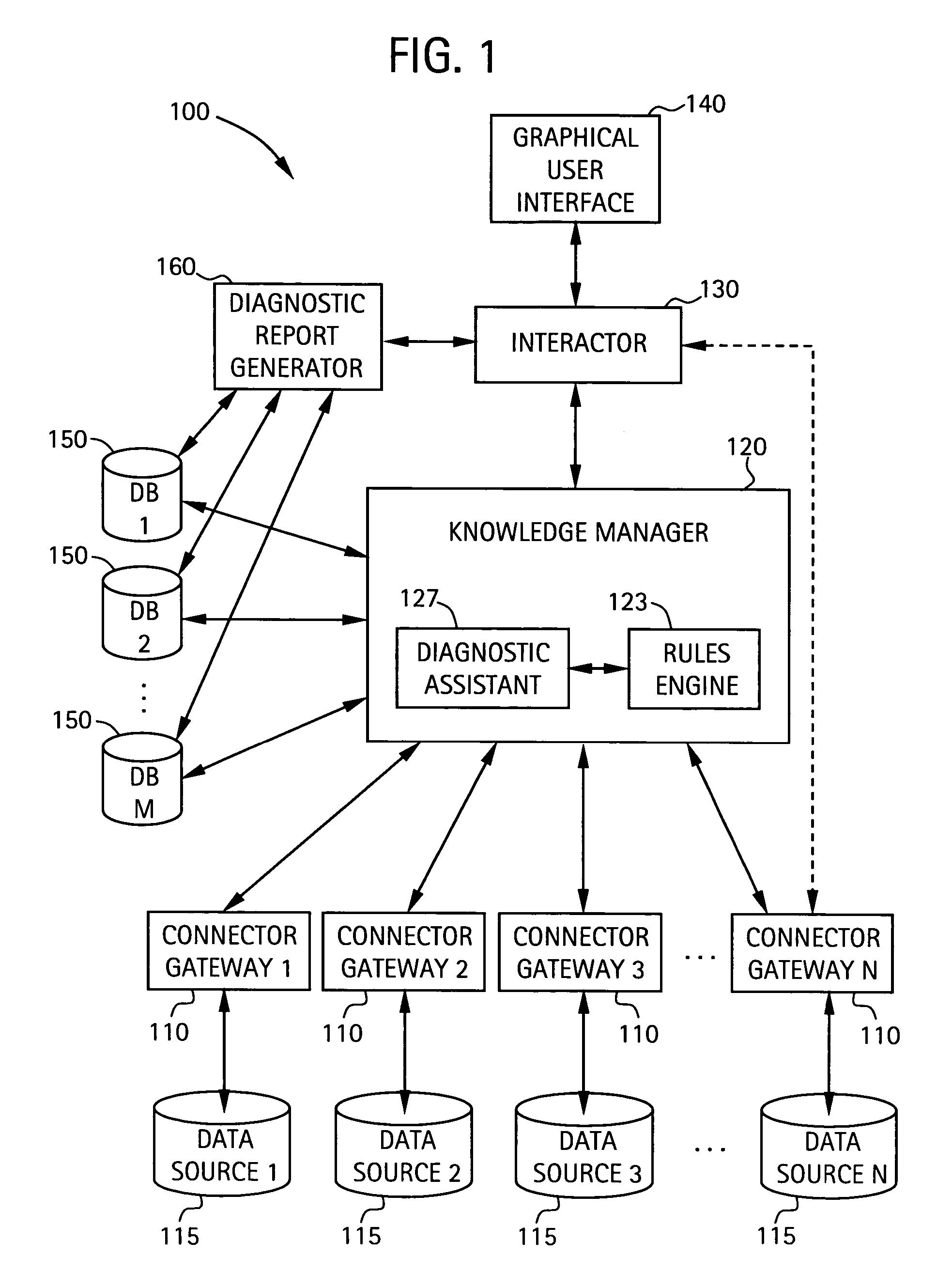
System and method for integrating multiple data sources into service-centric computer networking services diagnostic conclusions
InactiveUS20060156086A1Electronic circuit testingNetwork connectionsGraphicsGraphical user interface
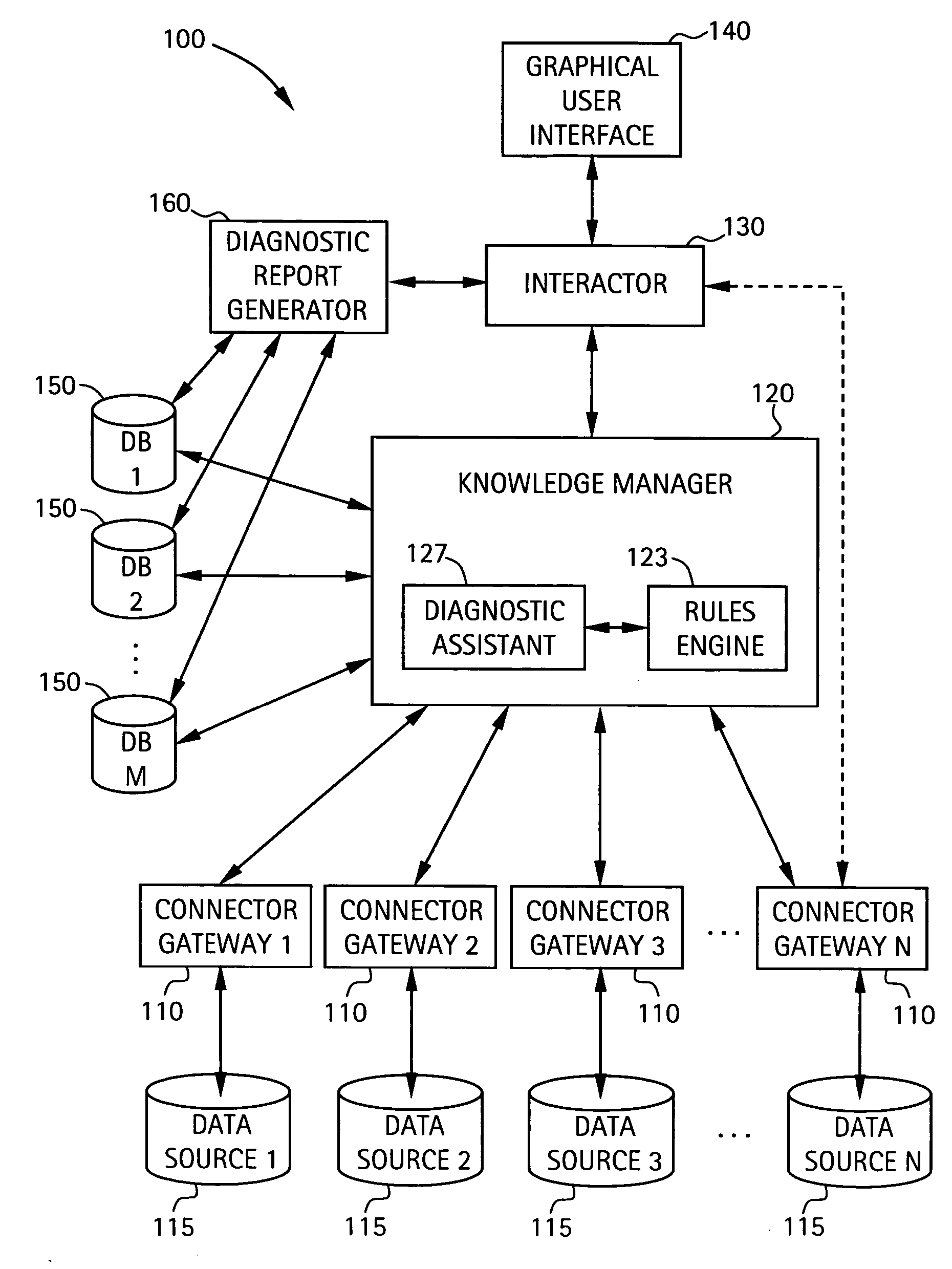
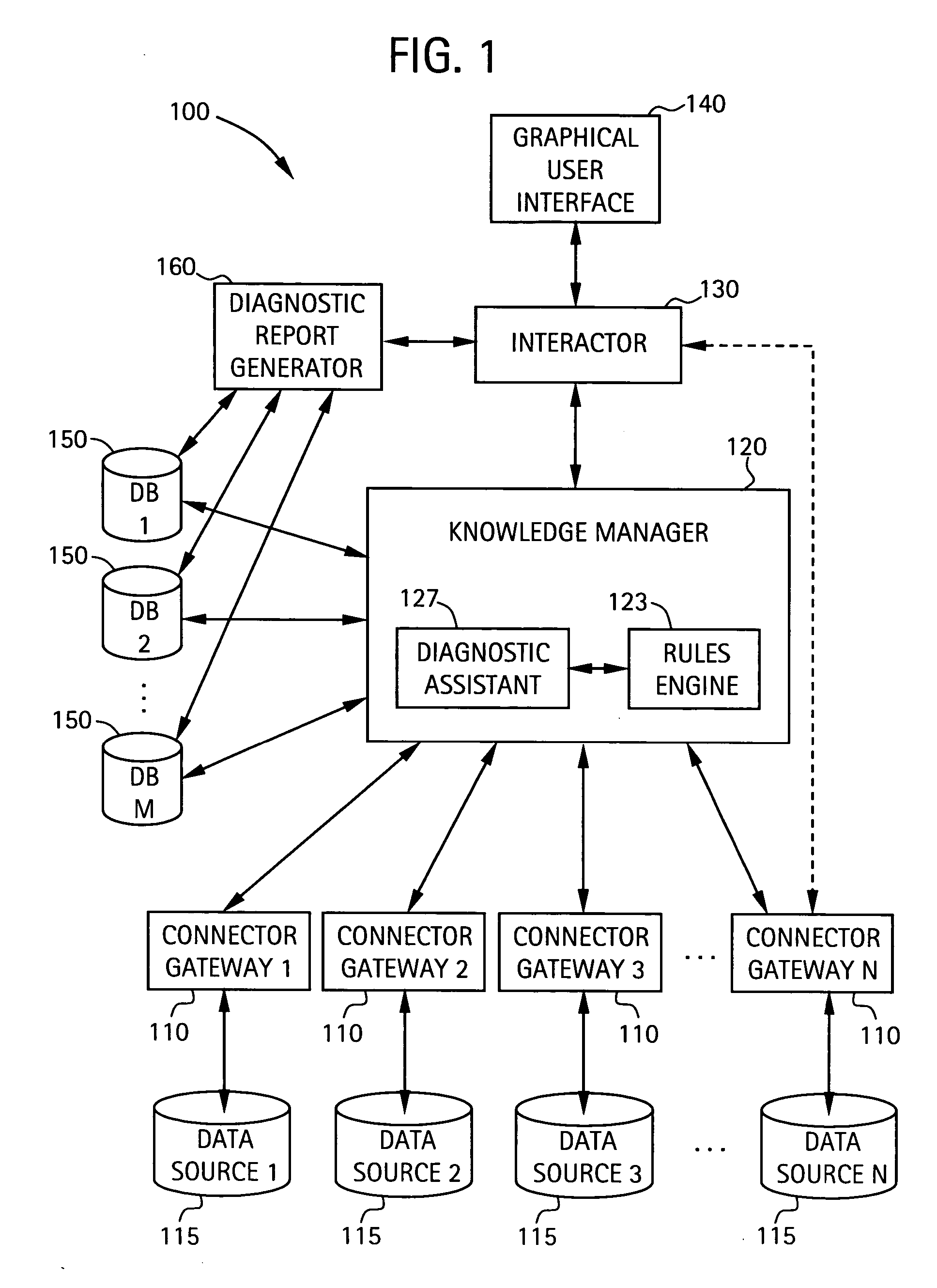
A system for diagnosing impairments in computer networking services a plurality of connector gateways for interfacing to a plurality of data sources to provide access by the system to computer network data, business-related data, and extra-enterprise environmental data in the plurality of data sources. The system includes a knowledge manager in communication with each of the plurality of connector gateways for analyzing data from the plurality of data sources to generate diagnostic conclusions for resolving the impairments in the computer networking services. The system also includes an interactor in communication with the knowledge manager for generating format information for displaying at least the diagnostic conclusions to a user. The system can include a graphical user interface for displaying at least the diagnostic conclusions associated with edges of an edge model of the computer networking services of the user.
Owner:SPIRENT COMM
Methods and systems for providing programmable computerized interactors
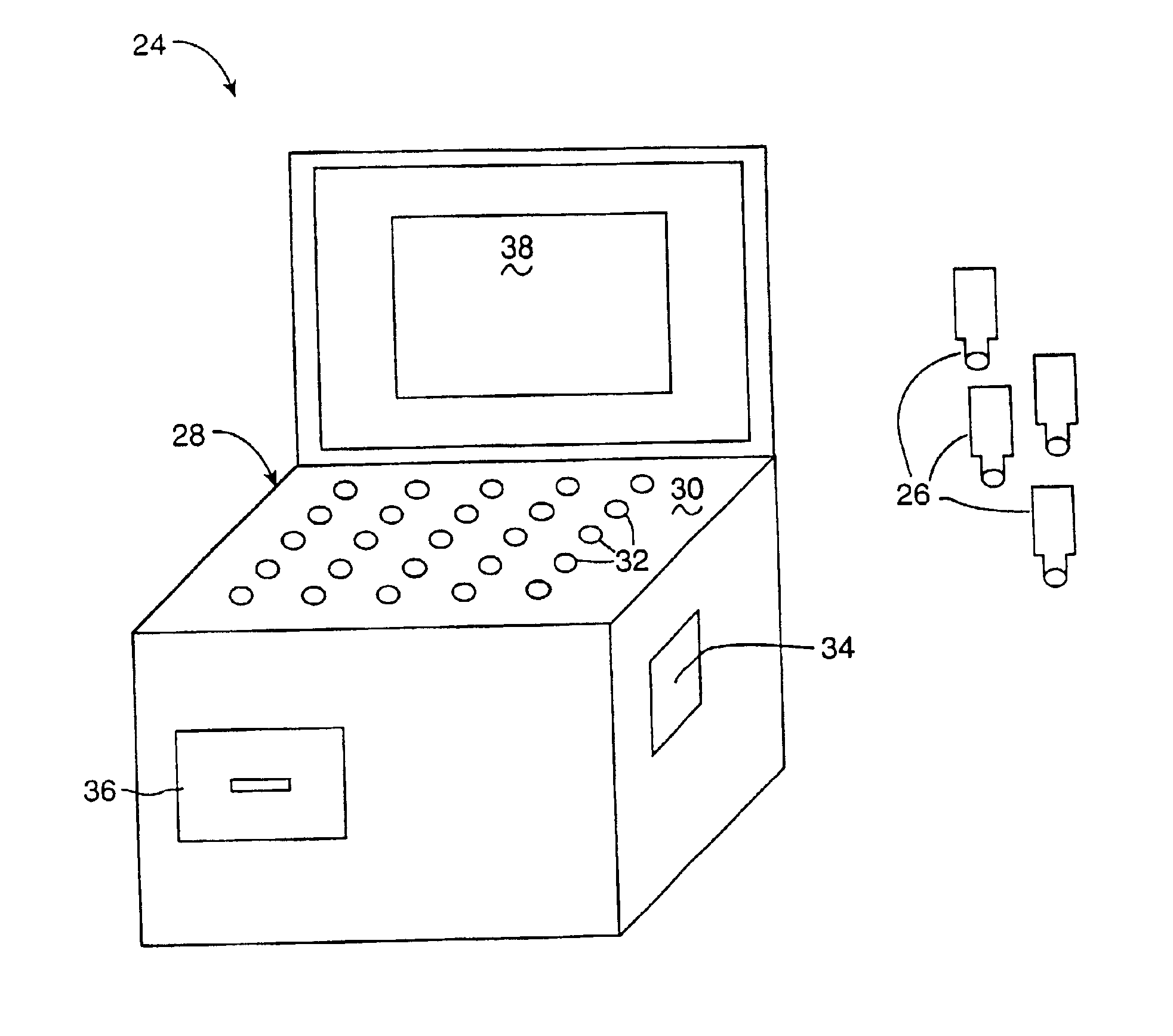
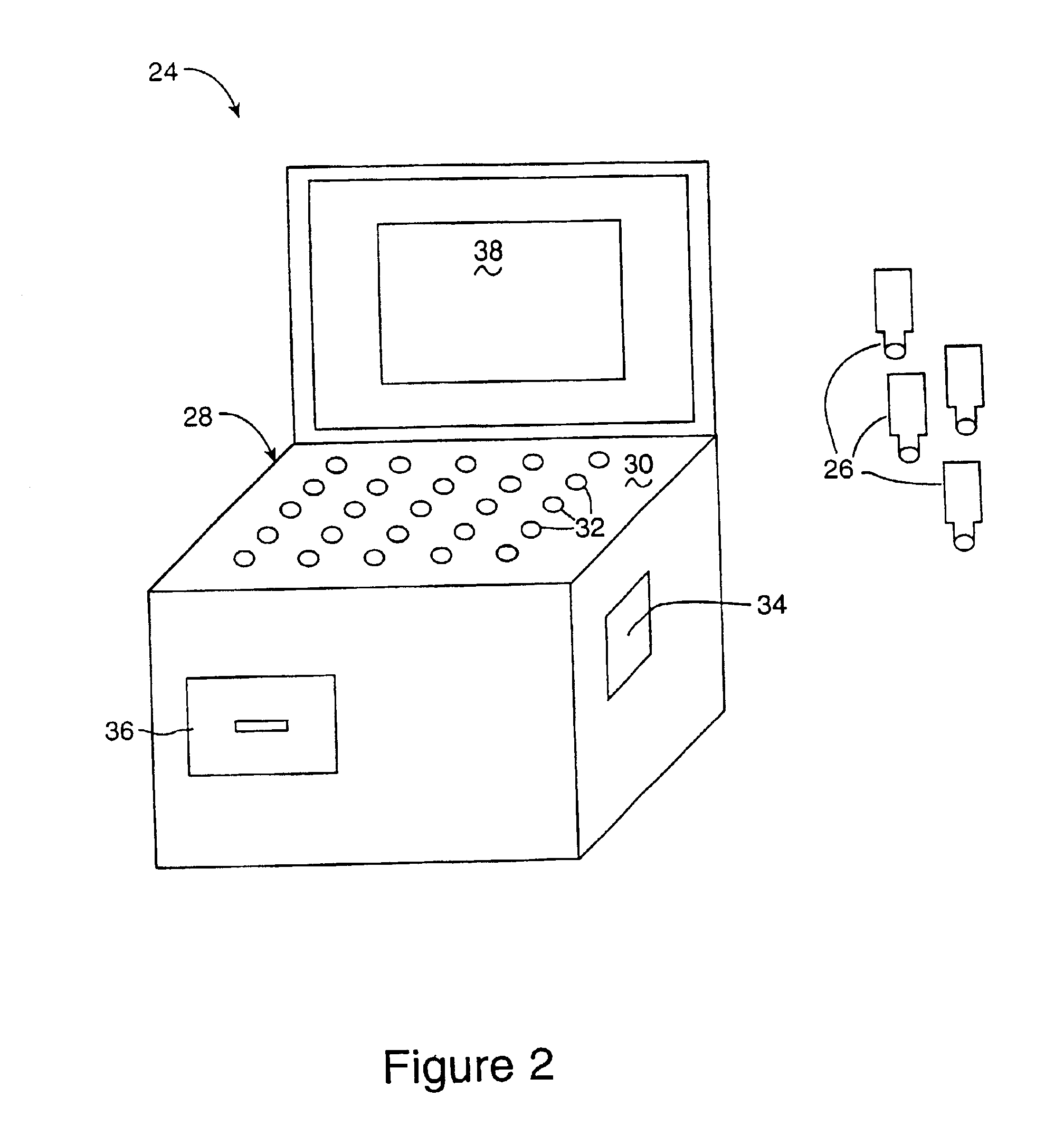
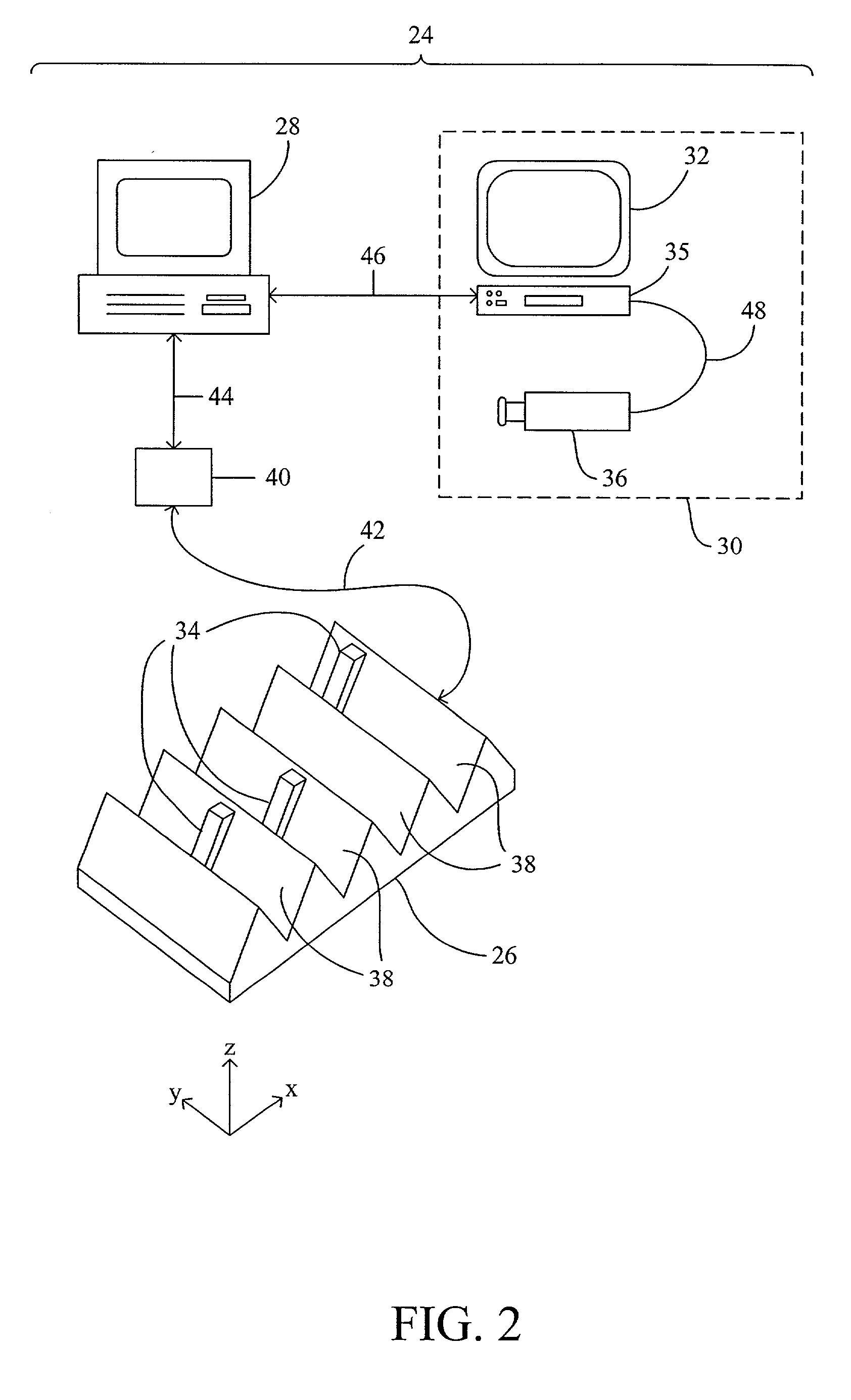
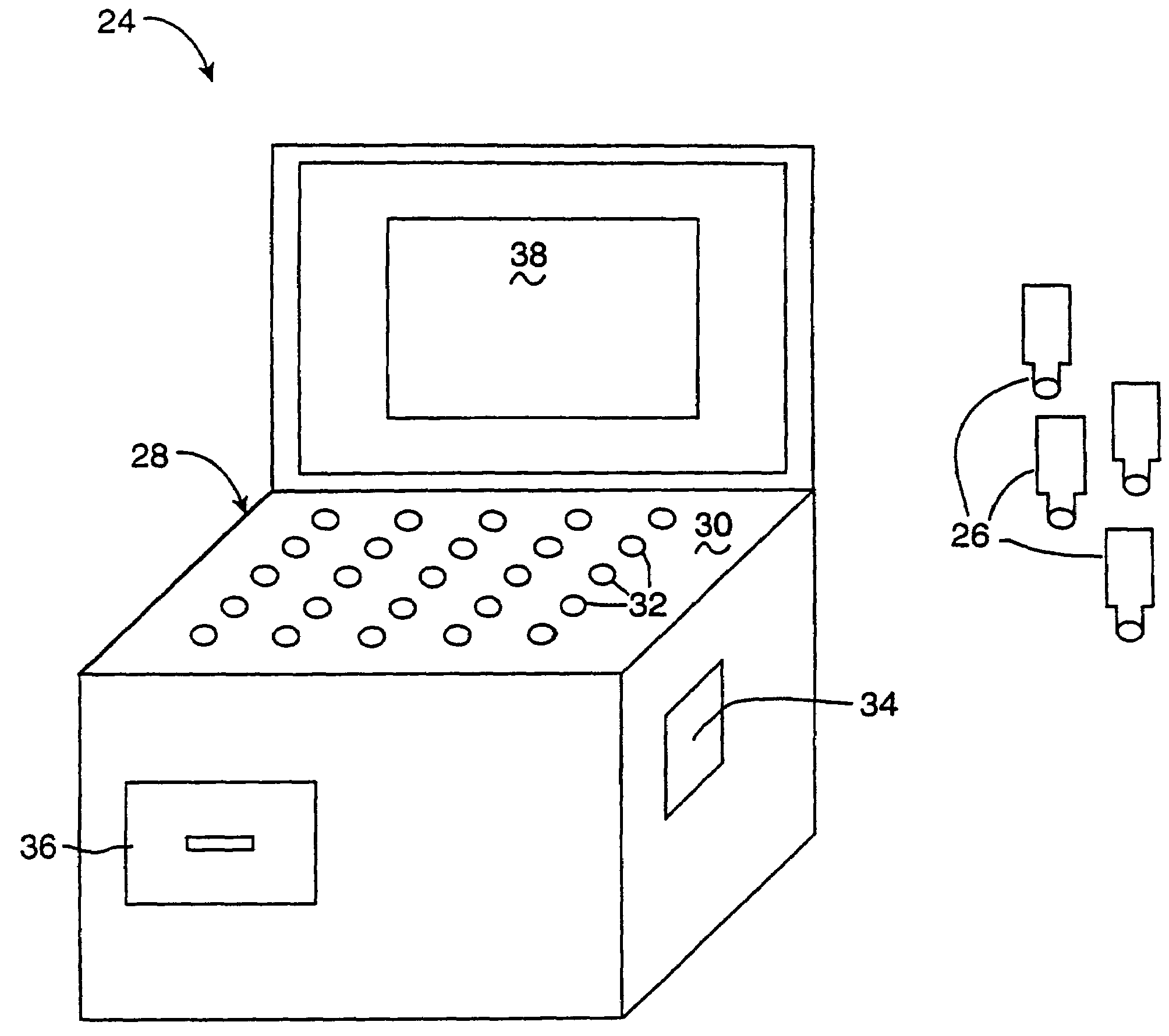
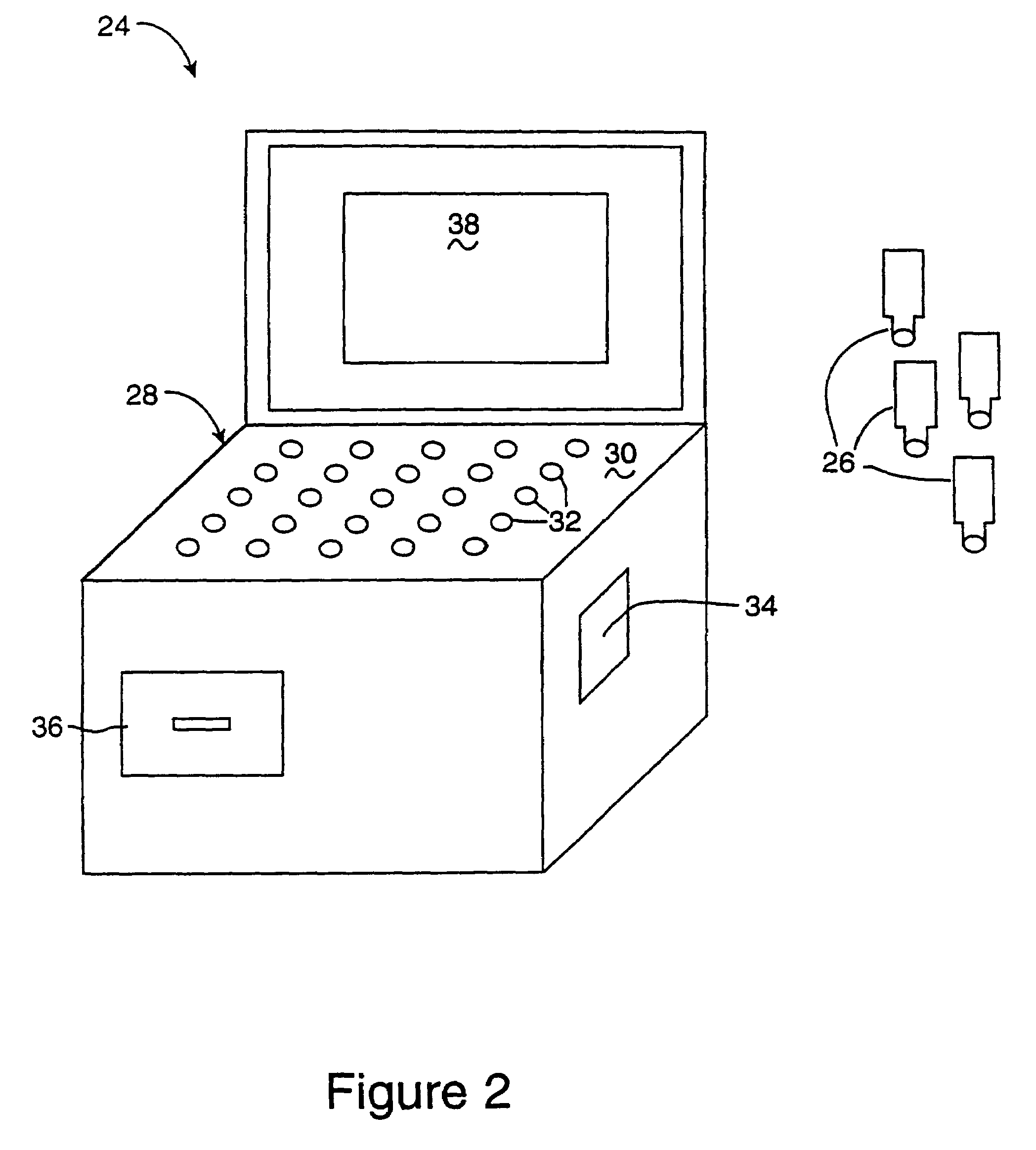
InactiveUS6952196B2Easy to optimizeSimple interfaceElectrophonic musical instrumentsCathode-ray tube indicatorsSemantic contextComputerized system
A computerized interactor system uses physical, three-dimensional objects as metaphors for input of user intent to a computer system. When one or more interactors are engaged with a detection field, the detection field reads an identifier associated with the object and communicates the identifier to a computer system. The computer system determines the meaning of the interactor based upon its identifier and upon a semantic context in which the computer system is operating. One specific embodiment of the present invention is a bead interactor system that is a user playable sound and light show system. When an interactor bead is positioned within the detection space of the bead interactor system, a sound sequence begins and continues to play unaltered until the bead interactor is removed or other bead interactors are positioned within or removed from the detection space. Each bead represents a different sound and the row and column location of the bead within the detection space controls how the sound is modified, e.g., louder or softer, higher pitched or lower pitched, the period of play, etc. In some embodiments, the beads are translucent in order to conduct light from light sources located under each bead receptacle. The available sounds are determined not only by the identity of the beads and their positioning within the detection space and their associated states or orientations, but also by sound data stored in an accompanying computer readable medium. Hence a user can access a variety of sound collections by simply installing a different computer readable medium.
Owner:VULCAN PATENTS
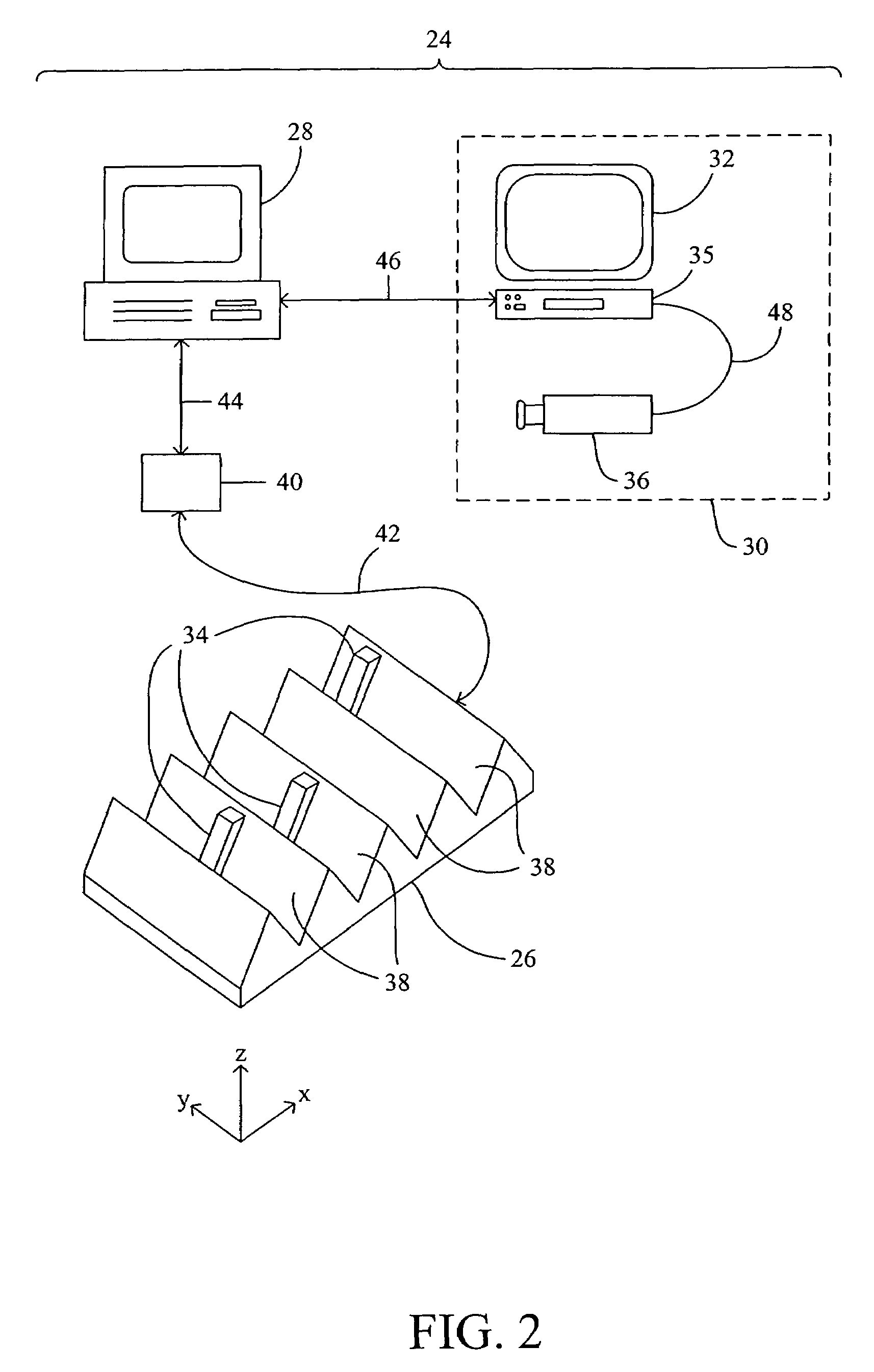
Infrared multipoint interactive electronic whiteboard system and whiteboard projection calibration method
ActiveCN102880360APowerfulEnhanced handlingInput/output for user-computer interactionGraph readingInfraredWireless data
The invention discloses an infrared multipoint interactive electronic whiteboard system and a whiteboard projection calibration method and belongs to the field of electronic whiteboards. The system comprises a computer, a projector, a projection screen, an infrared camera, an infrared pen, a laser positioning device, the computer internally comprises a background wireless data controller and a foreground multipoint interactor, the background wireless data controller comprises a calibration module, a Bluetooth wireless communication module and a wireless data processing module, and the foreground multipoint interactor comprises a gesture recognition module, a drawing function module, a picture display function module, a video display function module, a PPT (power point) file playing operation module, a PDF (portable document format) file playing operation module, a handwritten input recognition module and a small-window display module. By the system and the method, except for basic functions, control functions of opening, full-screen playing, page turning, free writing and the like of PPT and PDF files are further increased.
Owner:NORTHEASTERN UNIV
System and method for integrating multiple data sources into service-centric computer networking services diagnostic conclusions
InactiveUS7434099B2Electronic circuit testingError detection/correctionGraphicsGraphical user interface
A system for diagnosing impairments in computer networking services a plurality of connector gateways for interfacing to a plurality of data sources to provide access by the system to computer network data, business-related data, and extra-enterprise environmental data in the plurality of data sources. The system includes a knowledge manager in communication with each of the plurality of connector gateways for analyzing data from the plurality of data sources to generate diagnostic conclusions for resolving the impairments in the computer networking services. The system also includes an interactor in communication with the knowledge manager for generating format information for displaying at least the diagnostic conclusions to a user. The system can include a graphical user interface for displaying at least the diagnostic conclusions associated with edges of an edge model of the computer networking services of the user.
Owner:SPIRENT COMM
Screening systems utilizing RTP801
InactiveUS20070281326A1Useful in treatmentCompound screeningApoptosis detectionGene targetsUnique gene
RTP801 represents a unique gene target for hypoxia-inducible factor-1 (HIF-1). Down-regulation of the mTOR pathway activity by hypoxia requires de novo mRNA synthesis and correlates with increased expression of RTP801. The present invention relates to screening systems utilizing RTP801 and / or RTP801 interactors and / or RTP801 biological activity, to drug candidates identified by such screening systems, and to the use of such drug candidates in the treatment of various disorders.
Owner:QUARK FARMACUITIKALS INC
Money order matchmaking trading robot based on artificial intelligence
InactiveCN107862608AIncrease authenticityIncrease motivation to sign onlineFinanceMachine learningRelevant informationRapid identification
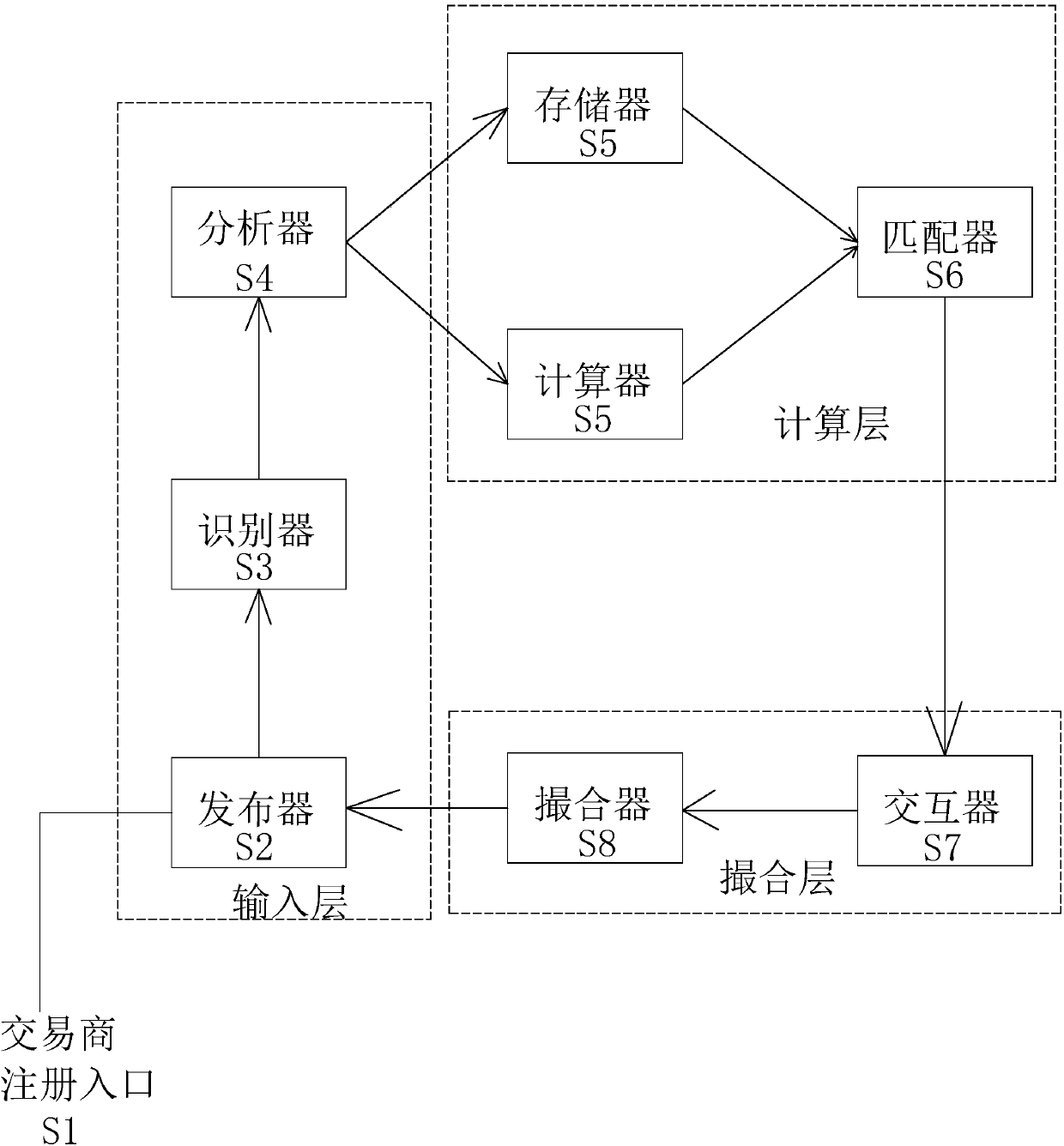
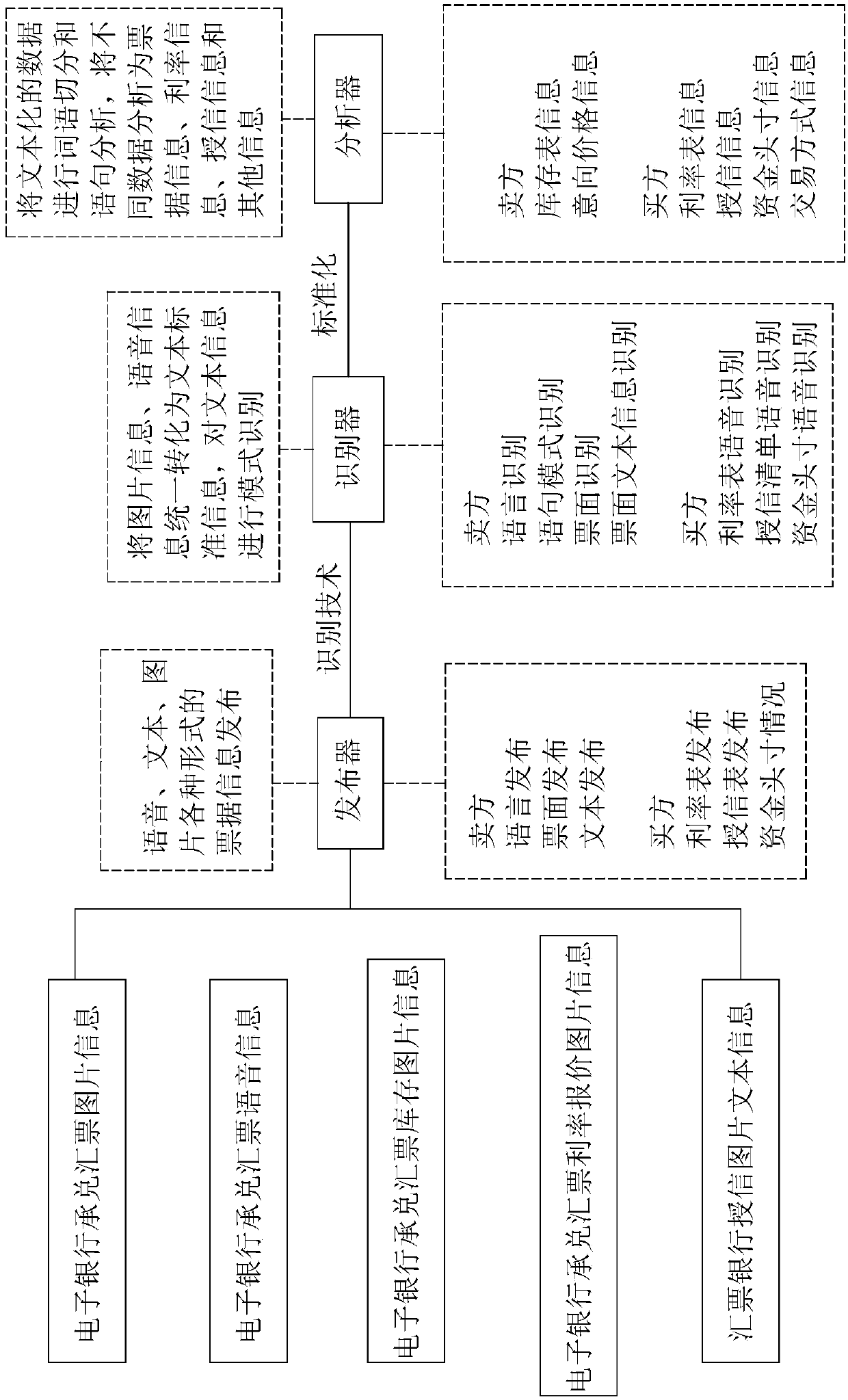
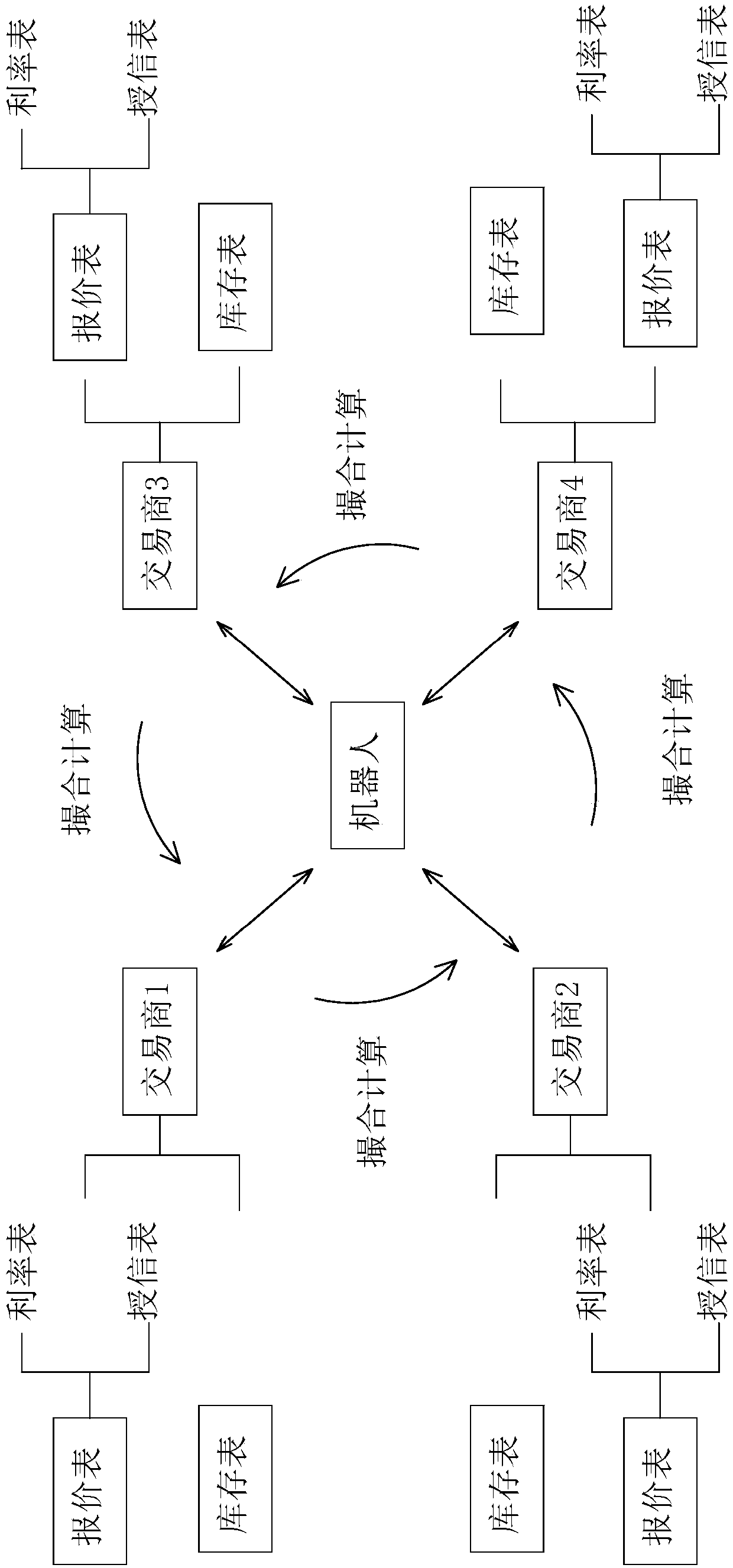
The invention provides a money order matchmaking trading robot based on artificial intelligence. The money order matchmaking trading robot based on the artificial intelligence comprises a publisher, used for receiving money order relevant information, and sending the information to an identifier; the identifier, used for identifying the money order relevant information and organizing into text information and sending to an analyzer; the analyzer, used for processing and classifying the text information, pushing the classified buying information to a storage for storing, sending the selling information and a calculation instruction to a calculator; the calculator, according to the stored data, used for calculating the received selling information; a matcher, used for summarizing calculatingresults of the calculator, screening an optimized quotation, and pushing to both the buyer and the seller; an interactor, used for re-communicating with a bill robot or the buyer of the correspondingprice by the seller, and pushing an optimal matching party result to a matchmaking device; and the matchmaking device, used for generating a standard trading contract. A traditional mode can be replaced by the trading robot, so the information rapid identification and calculation, storage, matching, matchmaking, contracting and trading of the buyer and the seller are realized.
Owner:田标
Computerized interactor systems and methods for providing same
InactiveUS6940486B2Simple interfaceReduce fatigueInput/output for user-computer interactionInterconnection arrangementsSemantic contextComputerized system
Owner:HANGER SOLUTIONS LLC
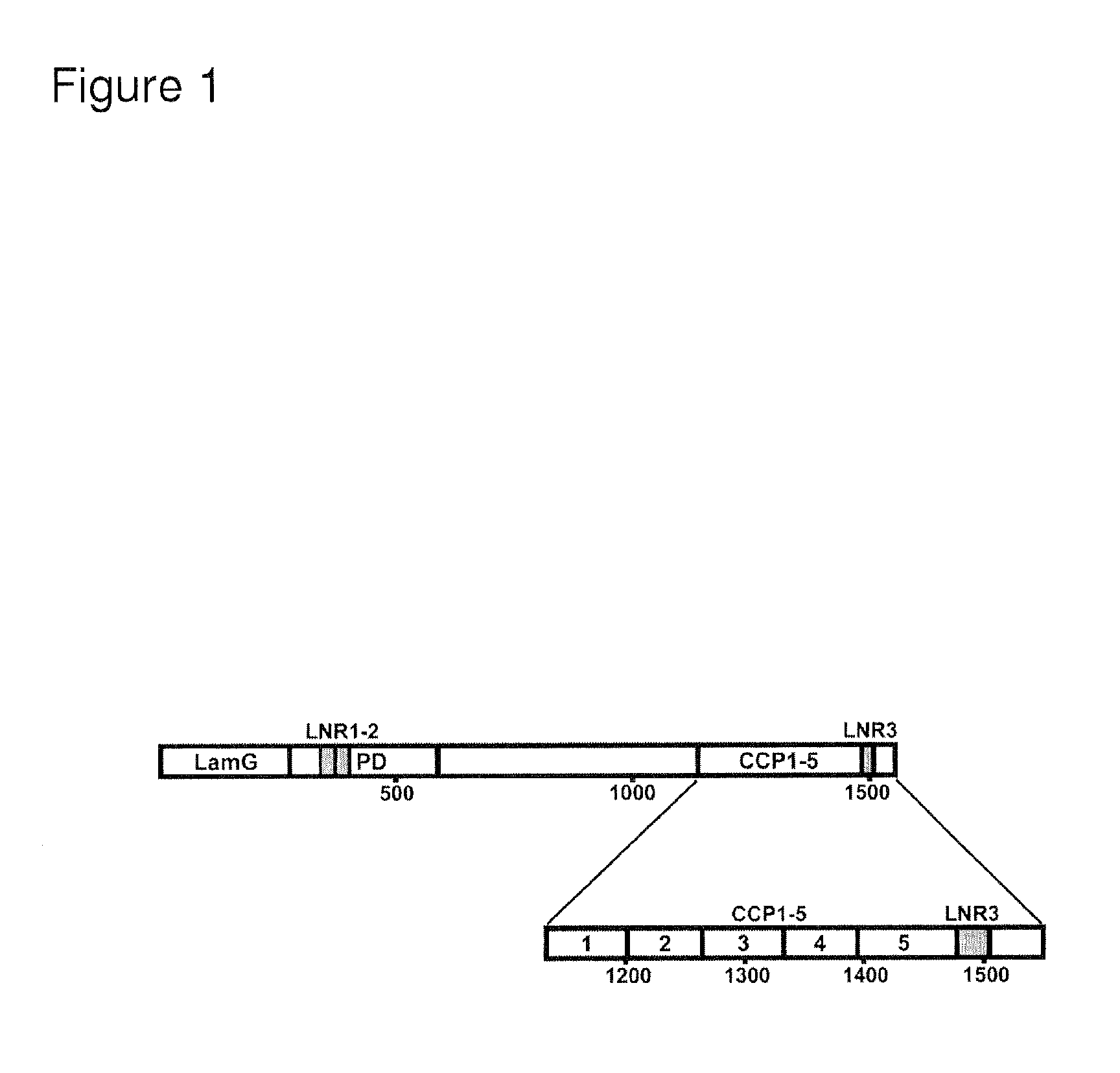
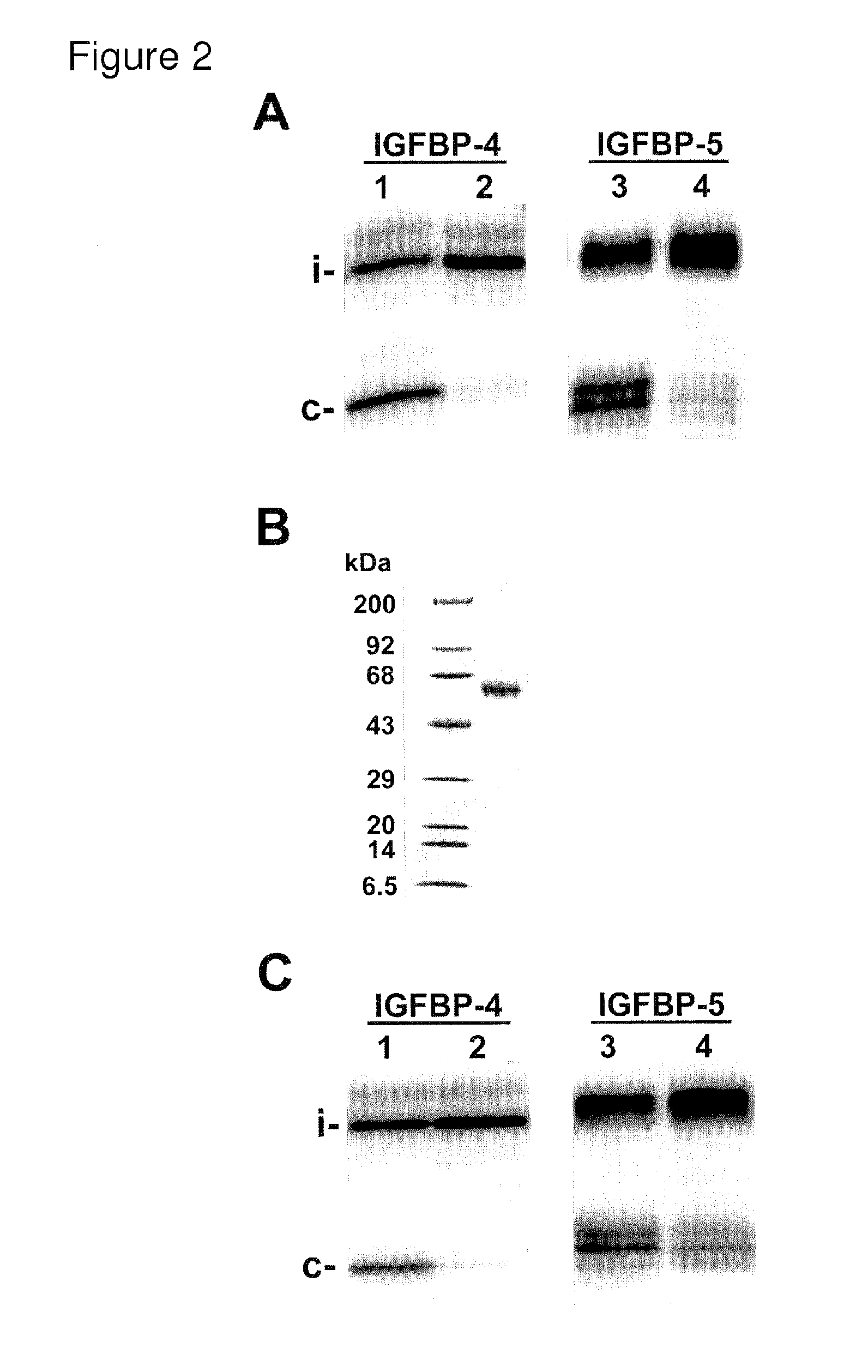
Selective exosite inhibition of papp-a activity against igfbp-4
ActiveUS20100310646A1Improve transcription efficiencyInhibit bindingAntibacterial agentsFungiDifferential inhibitionProteinase activity
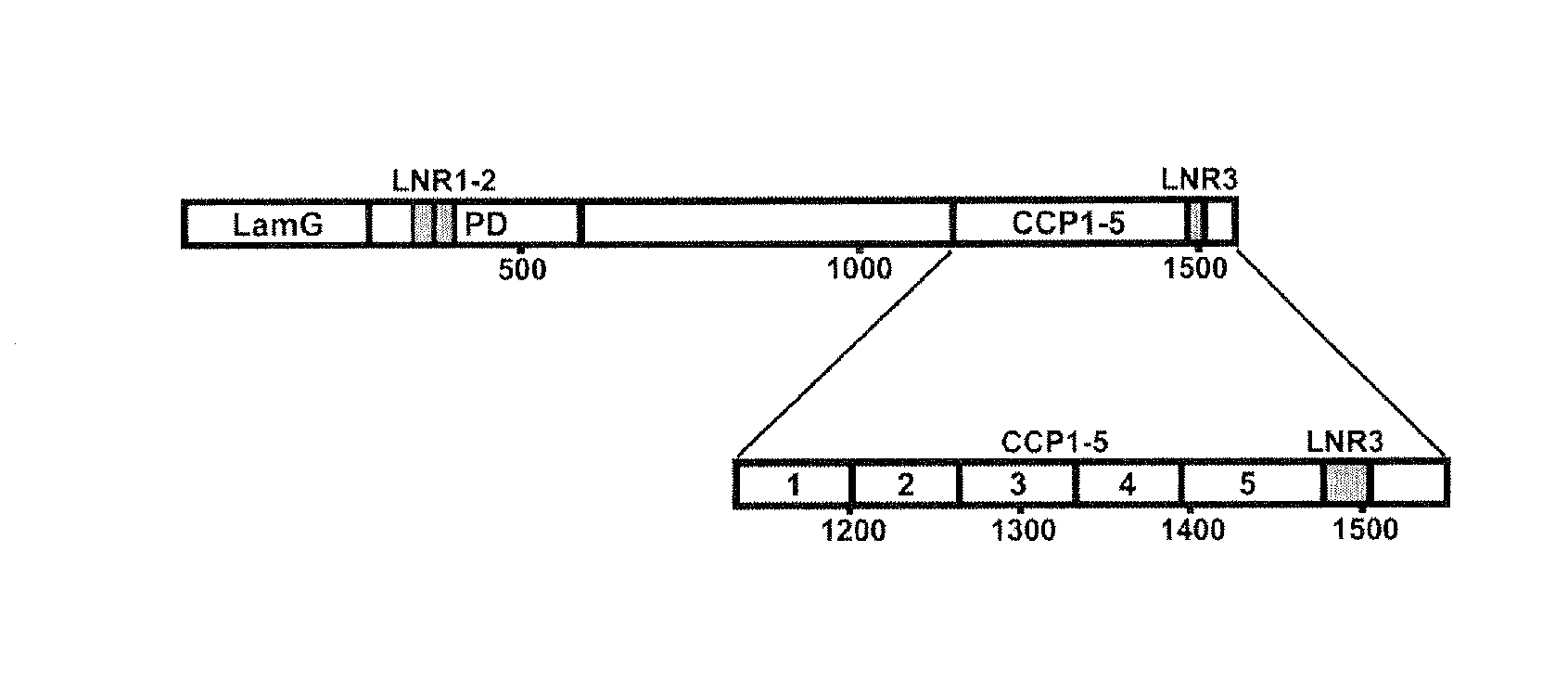
The present invention relates in one embodiment to PAPP-A exosite(s) interactors such as antibodies which bind to a region comprising LNR3 of PAPP-A and efficiently inhibit proteolysis of IGFBP-4, but not -5. The region comprising LNR3 represents a substrate binding exosite, which can be targeted for selective proteolytic inhibition. Accordingly, the present invention relates in one embodiment to differential inhibition of natural protease substrates by exosite targeting.
Owner:AARHUS UNIV
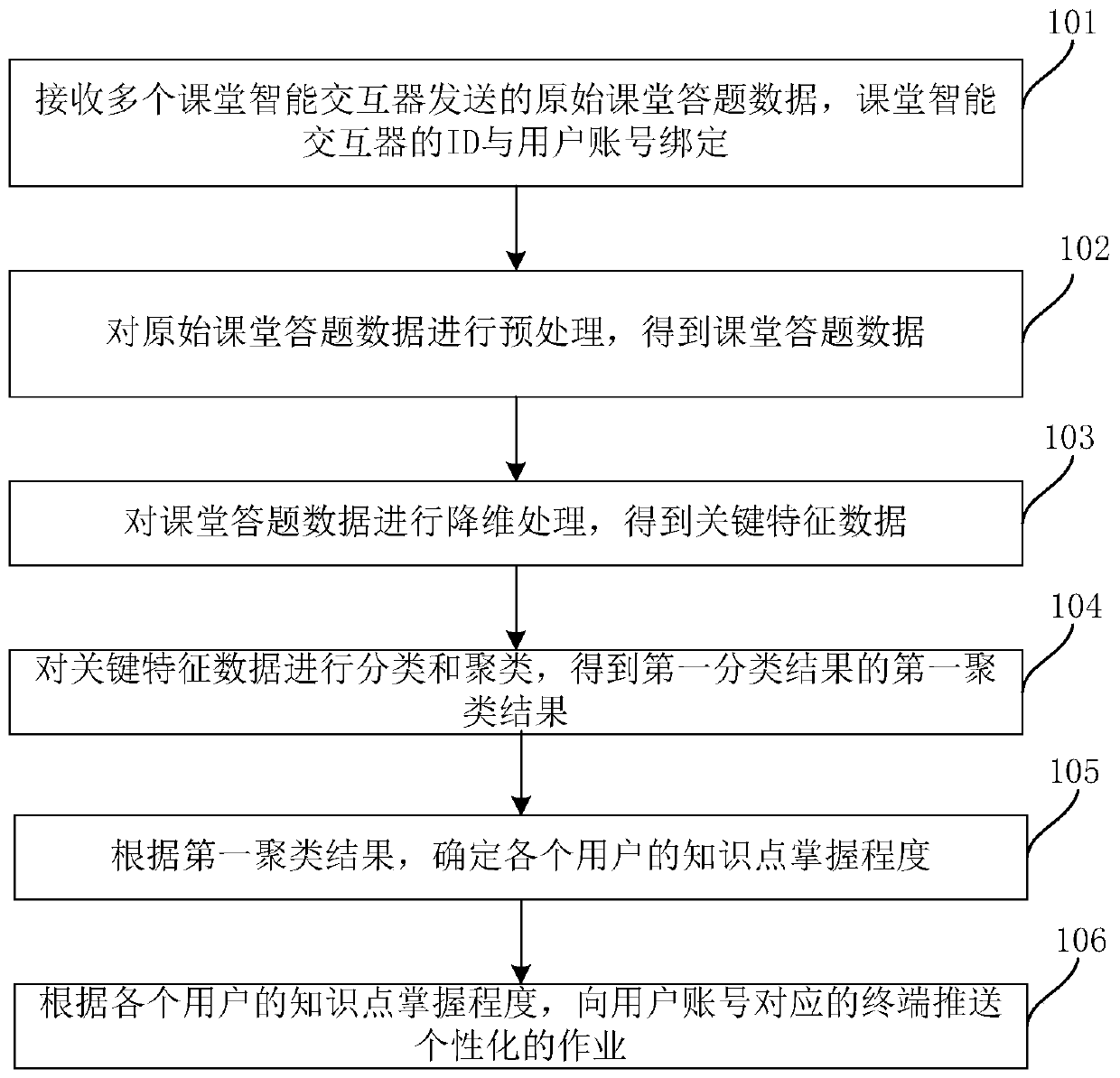
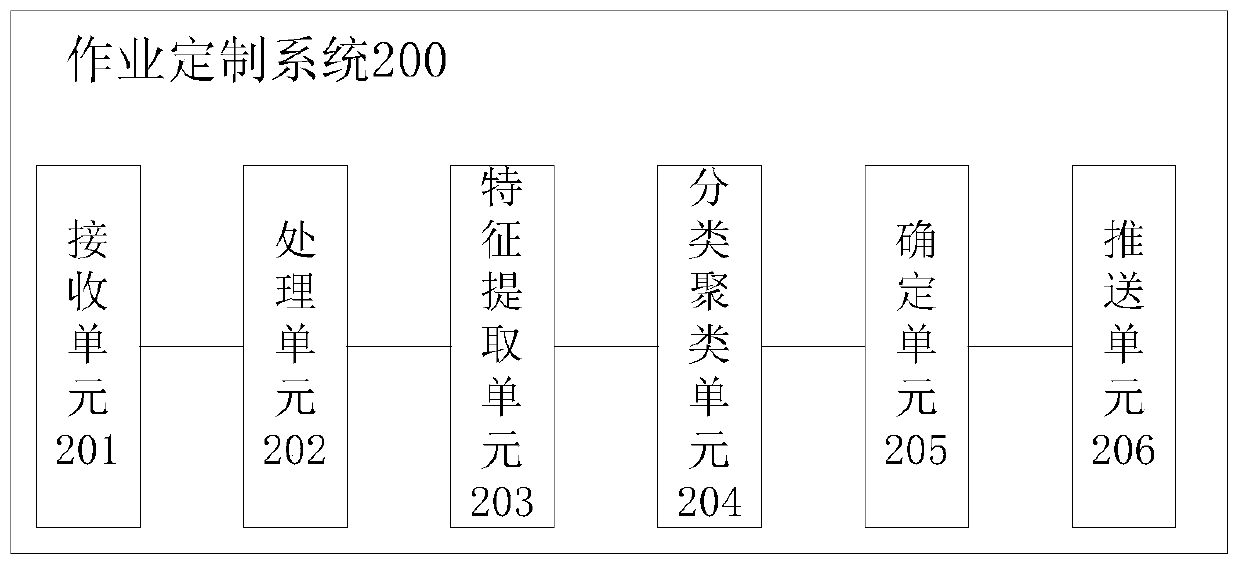
Homework customization method and system
PendingCN110309201ARealize personalized customizationData processing applicationsRelational databasesPersonalizationInteractor
The invention provides a homework customization method, which comprises the steps of receiving the original classroom answer data sent by a plurality of classroom intelligent interactors, and bindingthe IDs of the classroom intelligent interactors with a user account; preprocessing the original classroom answer data to obtain the classroom answer data; carrying out dimension reduction processingon the classroom answer data to obtain the key feature data of the classroom answer data; classifying and clustering the key feature data to obtain a first clustering result of the first classification result; determining the knowledge point mastering degree of each user according to the first clustering result; and pushing the personalized operation to a terminal corresponding to the user accountaccording to the knowledge point mastery degree of each user, so that the mastering conditions of different students on different knowledge points can be determined according to the original classroom answering data of the students in the classroom, the homework with different difficulty coefficients is pushed to the different students after the class, and the personalized customization of the students is realized.
Owner:广州云蝶科技有限公司
System and method for panning and selecting on large displays using mobile devices without client software
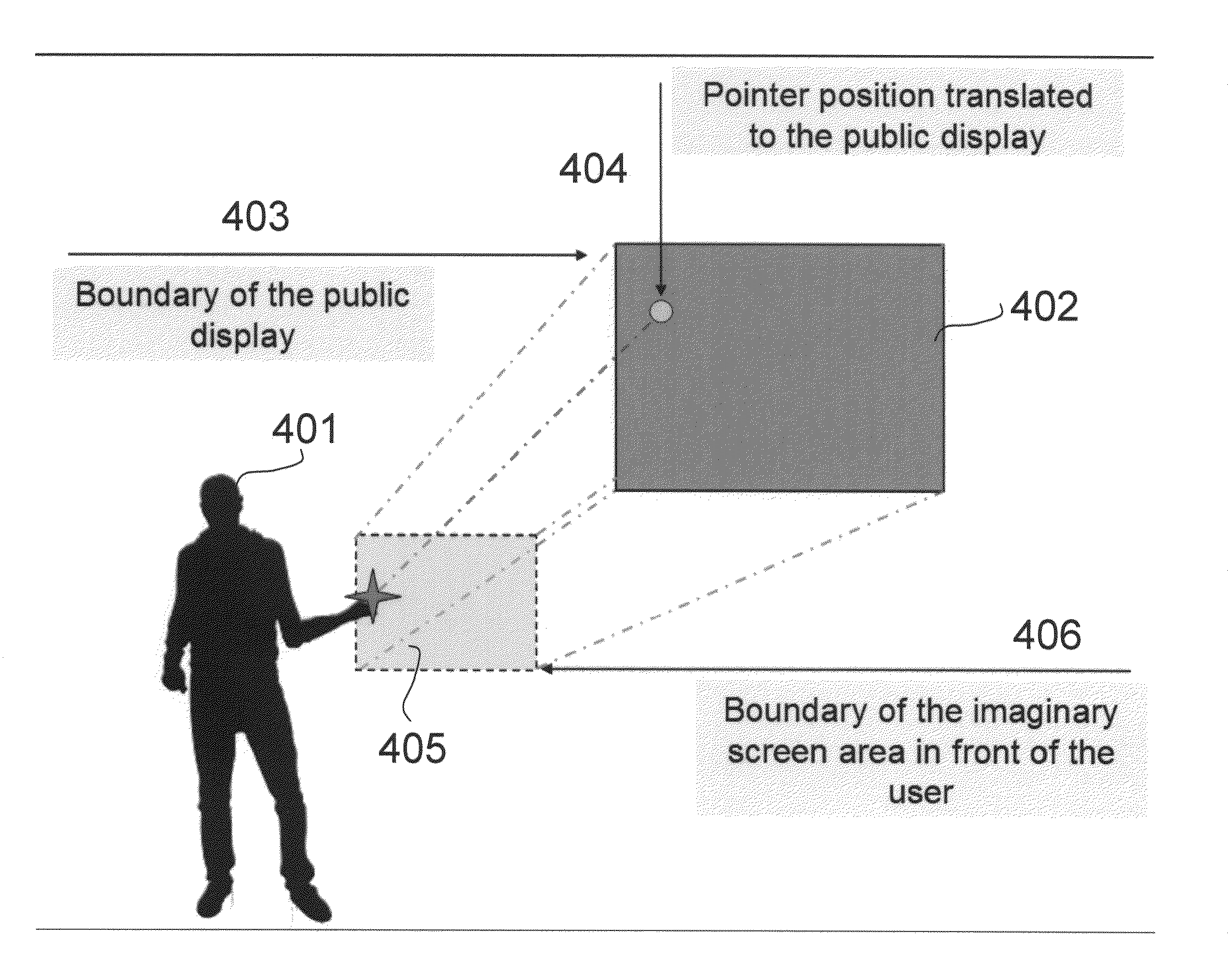
System and method that utilizes light sources, for example projectors embedded in mobile camera phones, to allow users to interact with documents on large displays without requiring users to install any third-party software. With this approach, multiple users can select interactors on the large display by displaying different images via their projector.
Owner:FUJIFILM BUSINESS INNOVATION CORP
Methods and Apparatus for Enabling a Dynamic Network of Interactors According to Personal Trust Levels Between Interactors
A system for causing routing of a communication event includes a sending platform for initiating and sending the communication event, a communications network for carrying the communication event, a receiving platform for receiving the communication event for final routing, and a router resident at least in the receiving platform for preparing and executing or forwarding for execution a routing instruction for handling the incoming communication event or notification thereof, the routing instruction thus executed, overriding a default routing instruction, the overriding routing instruction initiated upon discovery by the router of some level of trust metric between the sender and intended recipient of the event.
Owner:FORTE INTERNET SOFTWARE
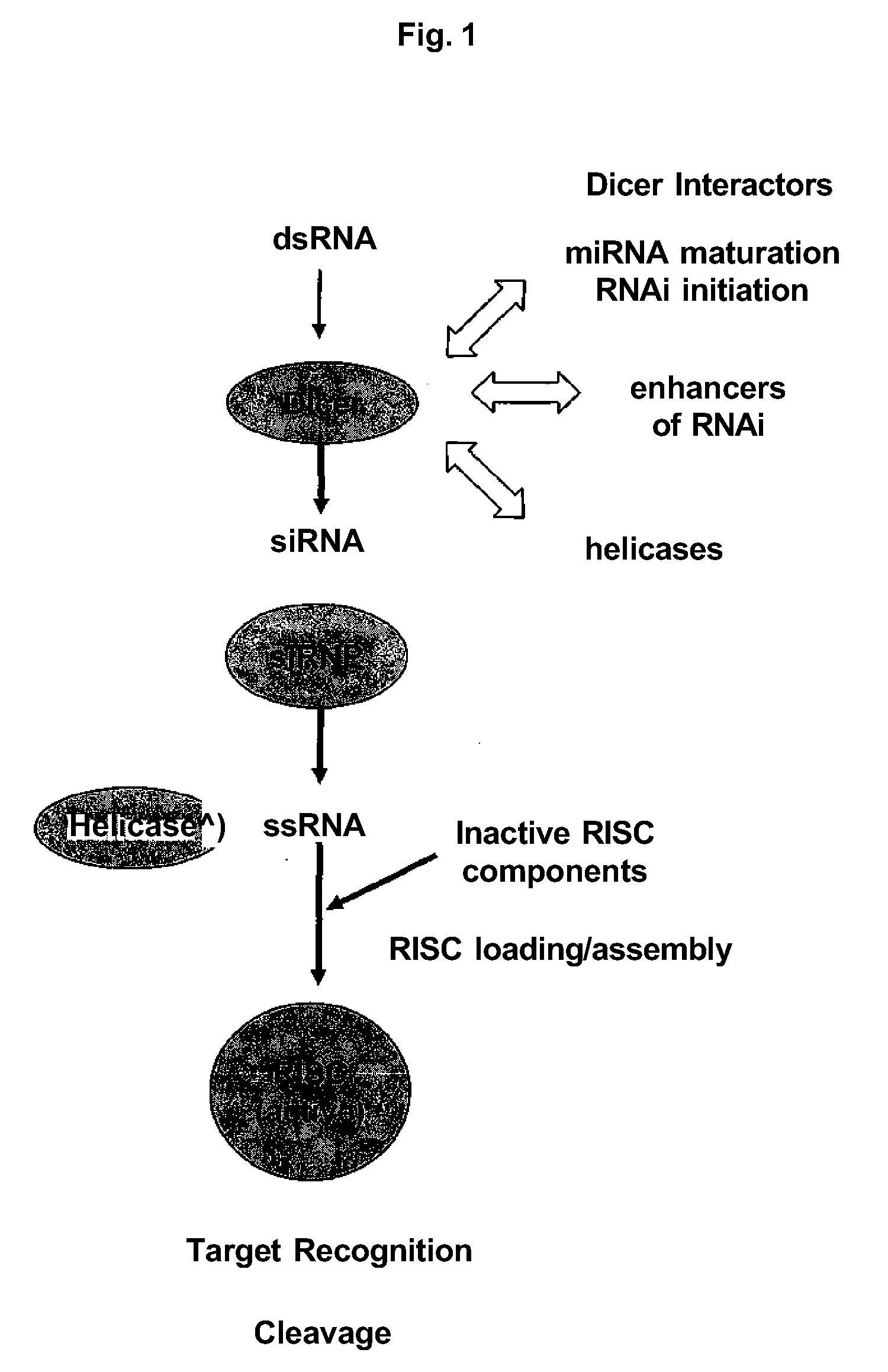
Dicer interacting proteins and uses therefor
InactiveUS20070031417A2Microbiological testing/measurementGenetic material ingredientsDicerInteractor
Abstract of the DisclosureABSTRACT Dicer (e.g., DCR-1) interactors are disclosed as are methods to positively or negatively modulate Dicer activity. Uses of Dicer interactors as drug targets are featured. Also featured are uses of Dicer interactors and modulators of same to modulate various Dicer functions in vitro, in cell cultures, and in vivo.
Owner:UNIV OF MASSACHUSETTS
Cyclotron actuator using a shape memory alloy
An actuator assembly for use within the vacuum field of a cyclotron, one embodiment of which comprises an interactor which is moveable between a first position and a second position, at least one support structure for supporting the interactor in the first and second positions, a shape memory alloy (SMA) element connected to the interactor and / or support structure and being adapted to exert a force on the interactor and / or support structure so as to urge the interactor from the first position to the second position, an electromagnetic activator operatively associated with the SMA element for causing the element to exert the force when the electromagnetic activator is selectably activated, and a return mechanism operatively connected to the interactor, the support structure and / or the SMA element so as to urge the interactor from the second position to the first position when the electromagnetic activator is deactivated.
Owner:GENERAL ELECTRIC CO
Use of the ENDO-180 gene and polypeptide for diagnosis and treatment
InactiveUS20090202566A1Slow paceReduce and limit formationOrganic active ingredientsMetabolism disorderDiseaseFibrosis
This application is directed to a process of identifying a compound capable of modulating activity of a human ENDO180 receptor that comprises the steps of measuring the binding of the ENDO180 receptor to an interactor with which the ENDO180 receptor interacts specifically in vivo, in the absence or presence of a compound, and determining whether the binding of the ENDO180 receptor to the interactor is affected by the compound. This application is also directed to use of a compound identified by that process in the preparation of a medicament for therapy of disease, in particular fibrosis. This application also relates to the use of ENDO180 modulators in treatment of disease.
Owner:QUARK FARMACUITIKALS INC
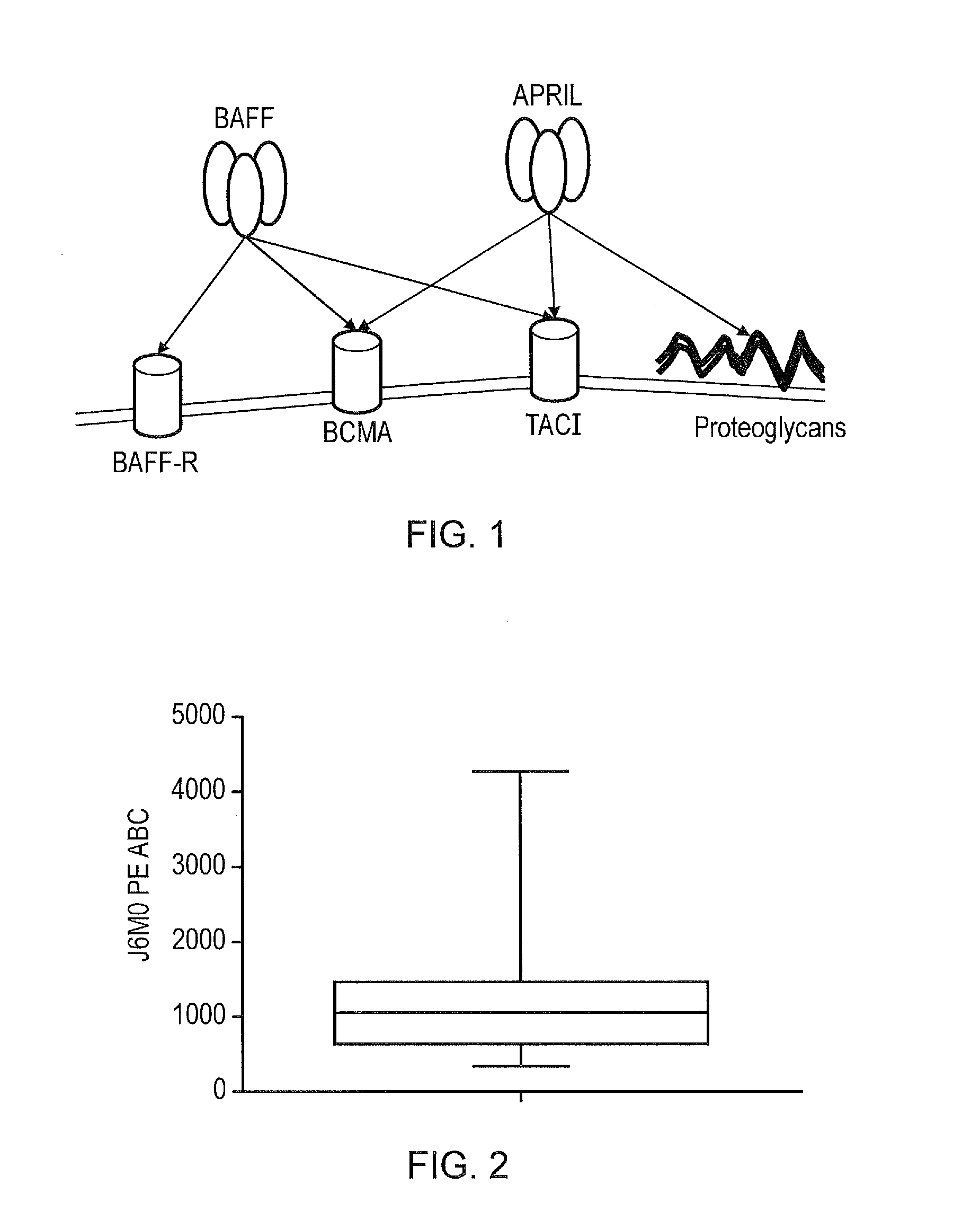
April variants
ActiveUS20160362467A1High BCMA : TACI binding ratioStrong specificityPolypeptide with localisation/targeting motifImmunoglobulin superfamilyBinding ratioWild type
The present invention provides a variant proliferation-inducing ligand (APRIL), which has a higher binding affinity to BCMA than wild-type APRIL; and / or altered binding kinetics compared with wild-type APRIL, and / or a higher BCMA:TACI (transmembrane activator and calcium modulator and cyclophilin ligand interactor) binding ratio than wild-type APRIL and which comprises mutations at one or more of the following positions: A125, V174, T175, M200, P201, S202, H203, D205 and R206.
Owner:UCL BUSINESS PLC
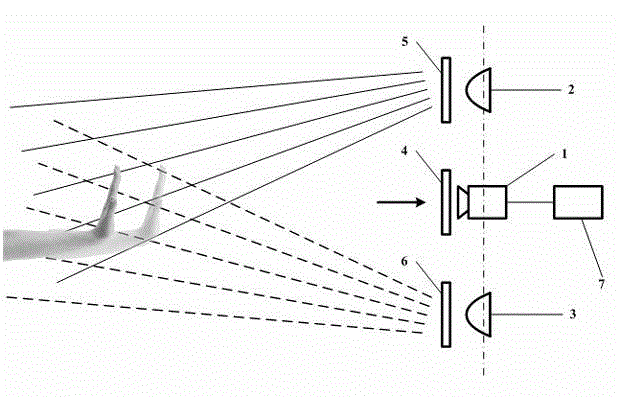
Quick three-dimensional display interacting device and method based on dual-structure light infrared identification
ActiveCN102750046AStrong expandabilityLower control costsInput/output for user-computer interactionGraph readingHardware structureComposite pattern
The invention discloses a quick three-dimensional display interacting device and method based on dual-structure light infrared identification. The quick three-dimensional display interacting device comprises an image capturing system additionally provided with an infrared filter, dual infrared light sources and dual pattern templates, wherein the image capturing system is arranged between the dual infrared light sources and is aligned to an overlapped region radiated by the dual light sources. The method comprises the following main steps of: projecting a composite pattern to a hand part of an interactor through respective pattern samples by the dual light sources; shooting and acquiring the composite pattern by the image capturing system; and processing an image acquired by the image capturing system in real time, distinguishing a foreground from a background to obtain the approximate outline of the hand part, matching the obtained outline with the shape prestored in a data bank, judging the outline which has highest matching degree and the similarity exceeding the threshold as gesture made by the current interactor and making corresponding response to the gesture by a computer. The quick three-dimensional display interacting device and method disclosed by the invention can be applied to three-dimensional display equipment so as to achieve the bare-handed three-dimensional interaction function at a quicker algorithm and a more compact hardware structure.
Owner:SHENZHEN KECHUANG DIGITAL DISPLAY TECH
Real-time interaction intelligent scene construction and system
InactiveCN108961881AImprove education efficiencyBreak the exam-oriented education modelData processing applicationsElectrical appliancesEngineeringInteractor
The invention discloses a real-time interaction intelligent scene construction and system. The real-time interaction intelligent scene system comprises designing a virtual image, recognizing the terminal interaction information of the virtual image, and determining the terminal interaction information authority; arranging on the terminal to virtualize the needed scene and object by a camera, interacting the entity information in the scene and uploading to a VR end, enriching and reconstructing the scene and object in the VR by the terminal, uploading the reconstructed scene and objects to construct the scene and objects, and displaying the VR scene through restoration and reproduction. The real-time interaction intelligent scene construction system is an interactive education system, and comprises interactive terminals and a server, the interactive terminals are divided into a teacher end and a student end; the server comprises a control module and an information identification module.According to the invention, the teacher is used as an education core, and the VR course design is used as an auxiliary, the education efficiency of the teacher end and the student end is improved, and the multi-person can interact on the local area network or the wide area network at the same time.
Owner:林墨嘉
Use of the endo-180 gene and polypeptide for diagnosis and treatment
InactiveUS20070072244A1Slow paceReduce and limit formationPeptide/protein ingredientsMetabolism disorderFibrosisInteractor
This application is directed to a process of identifying a compound capable of modulating activity of a human ENDO180 receptor that comprises the steps of measuring the binding of the ENDO180 receptor to an interactor with which the ENDO180 receptor interacts specifically in vivo, in the absence or presence of a compound, and determining whether the binding of the ENDO180 receptor to the interactor is affected by the compound. This application is also directed to use of a compound identified by that process in the preparation of a medicament for therapy of disease, in particular fibrosis. This application also relates to the use of ENDO180 modulators in treatment of disease.
Owner:QUARK FARMACUITIKALS INC
Configurable integration method and system for information system
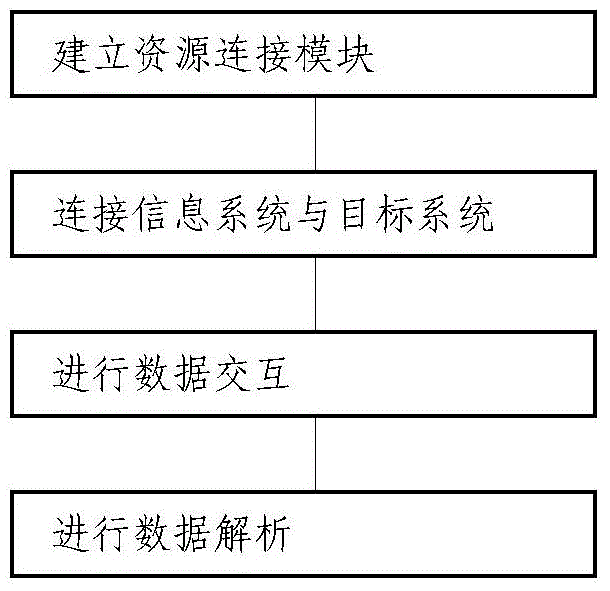
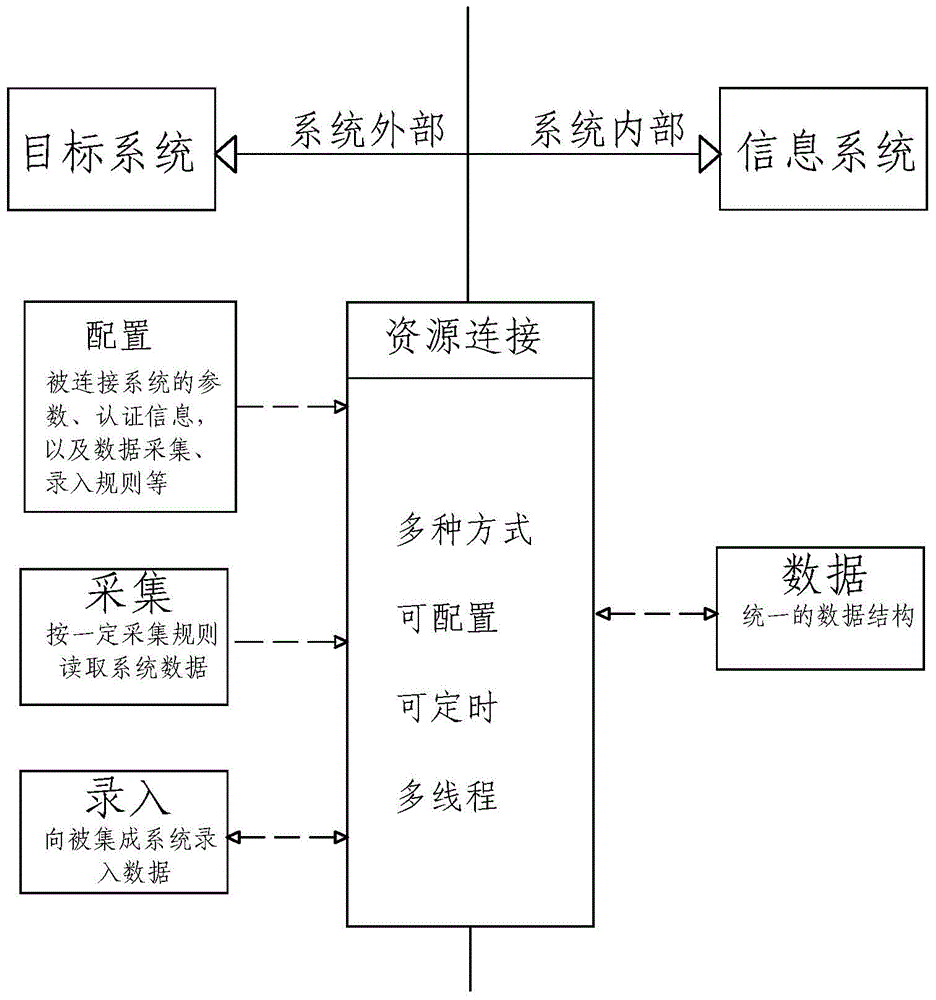
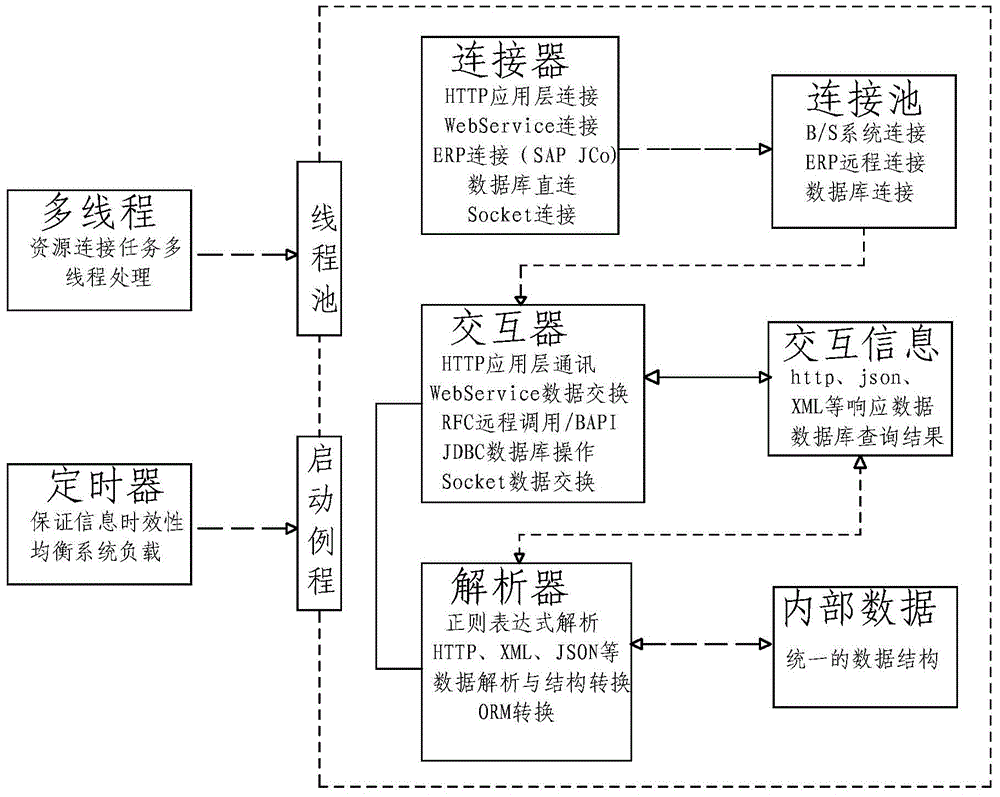
InactiveCN104991817AAchieve integrationOvercome deficienciesMultiprogramming arrangementsDigital data authenticationConnection poolGoal system
A configurable integration method and system for an information system are disclosed. The method comprises the following processes of: establishing a resource connection module and carrying out data interaction between the information system and a target system; connecting the information system with the target system, establishing links between the information system and the target system according to data flow demands, and forming a connection pool by all the links; performing data interaction, and performing data exchange between the information system and the target system; and performing data analysis, and converting data of the information system and the target system to be in a unified data format. The system comprises the resource connection module for data interaction between the information system and the target system; and the resource connection module comprises a connector, an interactor and an analyzer. According to the configurable integration method and system, various integration ways are comprehensively adopted, communication with development, implementation or operation and maintenance personnel of an integrated system is not required, and according to the specific situation and integration demands of the integrated system, the deficiencies of an existing integration scheme are effectively avoided and the non-coupled inter-system integration is realized.
Owner:STATE GRID SHANDONG ELECTRIC POWER
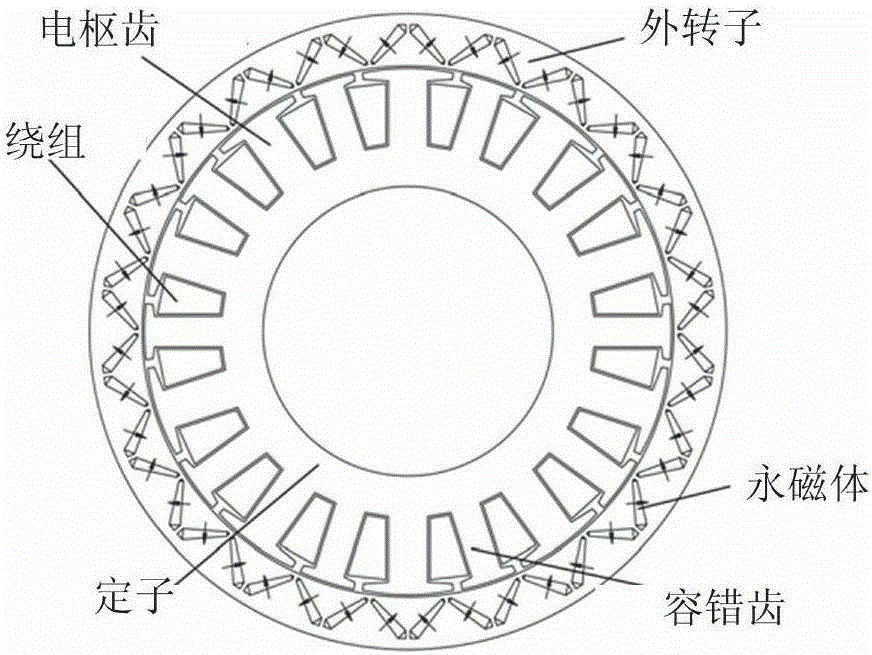
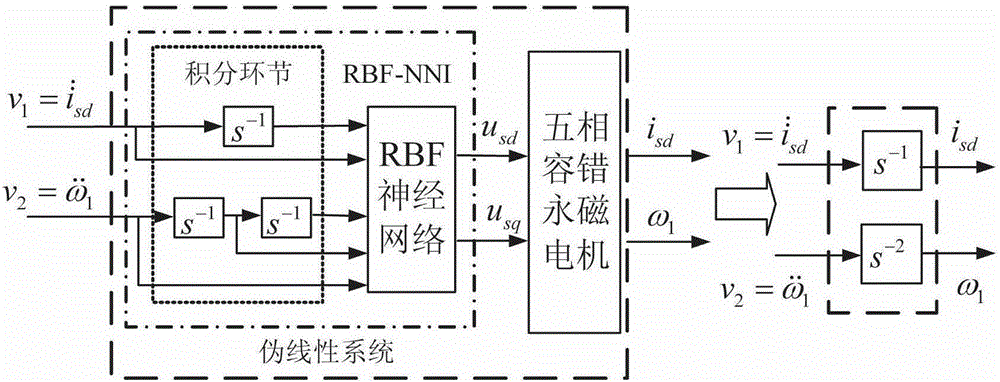
Neural network inverse system-based internal model control method for five-phase fault-tolerant permanent magnet motor
ActiveCN105186958AImprove generalization abilityImprove convergence accuracyElectronic commutation motor controlVector control systemsIntegratorAnti jamming
The invention discloses a neural network inverse system-based internal model control method for a five-phase fault-tolerant permanent magnet motor. The invertibility of the controlled five-phase fault-tolerant permanent magnet motor is proved according to the invertibility principle and the Interactor algorithm; a training sample set of a neural network is built; samples become standardized data for training a neural network; the neural network is trained in Matlab offline; when the training accuracy reaches a set value 0.001, training is stopped; the static neural network which is trained offline and a preposed integrator form a neural network inversion; the trained static neural network inverse system and the preposed integral link are connected with each other in series in front of an original system, so as to form a pseudo-linear composite system; and finally, supplementary controllers are designed for the obtained two pseudo-linear sub-systems to achieve closed-loop control on the overall system according to the internal model control principle. Therefore, the anti-jamming capability and the robust performance of the system are improved.
Owner:东台城东科技创业园管理有限公司
Computerized interactor systems and methods for providing same
InactiveUS7545359B1Simple interfaceReduce fatigueInput/output for user-computer interactionSpecial service for subscribersSemantic contextComputerized system
A computerized interactor system uses physical, three-dimensional objects as metaphors for input of user intent to a computer system. When one or more interactors are engaged with a detection field, the detection field reads an identifier associated with the object and communicates the identifier to a computer system. The computer system determines the meaning of the interactor based upon its identifier and upon a semantic context in which the computer system is operating. The interactors can be used to control other systems, such as audio systems, or it can be used as intuitive inputs into a computer system for such purposes as marking events in a temporal flow. The interactors, as a minimum, communicate their identity, but may also be more sophisticated in that they can communicate additional processed or unprocessed data, i.e. they can include their own data processors. The detection field can be one-dimensional or multi-dimensional, and typically has different semantic meanings associated with different parts of the detection field.
Owner:HANGER SOLUTIONS LLC
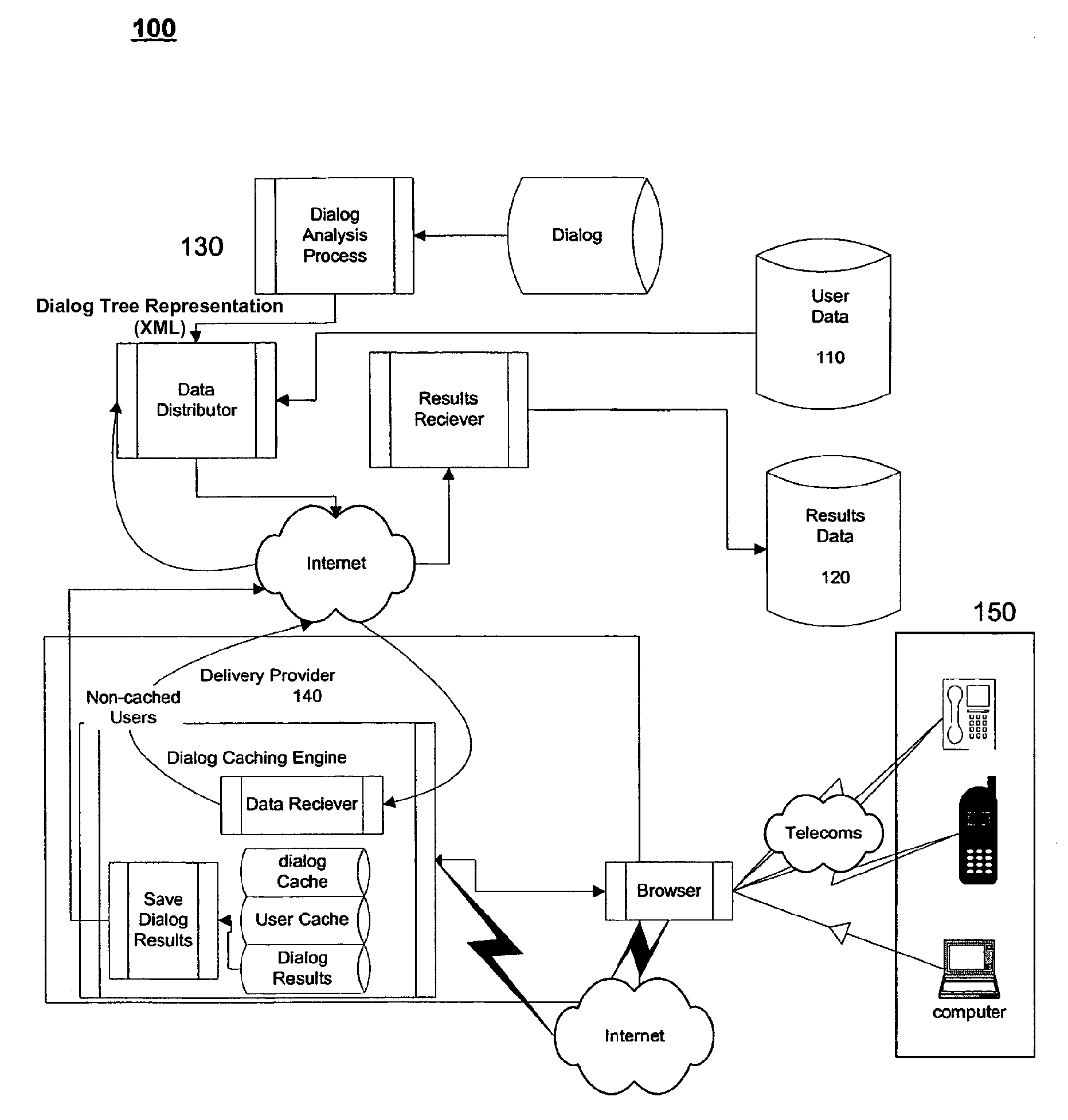
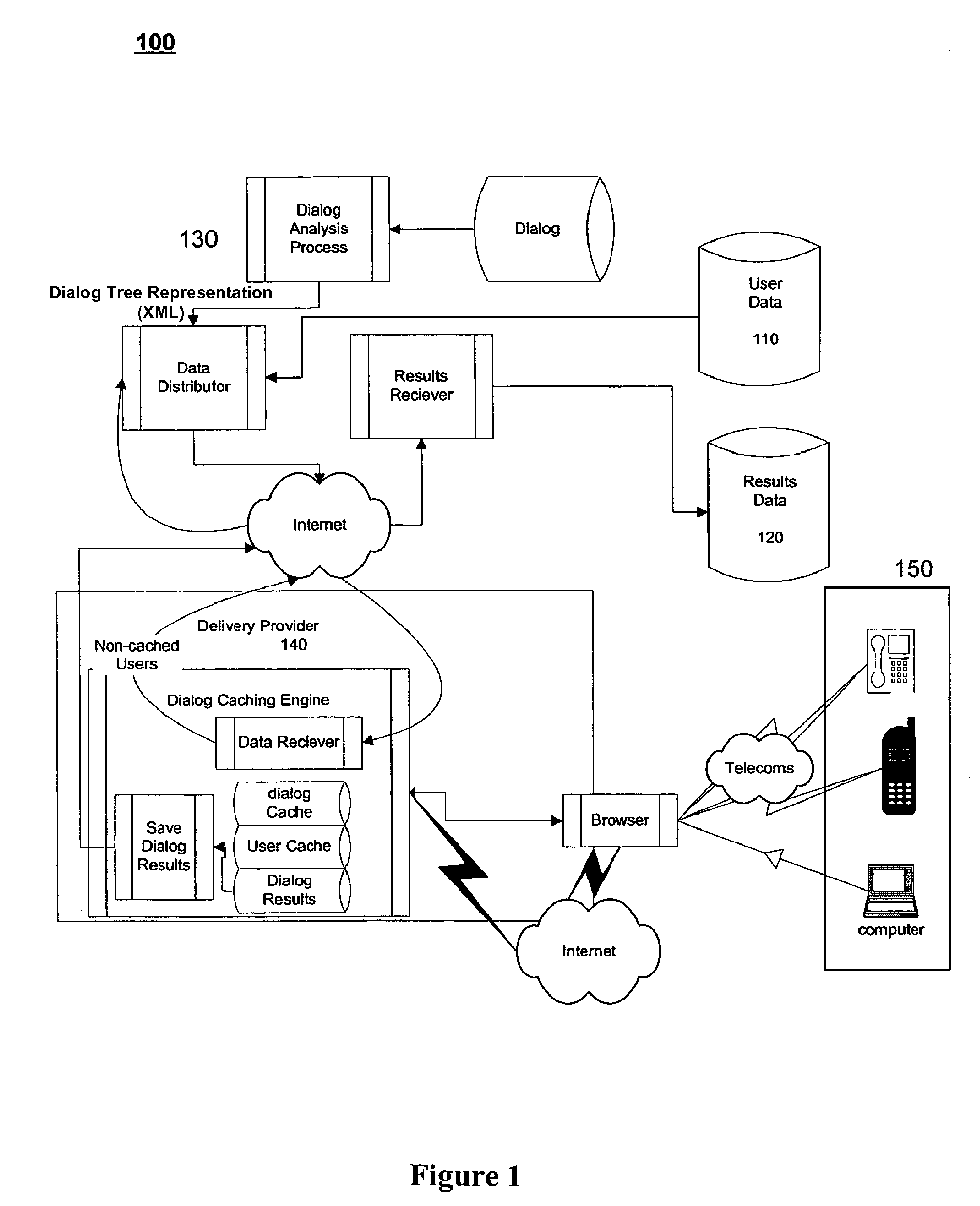
System and method for dialog caching
ActiveUS7558733B2Automatic call-answering/message-recording/conversation-recordingAutomatic exchangesData providerInteractor
A system for providing efficient data handling for automated dialogs and the resulting data is disclosed. The system for providing efficient data handling for automated dialogs and the resulting data includes a data provider, a dialog, a dialog delivery system suitable for delivering the dialog based on data from the data provider, and, a interactor suitable for interaction with the dialog delivered by the dialog delivery system, wherein the interactor provides dialog feedback efficiently stored locally until completion of the dialog and upon completion transmits the feedback to the dialog delivery system.
Owner:WELLTOK
Identification and comparison of protein-protein interactions that occur in populations and indentification of inhibitors of these interactors
Methods are described for detecting protein-protein interactions, among two populations of proteins, each having a complexity of at least 1,000. For example, proteins are fused either to the DNA-binding domain of a transcriptional activator or to the activation domain of a transcriptional activator. Two yeast strains, of the opposite mating type and carrying one type each of the fusion proteins are mated together. Productive interactions between the two halves due to protein-protein interactions lead to the reconstitution of the transcriptional activator, which in turn leads to the activation of a reporter gene containing a binding site for the DNA-binding domain. This analysis can be carried out for two or more populations of proteins. The differences in the genes encoding the proteins involved in the protein-protein interactions are characterized, thus leading to the identification of specific protein-protein interactions, and the genes encoding the interacting proteins, relevant to a particular tissue, stage or disease. Furthermore, inhibitors that interfere with these protein-protein interactions are identified by their ability to inactivate a reporter gene. The screening for such inhibitors can be in a multiplexed format where a set of inhibitors will be screened against a library of interactors.
Owner:CURAGEN CORP
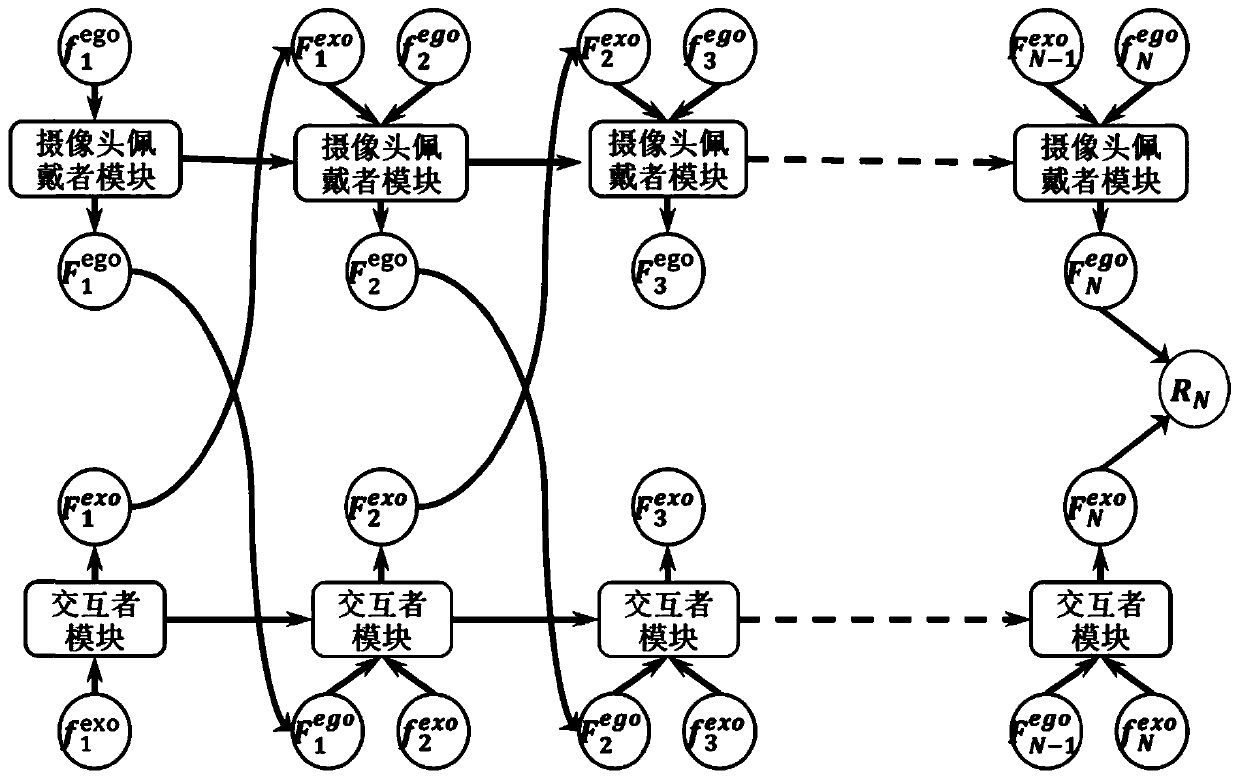
First-person perspective video interaction behavior identification method based on interaction modeling
PendingCN111241963ACharacter and pattern recognitionNeural architecturesInteractive modelingHuman body
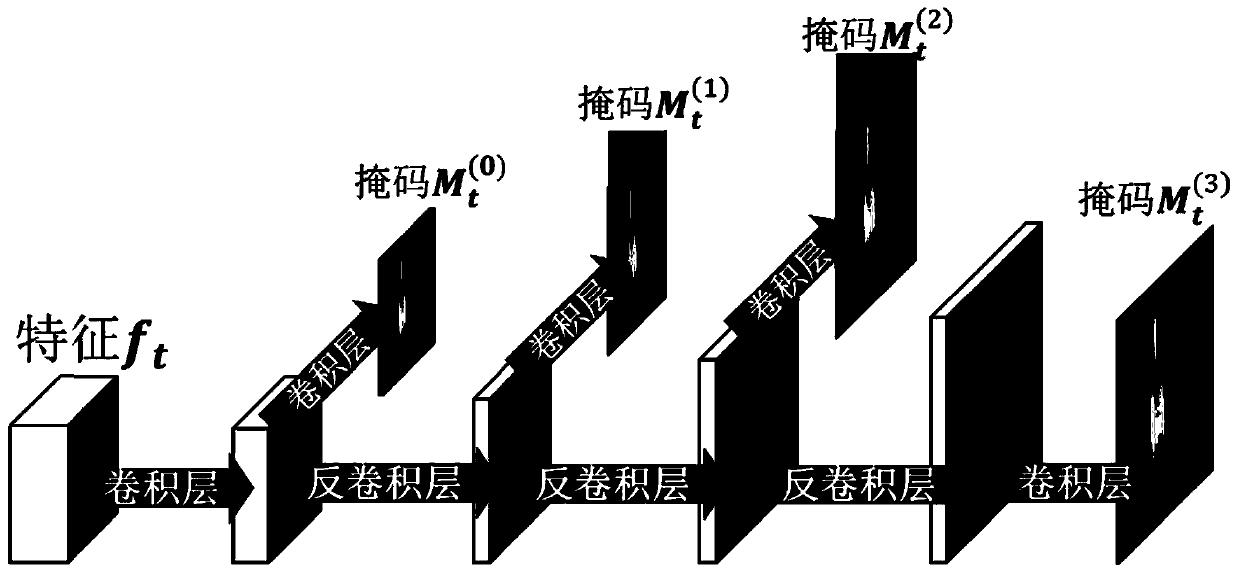
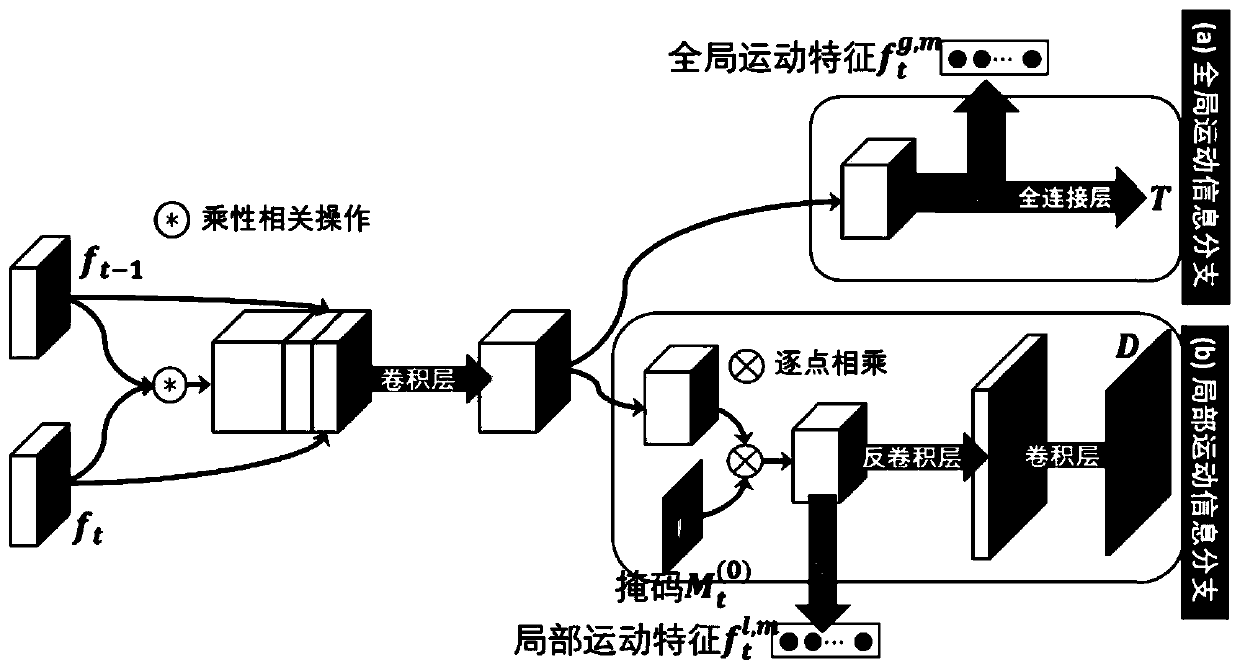
The invention discloses a first-person visual angle video interaction behavior identification method based on interaction modeling, and proposes to separate a camera wearer from an interactor, learn corresponding static appearance and dynamic motion characteristics respectively, and then explicitly model an interaction relationship between the camera wearer and the interactor. In order to separatethe interactor from the background, a mask is generated by using an attention model, and the learning of the attention model is assisted by using a human body analysis model; a motion module is provided to predict motion information matrixes corresponding to a camera wearer and an interactor respectively, and learning of the motion module is assisted through reconstruction of a next frame. And finally, a dual long-short-term memory module for interactive modeling is proposed, and an interactive relationship is explicitly modeled on the basis of the dual long-short-term memory module. According to the method, the interactive behavior of the first-person perspective can be well described and recognized, and a current optimal recognition result is obtained on a common first-person perspective interactive behavior research data set.
Owner:SUN YAT SEN UNIV
Hrpn interactors and uses thereof
InactiveUS20090044296A1Increased susceptibilityImprove scalabilityMicroorganismsFermentationDiseaseGrowth plant
The present invention relates to nucleic acid molecules configured to increase or decrease expression of a nucleic acid molecule that encodes a HrpN-interacting protein; nucleic acid constructs that include these nucleic acid molecules; host cells, transgenic plants, and transgenic plant seeds transformed thereby; and methods of increasing plant growth or imparting disease resistance to plants. Also disclosed are an isolated HIPM nucleic acid molecule and an isolated HIPM protein or polypeptide.
Owner:CORNELL RES FOUNDATION INC
Computerized interactor systems and methods for providing same
InactiveUS20020126085A1Easy to optimizeSimple interfaceInput/output for user-computer interactionInterconnection arrangementsComputer hardwareComputerized system
The present invention teaches a variety of methods and systems for providing computer / human interfaces. According to one method, the user interfaces with an electronic device such as a computer system by engaging a sensor with desired regions of an encoded physical medium. the encoded physical medium is preferably chosen to provide intuitive meaning to the user and is thus an improved metaphor for interfacing with the computer system. The sensor may have at least one identification number (ID) providing information such as user identity, sensor type, access type, or language type. The sensor can transmit the certain decoded information together with the at least one ID to the computer system.
Owner:HANGER SOLUTIONS LLC
Methods and systems for providing programmable computerized interactors
InactiveUS7724236B2Simple interfaceElectrophonic musical instrumentsTransmission systemsSemantic contextComputerized system
A computerized interactor system uses physical, three-dimensional objects as metaphors for input of user intent to a computer system. When one or more interactors are engaged with a detection field, the detection field reads an identifier associated with the object and communicates the identifier to a computer system. The computer system determines the meaning of the interactor based upon its identifier and upon a semantic context in which the computer system is operating.
Owner:VULCAN PATENTS
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com