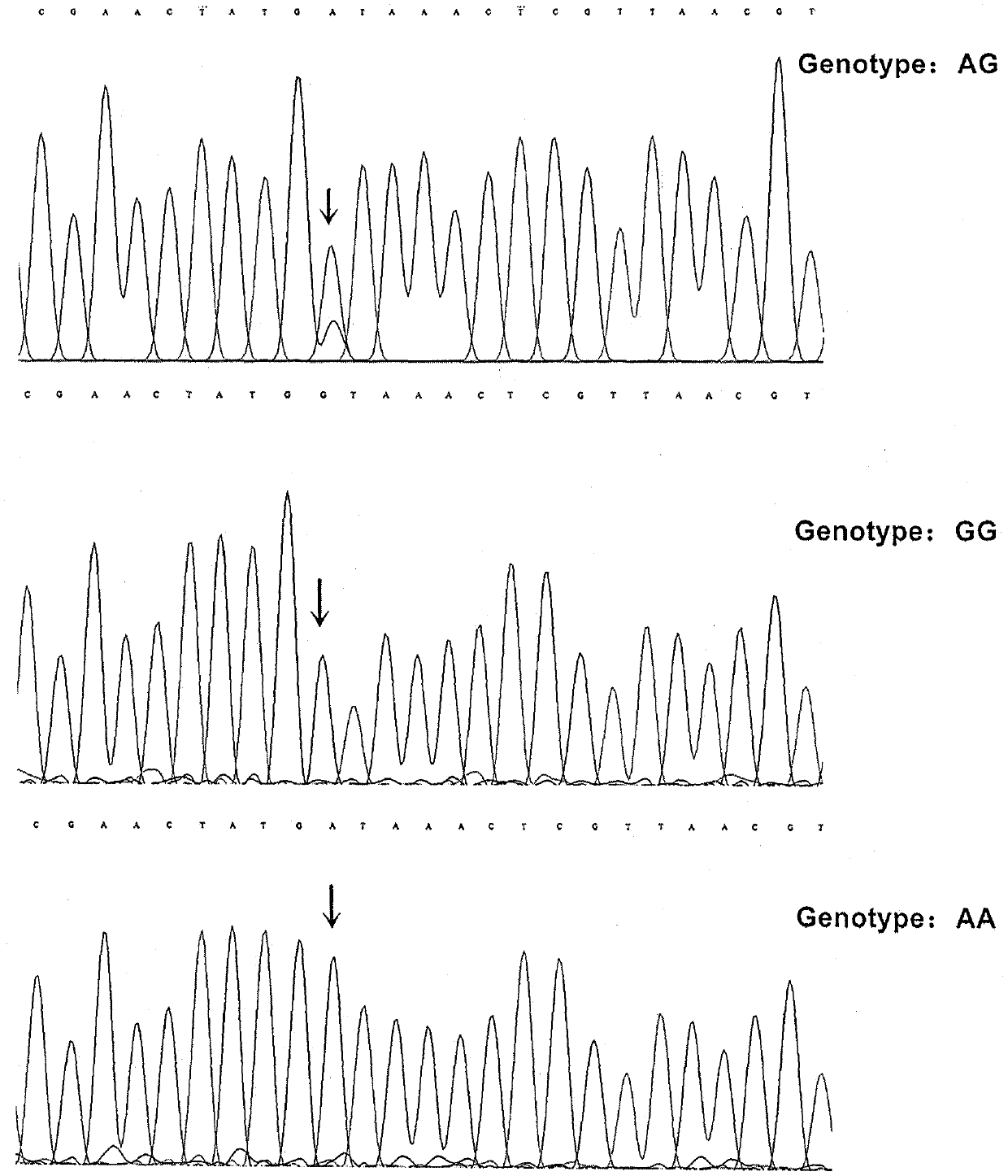
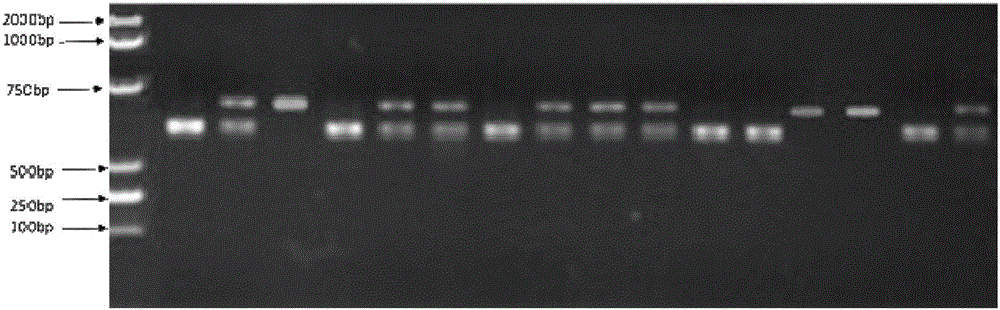
Patents
Literature
69 results about "CO-DOMINANT" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Co-dominance InDel molecular marker for identifying single embryo and multiple embryos of citrus and application thereof
ActiveCN105603096AImprove breeding efficiencyOvercoming time consumingMicrobiological testing/measurementDNA preparationGenomicsRe sequencing
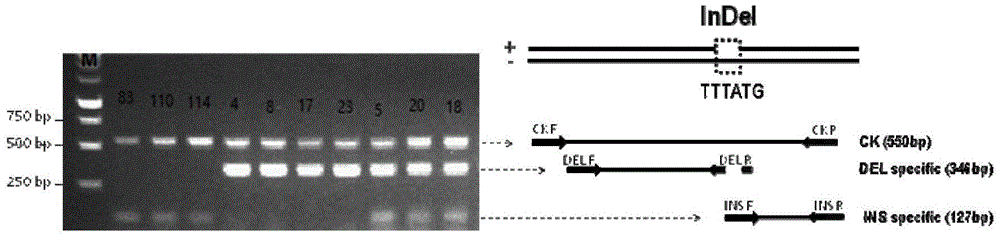
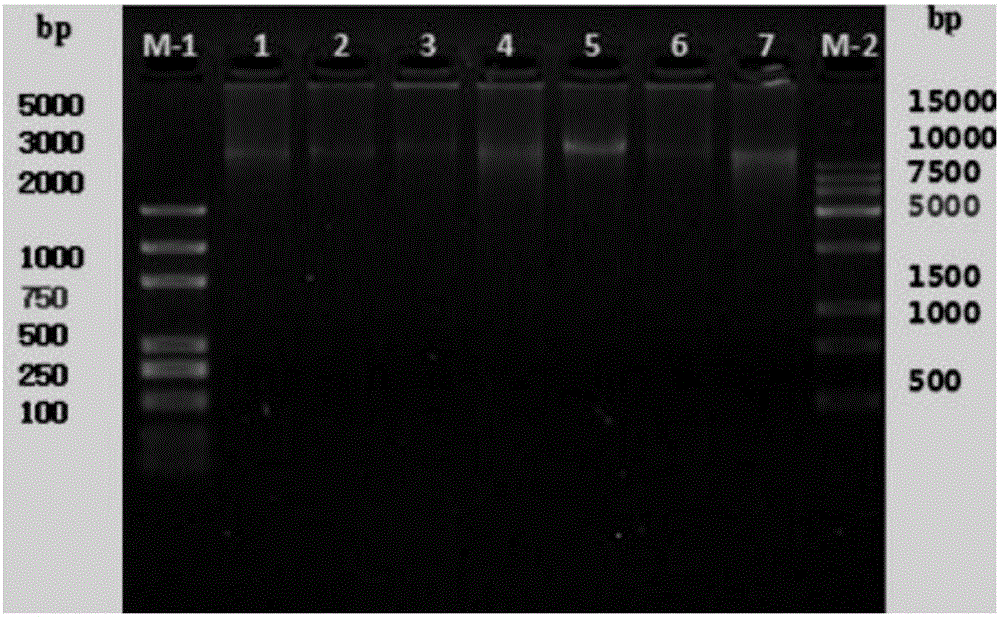
The invention belongs to the field of citrus genomics and discloses a co-dominance InDel molecular marker for identifying single embryo and multiple embryos of citrus and application thereof. By carrying out depth re-sequencing on whole genome DNA of 107 materials including tangelos and oranges, whole genome sequence information is obtained, by means of comparing and screening, the gene sequence information in single-embryo and multi-embryo candidate areas is obtained, the sequence with the highest relevance degree to phenotype is screened out, and the sequence is an InDel sequence of 6bp. Accordingly, a set of simple, fast and co-dominant PCR markers are developed, and primers are used for carrying out PCR amplification on CK F / R, INS F / R and DEL F / R, so that the co-dominance InDel molecular marker for identifying the single embryo and the multiple embryos of the citrus is obtained. Finally, the marker is utilized for testing loci in 115 single-plant natural groups and 100 single-plant hybridized groups. The marker has the biggest advantages that cost is low, accuracy is high, and embryonic early selection can be achieved during the citrus seedling stage.
Owner:HUAZHONG AGRI UNIV
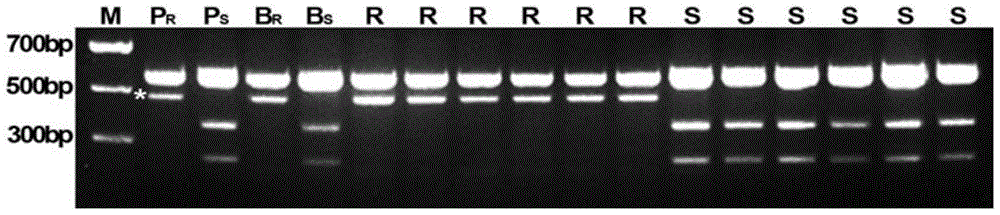
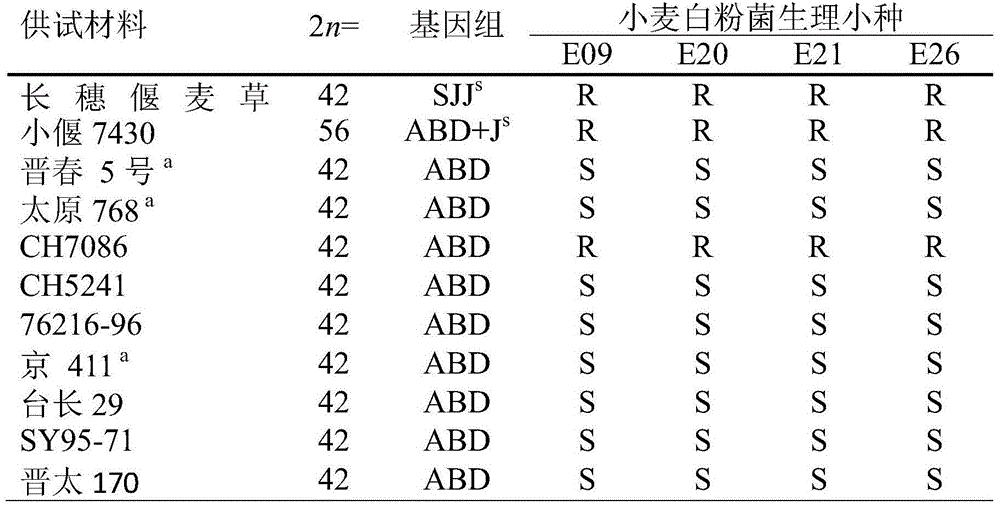
Molecular marker of wheat powdery mildew disease-resistant genes Pm51 and application of molecular marker
ActiveCN104313021AImprove utilizationRapid identificationMicrobiological testing/measurementDNA/RNA fragmentationDiseaseBiotechnology
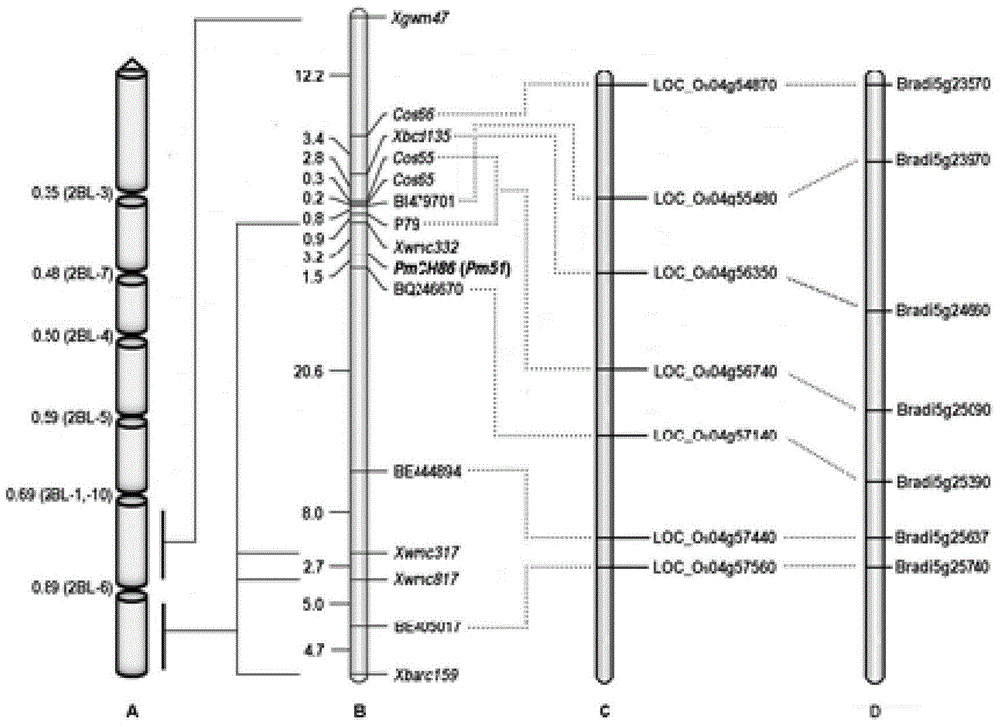
The invention relates to plant molecular genetics and wheat breeding and in particular relates to a molecular marker of novel wheat powdery mildew disease-resistant genes Pm51 and application of the molecular marker. A functional molecular marker BQ246670 linked with powdery mildew disease-resistant genes Pm51 is obtained. The marker is a co-dominant marker, the genetic distance between the marker and the Pm51 is 1.5cM, the existence and existential state of wheat genes Pm51 can be rapidly and accurately identified, and the resistance of wheat powdery mildew disease is predicted, so that the utilization of the wheat powdery mildew disease-resistant genes Pm51 is accelerated.
Owner:SHANXI UNIV OF CHINESE MEDICINE
CAPS molecular marker primer for detecting CMS (cytoplasmic male sterility) restoring genes of capsicum annuum and application of CAPS molecular marker primer
InactiveCN108411027AImprove accuracyVerify validityMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCO-DOMINANT
The invention relates to the field of biotechnology-assisted seed breeding, and particularly discloses a CAPS molecular marker primer for detecting CMS (cytoplasmic male sterility) restoring genes ofcapsicum annuum and application of the CAPS molecular marker primer. CAPS molecular markers are tightly linked with the CMS restoring genes of the capsicum annuum and are M1 markers. The CAPS molecular marker primer and the application have the advantages that detection can be facilitated by the CAPS molecular markers, stable and reliable results can be obtained, the CAPS molecular markers are co-dominant markers, and accordingly sterility restoring genes of capsicum annuum strains can be detected on a large scale only by means of simple conventional experiment operation such as hereditary substance extraction, PCR (polymerase chain reaction), restriction incision enzyme digestion and routine agarose electrophoresis; the selective breeding ranges of restoring line materials for the capsicum annuum can be expanded by development and application of the CAPS molecular markers, the seed breeding efficiency can be improved, seed breeding progresses can be accelerated, and the CAPS molecularmarker primer and the application have important significance in molecular marker-assisted seed breeding for CMS three-line matched seed production and restoring line materials for the capsicum annuum.
Owner:WUHAN ACADEMY OF AGRI SCI
Molecular marker of brassica napus dominant nucleic sterility restoring line and preparation method and application thereof
InactiveCN101701263AOvercome the downside of choosingReduce breeding workloadMicrobiological testing/measurementFermentationBrassicaMarker-assisted selection
The invention belongs to the technical field of breeding of rape molecules, in particular to a preparation method of a co-dominant SCAR molecular marker of a brassica napus dominant nucleic sterility restoring line and application of the molecular marker as marker-assisted selection on selectively breeding the brassica napus dominant nucleic sterility restoring line. The genome DNA of a sterile line 4185A separated by the first generation after a dominant nucleic sterility line brassica napus material homozygous two-type line GMS3 is planted and a brassica napus dominant nucleic sterility restoring line 4185B-2 is amplified by a primer YQ-Rf1 and a primer YQ-Rf2, and four amplified DNA segments are respectively obtained; then the cloning, the sequence test and the nucleotide sequence comparison are carried out on the DNA amplified segments, and the primer YQ-Rfa and the primer YQ-Rfb are designed according to the difference of sequences; and the PCR amplification and selective effect inspection are carried out, thereby obtaining the co-dominant SCAR molecular marker of the brassica napus dominant nucleic sterility restoring line. The invention provides a new marker for the breeding of the rape molecules. The invention also discloses a preparation method and application of the molecular marker.
Owner:HUAZHONG AGRI UNIV
Molecular breeding method for new rice variety carrying gene Pi65(t) with resistance to rice blast
ActiveCN104774945AMicrobiological testing/measurementPlant genotype modificationDiseaseAgricultural science
The invention belongs to the field of molecular biology and particularly relates to a molecular marker assisted breeding method of a new rice variety with resistance to rice blast and special primers thereof. The molecular mark is a co-dominant molecular mark Indel-1 of the rice gene Pi65(t) with resistance to rice blast and is a nucleotide sequence which is amplified from total DNA of rice by using a primer pair SEQ ID NO: 1 and SEQ ID NO: 2. The method can be applied to molecular marker assisted selection of Pi65(t) in resistance breeding of the resistance to blast of rice, so that the efficiency of the anti-disease variety breeding is improved and the workload of field identification is reduced.
Owner:LIAONING ACAD OF AGRI SCI +2
Molecular marker tightly linked with Cucumis sativus tumor gene Tu
InactiveCN101481737ASpeed up the process of quality breedingMicrobiological testing/measurementDNA/RNA fragmentationCO-DOMINANTGenetic engineering
The invention discloses a molecular marker which is closely linked with tuberculate trait gene Tu in cucumis sativus, belonging to the technical field of genetic engineering. Two SRAP markers are firstly obtained, wherein, the marker ME6EM9 has the sequence in SEQ ID No.1 in a sequence table, and the marker ME2EM4 has the sequences in SEQ ID No.3 and SEQ ID No.4 in the sequence table. The ME6EM9 and ME2EM4 are transformed into stable SCAR markers which respectively are C_SC69 and C_SC24. The C_SC69 is dominant marker, and 218bp products in SEQ ID No.2 are amplified; C_SC24 is co-dominant marker which has two amplified products that are respectively 514bp in SEQ ID No.5 and 455bp in SEQ ID No.6. The molecular marker of the invention can be applied to assisted selection of tuberculate trait in cucumis sativus and can speed up the breeding process of cucumber varieties.
Owner:SHANGHAI JIAO TONG UNIV
Pepper sterility restoring gene closely linked molecular marker, method and application
ActiveCN107312870AValid identificationReliable resultsMicrobiological testing/measurementDNA/RNA fragmentationHybrid seedAgricultural science
The invention belongs to the technical field of molecular biology and discloses a pepper sterility restoring gene closely linked molecular marker, a method and application. The molecular marker is scd06-17 and comprises a nucleotide sequence shown as SEQ ID NO.1 and a nucleotide sequence shown as SEQ ID NO.2. The molecular marker is stable and reliable in result and is a co-dominant molecular marker, whether a pepper material contains a cytoplasmic male sterility restoring gene or not can be determined in a pepper steeling stage, and accordingly breeding efficiency is improved, and a breeding process is accelerated. Compared with other domestic and international published pepper restoring gene molecular markers, the pepper sterility restoring gene closely linked molecular marker is significant to pepper three-line hybrid seed production and restoring material molecular marker assisted selection breeding.
Owner:河南省农业科学院园艺研究所
Rice flooding-tolerant gene Sub1 co-dominant molecular marker and application
ActiveCN109468315AShorten the breeding cycleLow costMicrobiological testing/measurementDNA/RNA fragmentationCO-DOMINANTAgricultural science
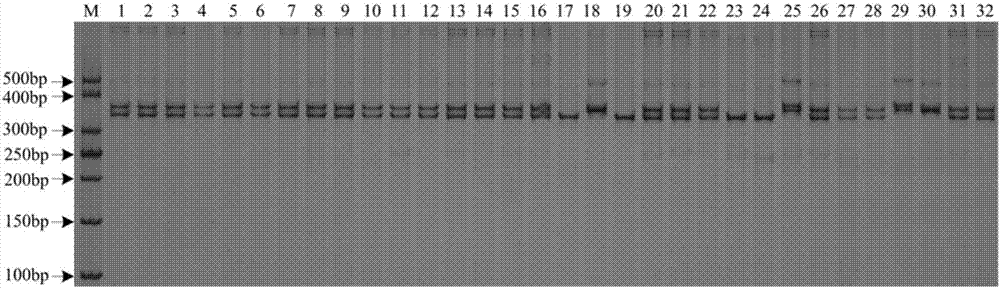
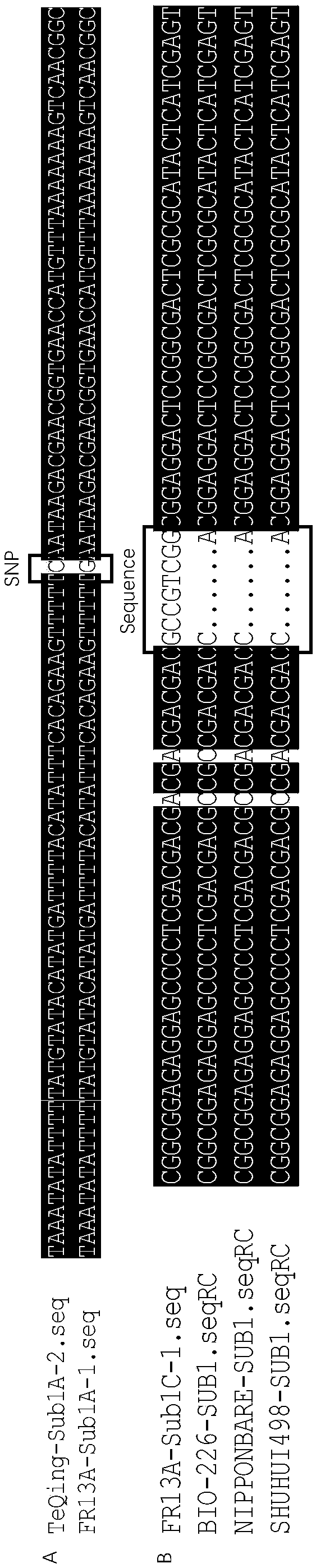
The invention provides a rice flooding-tolerant gene Sub1 co-dominant molecular marker and application. The molecular marker comprises an SNP marker and an InDel marker which are separately located onrice No. 9 chromosome genes Sub1A and Sub1C, wherein the polymorphic site of the SNP marker on the gene Sub1A is G / C; and the polymorphic site of the InDel marker on the gene Sub1C is 5'-GCCGTCG-3' / 5'-CA-3'. For the molecular marker, specific amplification primers are designed, and Sub1 genotype detection is carried out through PCR amplification. The provided molecular marker and an amplificationprimer thereof for Sub1 can be used for identifying the genotype of rice Sub1, a flooding-tolerant rice resource is bred, the rice flooding-tolerant gene Sub1 co-dominant molecular marker has the advantages of high identification accuracy, simplicity in operation, low cost and the like, the breeding period of the flooding-tolerant rice can be shortened, and the breeding cost is reduced.
Owner:YUAN LONGPING HIGH TECH AGRI CO LTD +2
Application of SNP molecular marker closely linked to aphid resistance gene of prunus persica
ActiveCN106434944AGuaranteed accuracyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCO-DOMINANT
The invention discloses application of an SNP molecular marker closely linked to an aphid resistance gene of prunus persica, wherein the molecular marker comprises KyHRM-17-45.71 and KyHRM-3-46.12, which are respectively located at 45.713 Mb and 46.121 Mb of prunus persica genome (Version 2.0) Scaffold 1; alleles KyHRM-17-45.71 are T and G, and the sequence is shown in SEQ ID NO.1; the alleles of KyHRM-3-46.12 are C and G, and the sequence is shown in SEQ ID NO.2. In the invention, the third generation of markup technology based on SNP is used in combination with a constructed isolated hybrid population, and a map-based cloning method is adopted to finely locate the aphid resistance gene of prunus persica; stable, co-dominant and pleomorphic SNP markers are developed in a fine location region, to obtain the marker closely linked to a target trait.
Owner:ZHENGZHOU FRUIT RES INST CHINESE ACADEMY OF AGRI SCI
Breeding and identifying method of soft and powdery mildew resistant triticum aestivum-Dasypyrum villosum translocation line
ActiveCN104365471AValid identificationExcellent agronomic traitsMicrobiological testing/measurementPlant genotype modificationAgricultural scienceTriticeae
The invention discloses a breeding and identifying method of a soft and powdery mildew resistant triticum aestivum-Dasypyrum villosum translocation line. The method comprises the steps of hybridizing and backcrossing a triticum aestivum-Dasypyrum villosum 5V alien addition line DA5V and a Chinese spring ph1b1b mutant, identifying individual plants relevant to 5VS and ph1b1bph1b1b from BC1F1 by use of a molecular marker, identifying individual selfed seeds by use of GISH (genomic in situ hybridization) technology, and analyzing individual plants containing translocated chromosomes by use of C-zoning, GISH and molecular markers to prove that the translocated chromosome is T5VS.5AL. A PCR (polymerase chain reaction) primer is designed by use of an EST (Expressed Sequence Tag) located on the homologous chromosome group of the fifth part of triticum, a co-dominant marker capable of identifying the translocated chromosome specially can be screened, and homozygosis and heterozygosis translocations can be distinguished. The breeding utilization value of the translocation line can be disclosed through agronomic traits, kernel hardness analysis and powdery mildew resistance identification.
Owner:NANJING AGRICULTURAL UNIVERSITY
Molecular marker closely linked with male and female plants of spinach (Spinacia oleracea), and application thereof
ActiveCN110438252AAccurate identificationImprove throughputMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCO-DOMINANT
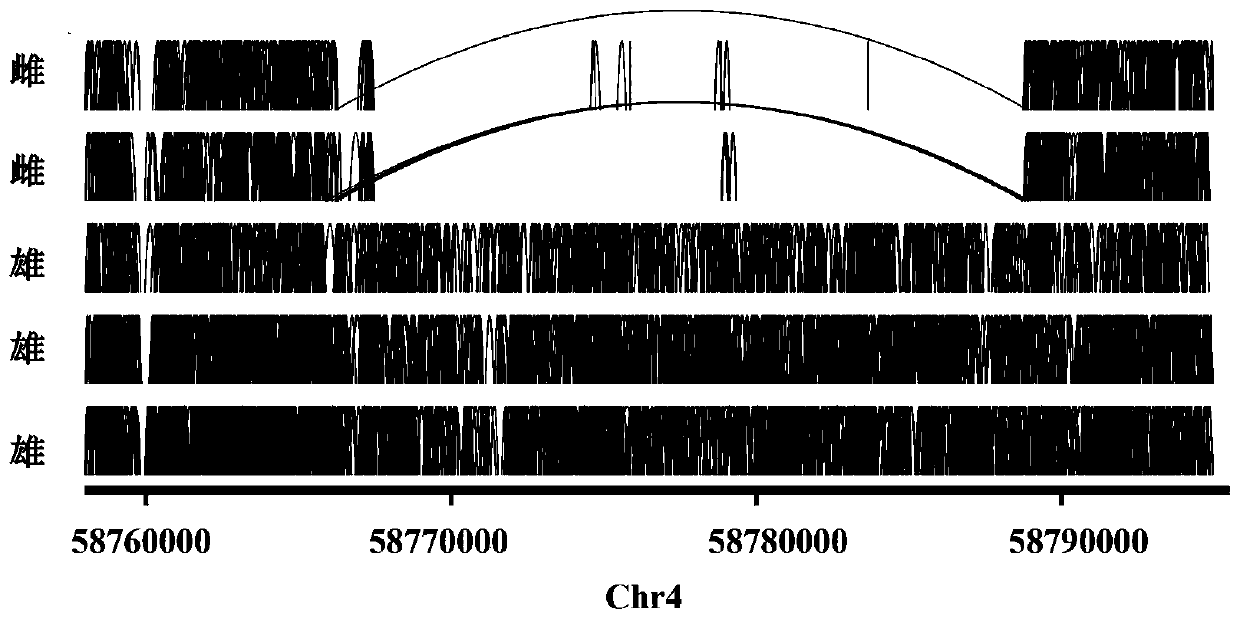
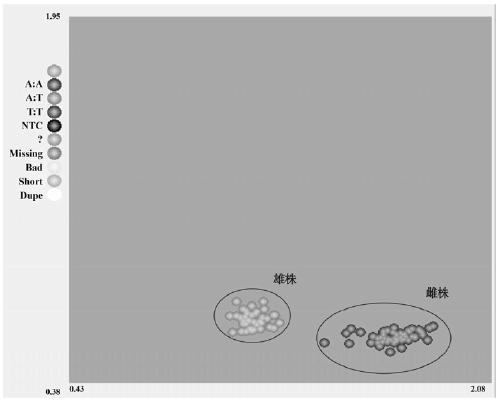
The invention provides a molecular marker closely linked with male and female plants of spinach (Spinacia oleracea) as well as application thereof. The molecular marker closely linked with male and female plants of spinach contains nucleotide sequence, of which the polymorphism is T / A, at locus 58764574 on chromosome 4 of spinach genome. According to the invention, the molecular marker closely linked with spinach sex gene is developed on basis of a KASP technology; and the molecular marker has been validated in a plurality of spinach populations to be closely linked with spinach sex gene. Thus, the molecular marker closely linked with spinach sex gene can be used for accurately identifying male and female plants of spinach. Being used for performing sex detection on spinach, the specificKASP primer provided by the invention is capable of achieving high analysis throughput and high accuracy; so that, the specific KASP primer is suitable for detection of large number of samples. The co-dominant molecular marker provided by the invention has important practical significance for improving female lines and inbred lines of spinach, shortening breeding time and improving breeding efficiency.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
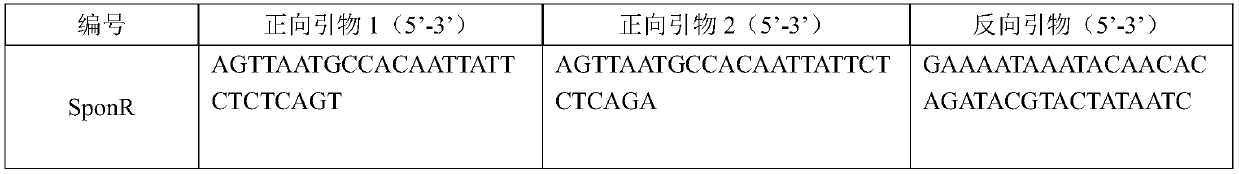
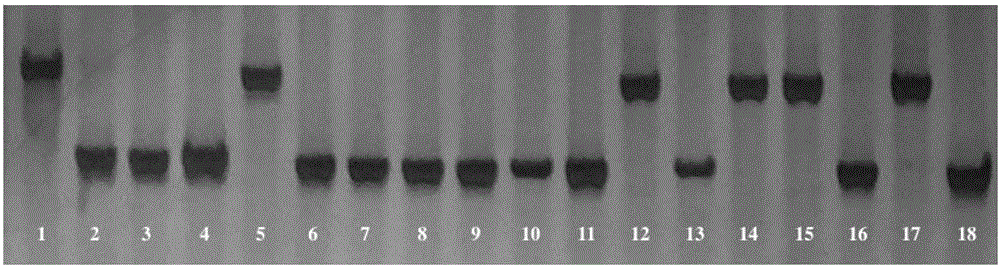
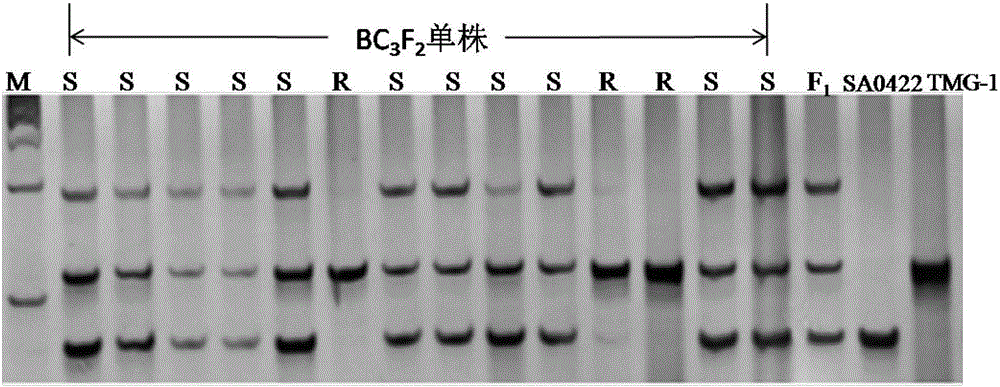
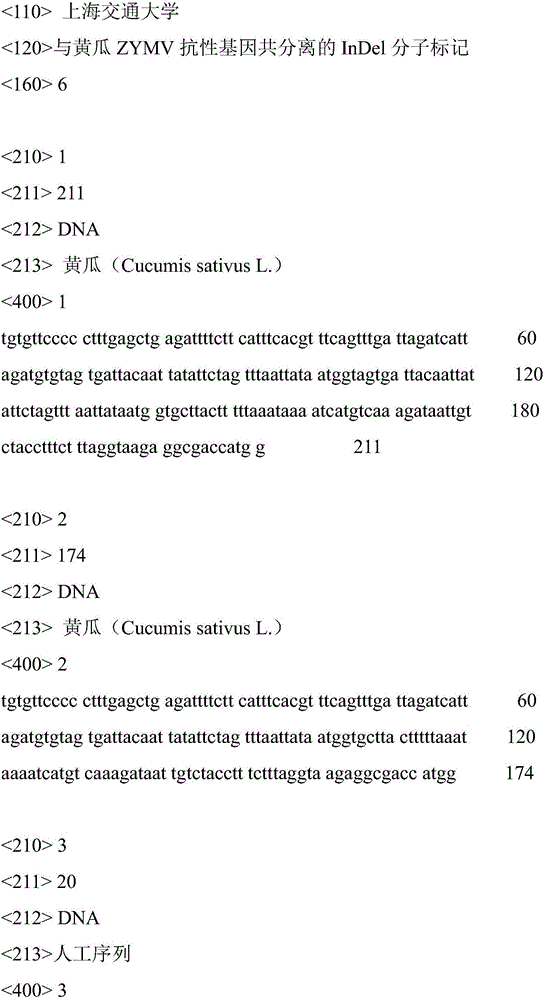
InDel molecular marker co-segregated from ZYMV resistance gene of cucumber
InactiveCN106811536AImprove throughputOmit the step of selfingMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceResistant genes
The invention relates to an InDel molecular marker co-segregated from a ZYMV resistance gene. The InDel molecular marker named InDel-zym1 and is composed of a nucleotide sequence fragment shown in SEQ ID NO. 1 and a nucleotide sequence fragment shown in SEQ ID NO. 2, wherein the nucleotide sequence fragment shown in SEQ ID NO. 1 is co-segregated from a resistance gene; the nucleotide sequence fragment shown in SEQ ID NO. 2 is co-segregated from a disease-susceptible gene. The InDel molecular marker provided by the invention has high stability and can be easily and rapidly applied to the auxiliary screening of ZYMV resistant individual plants in a cucumber seedling stage at high throughput; according to SEQ ID NO. 5 and SEQ ID No. 6, multiple pairs of co-dominant InDel markers can also be designed as molecular markers for ZYMV resistance screening on account of difference in DNA length of resistance candidate genes caused by insertion / deletion mutations. The InDel marker provided by the invention lays a foundation for ZYMV resistance molecular marker-assisted breeding, and greatly accelerates progress of breeding of ZYMV resistance molecules of cucumber.
Owner:SHANGHAI JIAO TONG UNIV
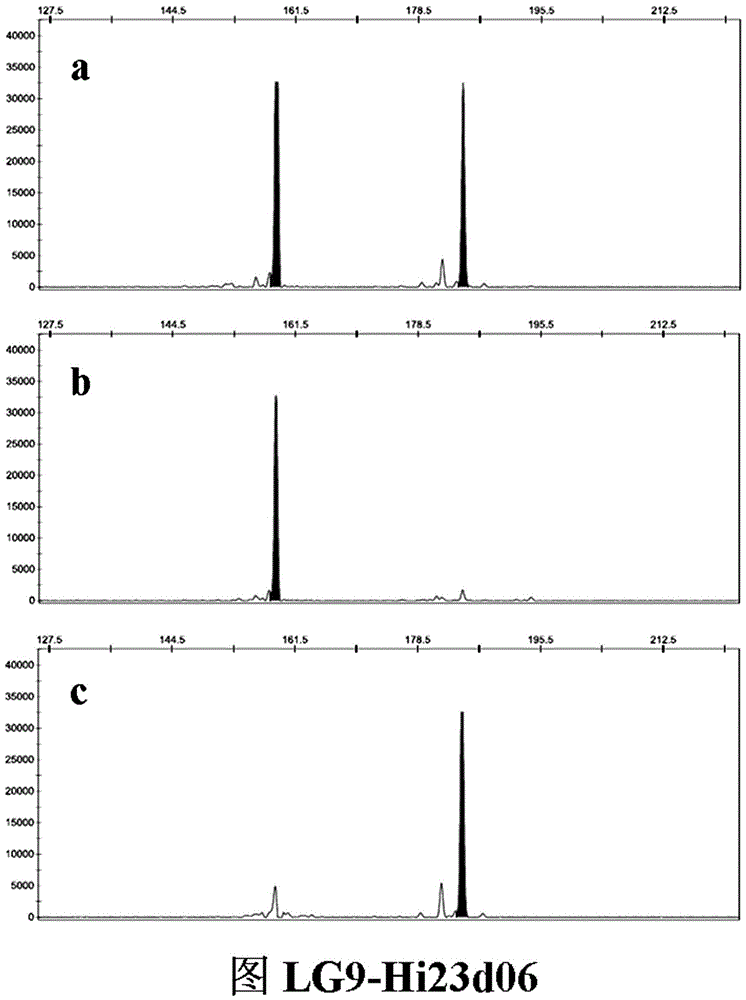
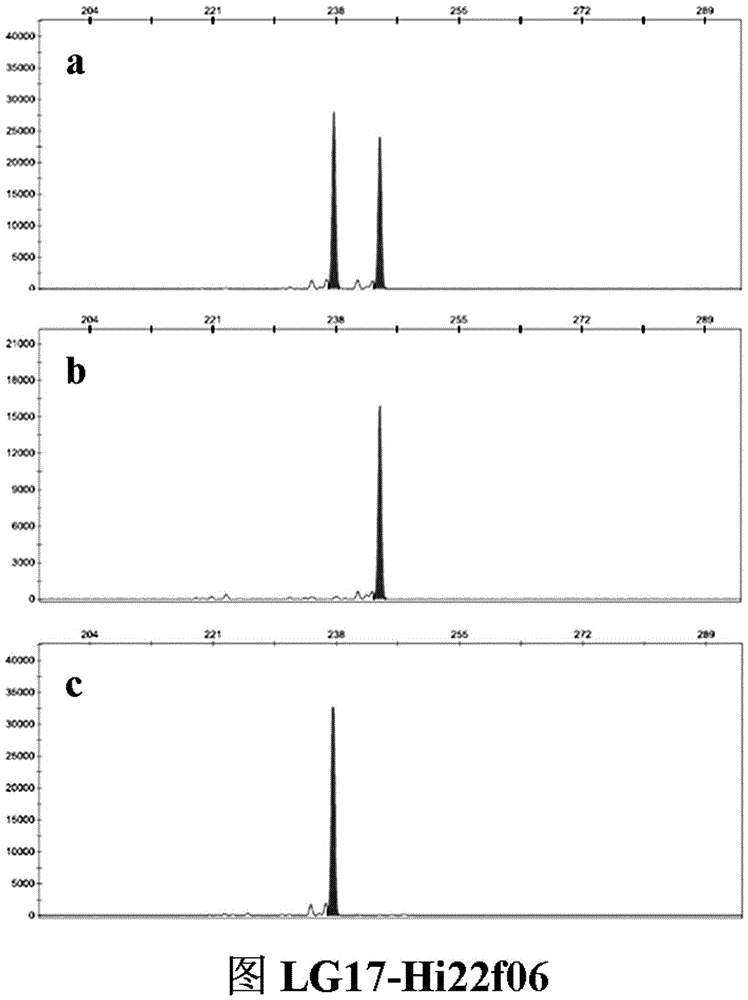
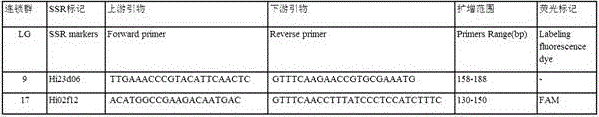
SSR (simple sequence repeat) molecular marker V for identifying descendant plants of Gala apple and application thereof
InactiveCN106755479ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCO-DOMINANT
The invention belongs to the field of innovation and research of plant genetics breeding and apple germplasm, in particular to an SSR (simple sequence repeat) molecular marker V for identifying descendant plants of Gala apple and application thereof. The SSR molecular marker V is characterized in that in the detection process, the SSR molecular markers linked with No.9 and No.17 chromosomes are simultaneously used, DNA (deoxyribonucleic acid) of an apple gene group is used as a PCR (polymerase chain reaction) amplification template, the SSR molecular markers are respectively used as primer pairs, and the PCR amplification and product detection are performed, so as to identify the genetics linkage, anther culture plants and variety sources. The SSR molecular marker V has the advantages that the SSR molecular marker is used for identifying the apple linkage group of the identifying material for the first time, and the SSR molecular markers linked with No.9 and No.17 chromosomes of the apple are disclosed; the molecular marker is a co-dominant marker, so that the Gala apple genetic linkage group, the anther culture plant and the Gala source plant are quickly and accurately identified; the molecular level support is provided for further accelerating the utilization of the Gala apple and important agronomic trait linkage gene, and the genetic breeding of apple homozygous plants.
Owner:AGRI BIOTECH RES CENT OF SHANXI PROVINCE +2
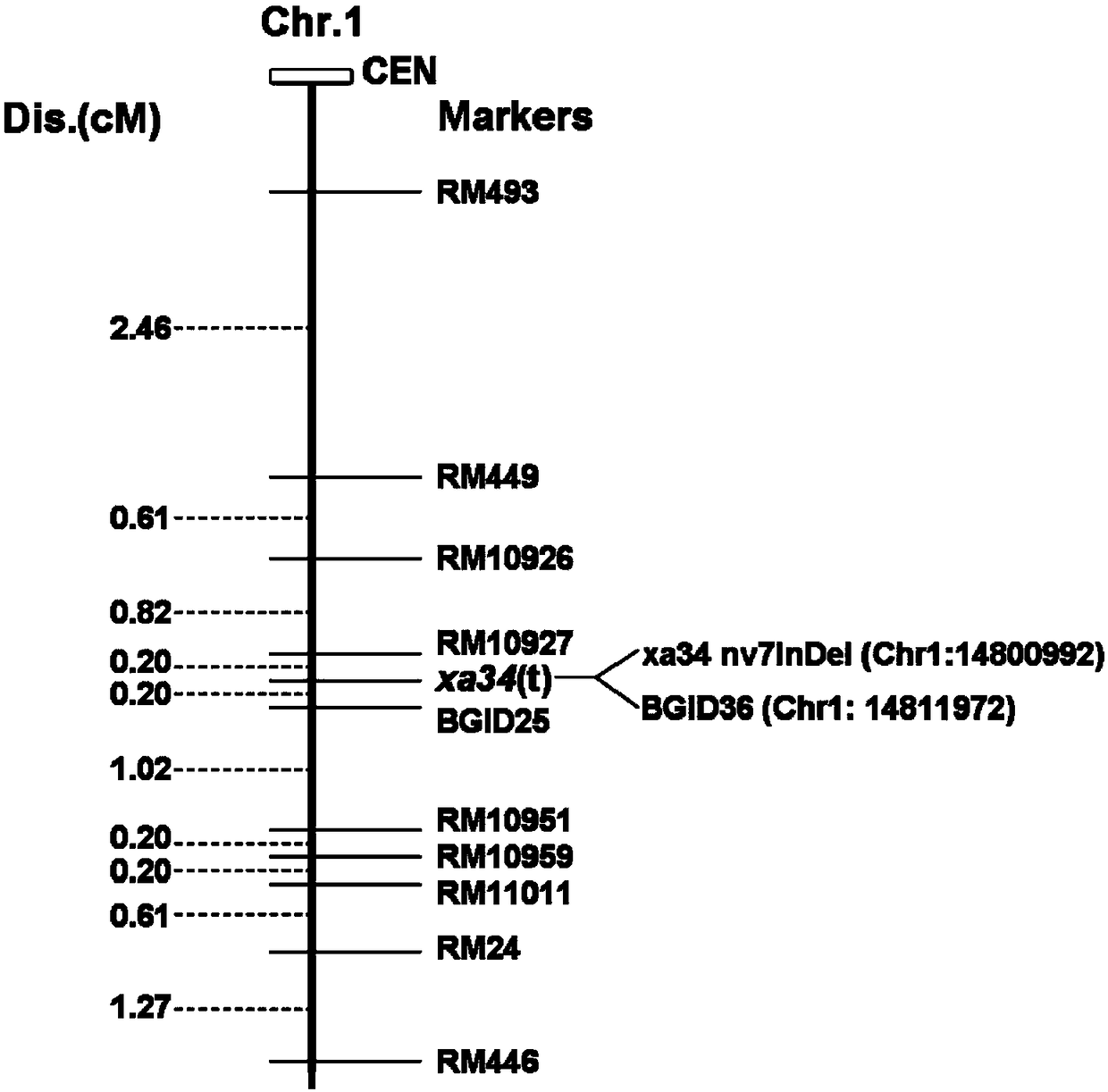
InDel molecular marker realizing coseparation with rice xanthomonas oryzae resistant gene xa34(t) as well as detection primers and applications of InDel molecular marker
ActiveCN108103237AOmit the step of selfingStrong specificityMicrobiological testing/measurementDNA/RNA fragmentationCO-DOMINANTAgricultural science
The invention discloses an InDel molecular marker realizing coseparation with the rice xanthomonas oryzae resistant gene xa34(t) as well as detection primers and applications of the InDel molecular marker. The InDel molecular marker has the nucleotide sequence as shown in SEQ ID NO.1. The InDel molecular marker is the co-dominant marker, can distinguish homozygotes from heterozygotes, and can be used for identifying the rice xanthomonas oryzae resistant variety. The InDel molecular marker is high in specificity, the band differences are great, and clear distinguishing can be realized through agarose gel electrophoresis. The invention further provides detection primers of the InDel molecular marker, which are composed of the primer as shown in SEQ ID NO.3 and the primer as shown in SEQ ID NO.4. The invention further provides a detection kit of the InDel molecular marker, which comprises the detection primers. With the detection primers or the detection kit, the molecular breeding process for the broad-spectrum xanthomonas oryzae resistance of rice is greatly accelerated.
Owner:PLANT PROTECTION RES INST OF GUANGDONG ACADEMY OF AGRI SCI
Indel (Insertion-deletion) molecular marker primer for exocarpium benincasae color gene and application thereof
ActiveCN108866226AStrong specificityAssisted breedingMicrobiological testing/measurementDNA/RNA fragmentationInsertion deletionCO-DOMINANT
The invention discloses an indel (Insertion-deletion) molecular marker primer for controlling an exocarpium benincasae color gene. The primer can produce a black skin material specific marker and a yellow skin material specific marker (co-dominant markers), and is good in repeatability and high in specificity. The invention also discloses an identification method of an exocarpium benincasae color.The method is accurate, quick, low in cost, short in identification period, and simple and convenient to operate. The invention further discloses application of the indel molecular marker primer controlling the exocarpium benincasae color gene in identifying the exocarpium benincasae color gene.
Owner:INST OF VEGETABLES GUANGDONG PROV ACAD OF AGRI SCI
SNP marker tightly linked with irregular stripe characters of cucumber fruits, and applications thereof
ActiveCN110205396AAccurate genetic lociEasy to identifyMicrobiological testing/measurementDNA/RNA fragmentationCO-DOMINANTCuticle
The invention provides an SNP marker tightly linked with irregular stripe characters of cucumber fruits. The SNP marker is characterized in that the SNP marker can be tightly linked with the irregularstripe genes of cucumber fruits; the forward and reverse primer sequences of the marker respectively are 5'GACCTCCCAACCCCTGATTTTTT3' and 5'ATCGTTCGCCGTCTTACCTGACT3'; the amplification length of the marker is 675 bp, an SNP locus is located on a forward-amplified 440bp locus, and the marker has A / G polymorphism; and a GG genotype is cucumbers with irregular stripes, and AA and AG genotypes are thecucumbers with no irregular stripes. The SNP molecular marker belongs to co-dominant markers, is reliable and fast in detection and can be used for the directed genetic breeding of the related characters of cucumber fruit epidermis; and the SNP marker has important theoretical and practical guiding significance on accelerating the genetic improvement process of cucumber varieties and enhancing breeding selection efficiency.
Owner:NANJING AGRICULTURAL UNIVERSITY
SSR (simple sequence repeat) molecular marker III for identifying descendant plants of Gala apple and application thereof
InactiveCN106755482ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGermplasm
The invention belongs to the field of innovation and research of plant genetics breeding and apple germplasm, in particular to an SSR (simple sequence repeat) molecular marker III for identifying descendant plants of Gala apple and application thereof. The SSR molecular marker III is characterized in that in the detection process, the SSR molecular markers linked with No.4, No.6, No.12 and No.14 chromosomes are simultaneously used, DNA (deoxyribonucleic acid) of an apple gene group is used as a PCR (polymerase chain reaction) amplification template, the SSR molecular markers are respectively used as primer pairs, and the PCR amplification and product detection are performed, so as to identify the genetics linkage, anther culture plants and variety sources. The SSR molecular marker III has the advantages that the SSR molecular marker is used for identifying the apple linkage group of the identifying material for the first time, and the SSR molecular markers linked with No.4, No.6, No.12 and No.14 chromosomes of the apple are disclosed; the molecular marker is a co-dominant marker, so that the Gala apple genetic linkage group, the anther culture plant and the Gala source plant are quickly and accurately identified; the molecular level support is provided for further accelerating the utilization of the Gala apple and important agronomic trait linkage gene, and the genetic breeding of apple homozygous plants.
Owner:POMOLOGY INST SHANXI ACAD OF AGRI SCI +2
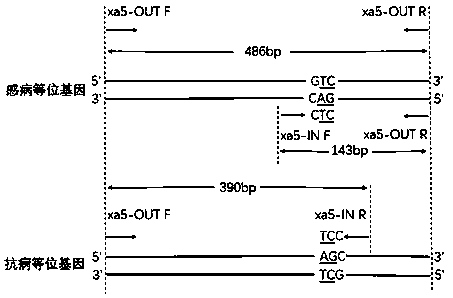
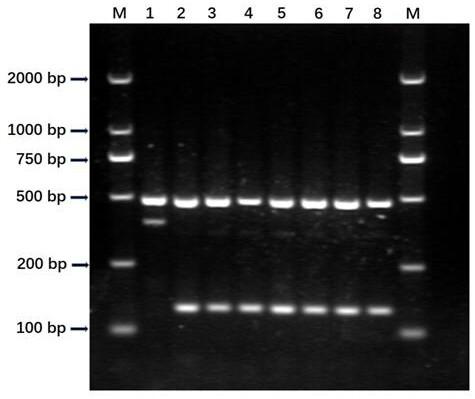
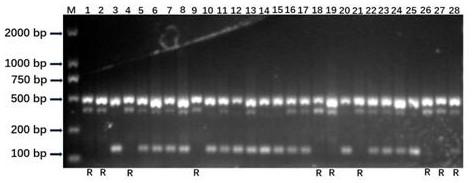
Rice bacterial blight resistance gene xa5 specific molecular marker and application thereof
InactiveCN109207631AImprove accuracyShorten the breeding cycleMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCO-DOMINANT
The invention discloses the development and the application of a rice bacterial blight resistance gene xa5 specific molecular marker. Four molecular marker primers were designed and added to the samePCR reaction system to amplify different rice genomic DNA. If there are two characteristic bands of 486bp and 390bp, it indicates that the sample is homozygous for xa5 gene. If there are two characteristic bands of 486 bp and 143 bp, the sample does not contain xa5 gene; If three characteristic bands of 486 bp, 390 bp and 143 bp are simultaneously present, it indicates that the sample is xa5 geneheterozygous. The four-primer PCR molecular marker is a co-dominant marker, which can effectively distinguish three different genotypes of rice bacterial blight resistance gene xa5, improve the selection efficiency of the gene, and accelerate the improvement and breeding process of rice bacterial blight resistance.
Owner:SHANGHAI AGROBIOLOGICAL GENE CENT
Specific SNP (Single Nucleotide Polymorphism) co-dominant molecular marker primer in rice brown planthopper resistance gene BPH9 gene and application
ActiveCN107488731AControl scaleImprove selection efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceCO-DOMINANT
The invention discloses a specific SNP (Single Nucleotide Polymorphism) co-dominant molecular marker primer in a rice brown planthopper resistance gene BPH9 and belongs to the field of plant molecular breeding. The marker primer and a sequence thereof are shown as follows: BPH9-IF: ACCATTGTTAGGCAGTTGTTCA; BPH9-ER: ATTCGACTCCCTTTCTTGTTATCT; BPH9-IR: CAGCCTCCTGAAGAGATCTTTCA; BPH9-EF: AATGTCGCACCCAGCAGC. The primer disclosed by the invention is used for carrying out PCR (Polymerase Chain Reaction) amplification on a rice DNA (Deoxyribonucleic Acid) sample and can be used for rapidly judging a gene type of rice BPH9 to be detected; the method is simple, convenient and rapid, is low in cost and can be widely applied to molecular marker assisted selective breeding or identification of BPH9 gene types of rice germplasm resources.
Owner:湖南隆平高科种业科学研究院有限公司 +2
Co-dominant SSR markers closely linked to tobacco TMV resistant gene N and application of co-dominant SSR markers
ActiveCN106498068ALow costMicrobiological testing/measurementDNA/RNA fragmentationCO-DOMINANTResistant genes
The invention discloses co-dominant SSR markers closely linked to tobacco TMV resistant gene N and application of the co-dominant SSR markers. Serial numbers of the co-dominant SSR markers are TM508-007 and TM508-118, and nucleotide sequences of amplified products of the co-dominant SSR markers are shown as SEQ ID No.1, SEQ ID No.2, SEQ ID No.3 and SEQ ID No.4 respectively. The application refers to application of the co-dominant SSR markers in detecting whether tobacco genome DNA contains the tobacco TMV resistance gene N or not. The co-dominant SSR markers have the advantages of stability, reliability, simplicity, convenience, quickness and low cost, thereby being applicable to N gene molecular marker assisted selection in tobacco TMV disease resistant breeding.
Owner:YUNNAN ACAD OF TOBACCO AGRI SCI
Primer pair, castanopsis hystrix SSR7 (Simple Sequence Repeat 7) marker and preparation method and application thereof
InactiveCN103740706AGood repeatabilityImprove reliabilityMicrobiological testing/measurementDNA preparationGenetic diversityCastanopsis hystrix
The invention discloses a primer pair, a castanopsis hystrix SSR7 (Simple Sequence Repeat 7) marker and a preparation method and application thereof. According to the preparation method, the total DNA (Deoxyribonucleic Acid) of castanopsis hystrix is subjected to PCR (Polymerase Chain Reaction) amplification by using the specific primer pair HZ7, thereby obtaining the SSR7 marker. The method is simple and feasible and is simple and convenient in operation; shown by tests which are carried out in members of two natural castanopsis hystrix populations and among the members, the obtained SSR has relatively high polymorphism. The castanopsis hystrix SSR7 marker is a co-dominant marker; compared with other molecular markers, the SSR marker is good in repeatability and high in reliability, can be applied to the genetic diversity evaluation of castanopsis hystrix, the genetic map construction of castanopsis hystrix, the research on the spatial distribution pattern of the genetic variation of castanopsis hystrix population and the origination and evolution of castanopsis hystrix and the standardization of target genes, and can also be applied to the molecular marker assisted breeding and QTL (Quantitative Trait Locus) research of castanopsis hystrix.
Owner:GUANGXI FORESTRY RES INST
SSR (simple sequence repeat) marker for tomato yellow leaf curl disease resisting character co-segregation and application thereof
InactiveCN106048089AMicrobiological testing/measurementMicroorganism based processesDiseaseAgricultural science
The invention discloses an SSR (simple sequence repeat) marker for tomato yellow leaf curl disease resisting character co-segregation and application thereof. The SSR marker includes primers: upstream primer T5-F: 5'-CAATCAATCGATGCACAAAACACC-3'; downstream primer T5-R: 5'-GACTGACTGCATTGGATTTGGCTT-3'. The marker can be amplified in CLN32120a-23 into a 580bp strip, can be amplified in Money marker into a 640bp strip and can be amplified in F1 into the above two strips, marking results are substantially consistent to that of field identification, it is proved that the marker can distinguish anti-disease materials, disease-sensitive materials and hybrid anti-disease materials and is a co-dominant marker in tight linkage with tomato yellow leaf curl disease virus resisting gene ty-5. Molecular marker detection is 90% matching with field detection in terms of detection results. The marker can be used in PCR (polymerase chain reaction) quick detection for tomato yellow leaf curl virus resisting ty-5 gene, and certain theoretical and practical basis is provided for the application of ty-5 molecular marker assisted selective breeding.
Owner:NORTHEAST AGRICULTURAL UNIVERSITY
Zelkova SSR1 marker and primer pair and preparation methods and application thereof
InactiveCN104450693AGood repeatabilityImprove reliabilityMicrobiological testing/measurementDNA/RNA fragmentationGenetic diversitySimple Repetitive Sequences
The invention discloses a Zelkova SSR1 marker and a primer pair and preparation methods and applications thereof. PCR amplification is carried out on Zelkova total DNAs of JS1 by an inventor utilizing a specific primer, so that the SSR1 marker with a simple and repeated sequence is obtained. The method is simple and practicable and simple and convenient to operate, the obtained simple and repeated sequence shows higher polymorphism for detection inside or among populations of three Zelkova natural colonies. The Zelkova SSR1 marker is a co-dominant marker; compared with other molecular markers, the SSR marker is good in repeatability and high in reliability, can be applied to Zelkova genetic diversity evaluation, Zelkova genetic map construction, Zelkova population heritable variation spatial distribution pattern and Zelkova origin and evolution research as well as target gene marking, and can also be applied to Zelkova molecular marker assistant breeding and QTL (quantitative Trait Locus) research.
Owner:CENTRAL SOUTH UNIVERSITY OF FORESTRY AND TECHNOLOGY
Primer pair, castanopsis hystrix SSR5 (Simple Sequence Repeat 5) marker and preparation method and application thereof
InactiveCN103740703AGood repeatabilityImprove reliabilityMicrobiological testing/measurementDNA preparationGenetic diversityCastanopsis hystrix
The invention discloses a primer pair, a castanopsis hystrix SSR5 (Simple Sequence Repeat 5) marker and a preparation method and application thereof. According to the preparation method, the total DNA (Deoxyribonucleic Acid) of castanopsis hystrix is subjected to PCR (Polymerase Chain Reaction) amplification by using the specific primer pair HZ5, thereby obtaining the SSR5 marker. The method is simple and feasible and is simple and convenient in operation; shown by tests which are carried out in members of two natural castanopsis hystrix populations and among the members, the obtained SSR has relatively high polymorphism. The castanopsis hystrix SSR5 marker is a co-dominant marker; compared with other molecular markers, the SSR marker is good in repeatability and high in reliability, can be applied to the genetic diversity evaluation of castanopsis hystrix, the genetic map construction of castanopsis hystrix, the research on the spatial distribution pattern of the genetic variation of castanopsis hystrix population and the origination and evolution of castanopsis hystrix and the standardization of target genes, and can also be applied to the molecular marker assisted breeding and QTL (Quantitative Trait Locus) research of castanopsis hystrix.
Owner:GUANGXI FORESTRY RES INST
Primer pair, castanopsis hystrix SSR3 (Simple Sequence Repeat 3) marker and preparation method and application thereof
InactiveCN103740705AGood repeatabilityImprove reliabilityMicrobiological testing/measurementDNA preparationGenetic diversityCastanopsis hystrix
The invention discloses a primer pair, a castanopsis hystrix SSR3 (Simple Sequence Repeat 3) marker and a preparation method and application thereof. According to the preparation method, the total DNA (Deoxyribonucleic Acid) of castanopsis hystrix is subjected to PCR (Polymerase Chain Reaction) amplification by using the specific primer pair HZ3, thereby obtaining the SSR3 marker. The method is simple and feasible and is simple and convenient in operation; shown by tests which are carried out in members of two natural castanopsis hystrix populations and among the members, the obtained SSR has relatively high polymorphism. The castanopsis hystrix SSR3 marker is a co-dominant marker; compared with other molecular markers, the SSR marker is good in repeatability and high in reliability, can be applied to the genetic diversity evaluation of castanopsis hystrix, the genetic map construction of castanopsis hystrix, the research on the spatial distribution pattern of the genetic variation of castanopsis hystrix population and the origination and evolution of castanopsis hystrix and the standardization of target genes, and can also be applied to the molecular marker assisted breeding and QTL (Quantitative Trait Locus) research of castanopsis hystrix.
Owner:GUANGXI FORESTRY RES INST
Method for identifying reality and purity of pepper male sterile three-line mating hybrids based on InDel (insertion-deletion) molecular markers
ActiveCN108893555AReduce filter rangeIdentification achievedMicrobiological testing/measurementDNA/RNA fragmentationInsertion deletionCO-DOMINANT
The invention provides a method for identifying reality and purity of pepper male sterile three-line mating hybrids based on InDel (insertion-deletion) molecular markers. The method includes the steps: culturing the pepper hybrids and parents to a bud and seedling stage; extracting DNA (deoxyribonucleic acid); screening the hybrids and the parents by core primers. Primers of co-dominant markers can serve as molecular markers for identifying the reality of the hybrids, and co-dominant markers of the pepper hybrids and the parents continues to be screened by combining a secondary core primer library until the co-dominant markers are screened out if the co-dominant markers of the pepper hybrids and the parents cannot be screened out in a core primer library. According to the method, a systemcapable of rapidly and simultaneously identifying the reality and the purity of the pepper male sterile three-line mating hybrids is built and cannot be limited by time and seasons, indoor rapid identification of the reality and the purity of the hybrids is achieved, and molecular biology bases are provided for protection and market supervision of the pepper male sterile three-line mating hybrids.
Owner:QINGDAO AGRI UNIV
SSR (simple sequence repeat) molecular marker IV for identifying descendant plants of Gala apple and application thereof
InactiveCN106755478ARapid identificationAccurate identificationMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGermplasm
The invention belongs to the field of innovation and research of plant genetics breeding and apple germplasm, in particular to an SSR (simple sequence repeat) molecular marker IV for identifying descendant plants of Gala apple and application thereof. The SSR molecular marker IV is characterized in that in the detection process, the SSR molecular markers linked with No.5 and No.10 chromosomes are simultaneously used, DNA (deoxyribonucleic acid) of an apple gene group is used as a PCR (polymerase chain reaction) amplification template, the SSR molecular markers are respectively used as primer pairs, and the PCR amplification and product detection are performed, so as to identify the genetics linkage, anther culture plants and variety sources. The SSR molecular marker IV has the advantages that the SSR molecular marker is used for identifying the apple linkage group of the identifying material for the first time, and the SSR molecular markers linked with No.5 and No.10 chromosomes of the apple are disclosed; the molecular marker is a co-dominant marker, so that the Gala apple genetic linkage group, the anther culture plant and the Gala source plant are quickly and accurately identified; the molecular level support is provided for further accelerating the utilization of the Gala apple and important agronomic trait linkage gene, and the genetic breeding of apple homozygous plants.
Owner:山西省农业科学院农业资源与经济研究所 +2
Specific molecular marker of Brassica rapa L.ssp pekinensis eIF (iso) 4E.a locus large fragment deletion mutation and application thereof
InactiveCN103103187ARich genetic backgroundImprove screening efficiencyMicrobiological testing/measurementDNA/RNA fragmentationCO-DOMINANTBrassica rapa
The invention discloses a locus specific co-dominant ASM (allele specific marker) directly related to identification of Brassica rapa L.ssp pekinensis eIF (iso) 4E.a wild type and mutant. The marker related to wild type locus detection is named as ASM-iso4E.a, has fragment size of 1636bp, and is shown as a SEQ ID No.1. The corresponding mutation locus detection related marker is named as ASM-iso4e.a, has fragment size of 640bp, and is shown as a SEQ ID No.2. In the invention, by making use of a homology based cloning technology, a mutant of the eIF (iso) 4E.a locus is discovered, and the molecular marker for identifying the locus is developed. The marker can be utilized for accurate selection of the genotype of backcrossing transformation progenies. Meanwhile, the marker also can be used for screening Brassica rapa L.ssp pekinensis germplasm resources so as to seek mutant materials with more abundant genetic backgrounds.
Owner:VEGETABLE RES INST OF SHANDONG ACADEMY OF AGRI SCI
Plant-anthocyanin synthesis control gene and application thereof
ActiveCN108018290AImprove detection accuracyEasy to operateMicrobiological testing/measurementPlant peptidesAgricultural scienceIntein
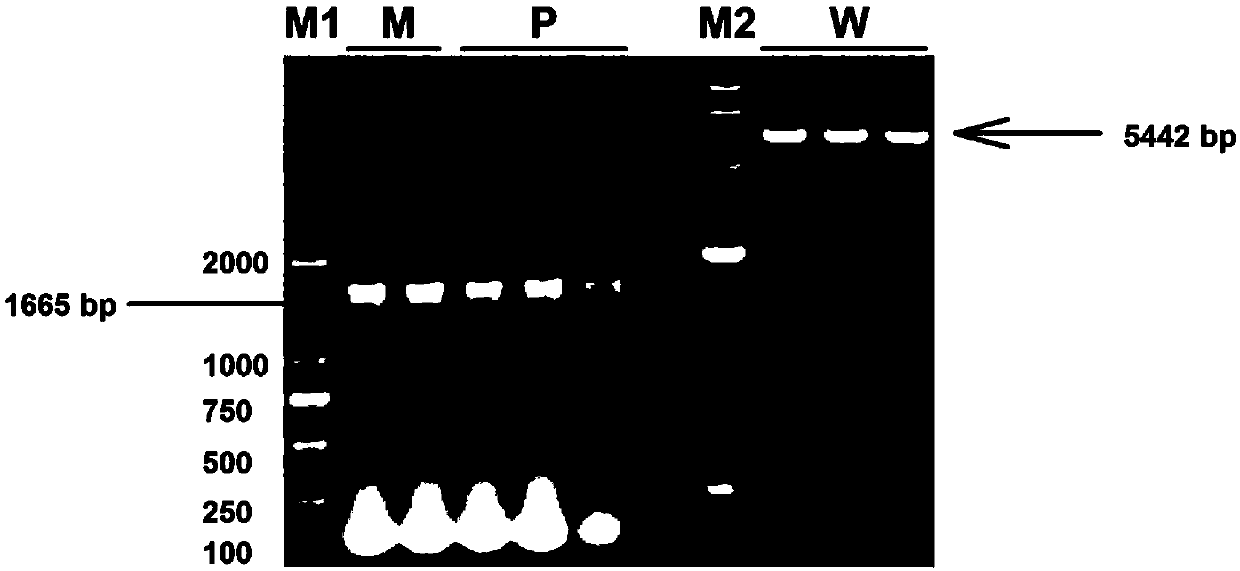
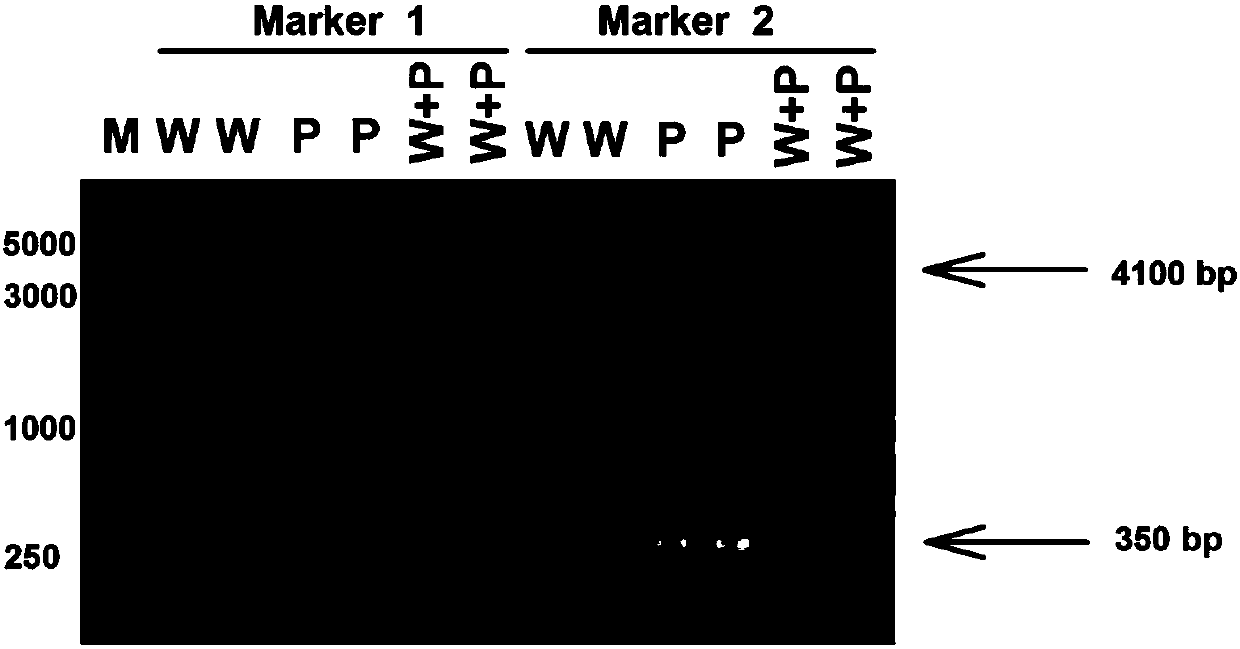
The invention discloses a plant-anthocyanin synthesis control gene and application thereof. The sequence of the disclosed gene is shown in the SEQ ID No.1. A 3,772-bp klenow-fragment insertion mutation of the gene exists in first intron of white heading brassica rapa, and the gDNA sequence of the mutation is shown in the SEQ ID No.5. According to the mutation in the intron in the gDNA nucleotide sequence of the gene, three specificity PCR primers of the Marker 1, the Marker 2 and the Marker 8 are designed, wherein the Marker 1 and the Marker 2 are co-dominant markers, and the Marker 8 is a maternal marker. The DNA of the target brassica rapa is subjected to PCR amplification and identified through agarose gel electrophoresis, drinamyl hybrid strains and homozygous strains in segregation population can be rapidly identified through the Marker 1 and the Marker 2, and the drinamyl hybrid strains and the homozygous strains in the segregation population can also be distinguished through theMarker 8 in cooperation with the heading characteristics.
Owner:NORTHWEST A & F UNIV
Primer pair, castanopsis hystrix SSR4 (Simple Sequence Repeat 4) marker and preparation method and application thereof
InactiveCN103740704AGood repeatabilityImprove reliabilityMicrobiological testing/measurementDNA preparationGenetic diversityCastanopsis hystrix
The invention discloses a primer pair, a castanopsis hystrix SSR4 (Simple Sequence Repeat 4) marker and a preparation method and application thereof. According to the preparation method, the total DNA (Deoxyribonucleic Acid) of castanopsis hystrix is subjected to PCR (Polymerase Chain Reaction) amplification by using the specific primer pair HZ4, thereby obtaining the SSR4 marker. The method is simple and feasible and is simple and convenient in operation; shown by tests which are carried out in members of two natural castanopsis hystrix populations and among the members, the obtained SSR has relatively high polymorphism. The castanopsis hystrix SSR4 marker is a co-dominant marker; compared with other molecular markers, the SSR marker is good in repeatability and high in reliability, can be applied to the genetic diversity evaluation of castanopsis hystrix, the genetic map construction of castanopsis hystrix, the research on the spatial distribution pattern of the genetic variation of castanopsis hystrix population and the origination and evolution of castanopsis hystrix and the standardization of target genes, and can also be applied to the molecular marker assisted breeding and QTL (Quantitative Trait Locus) research of castanopsis hystrix.
Owner:GUANGXI FORESTRY RES INST
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com