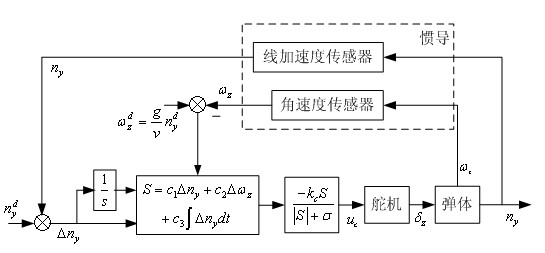
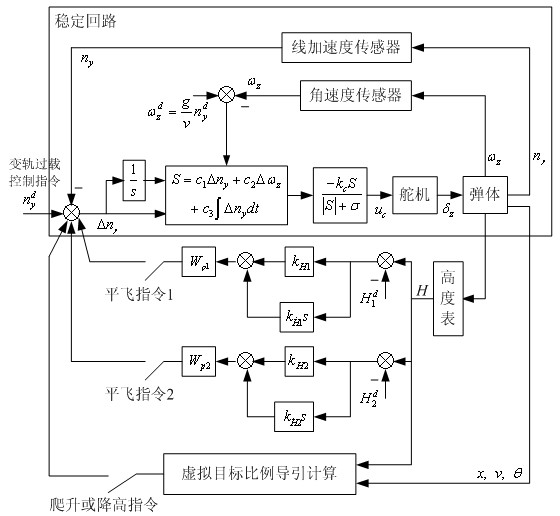
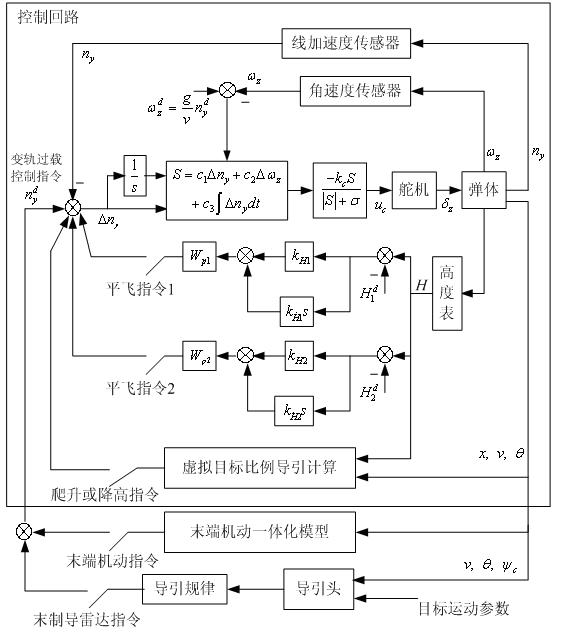
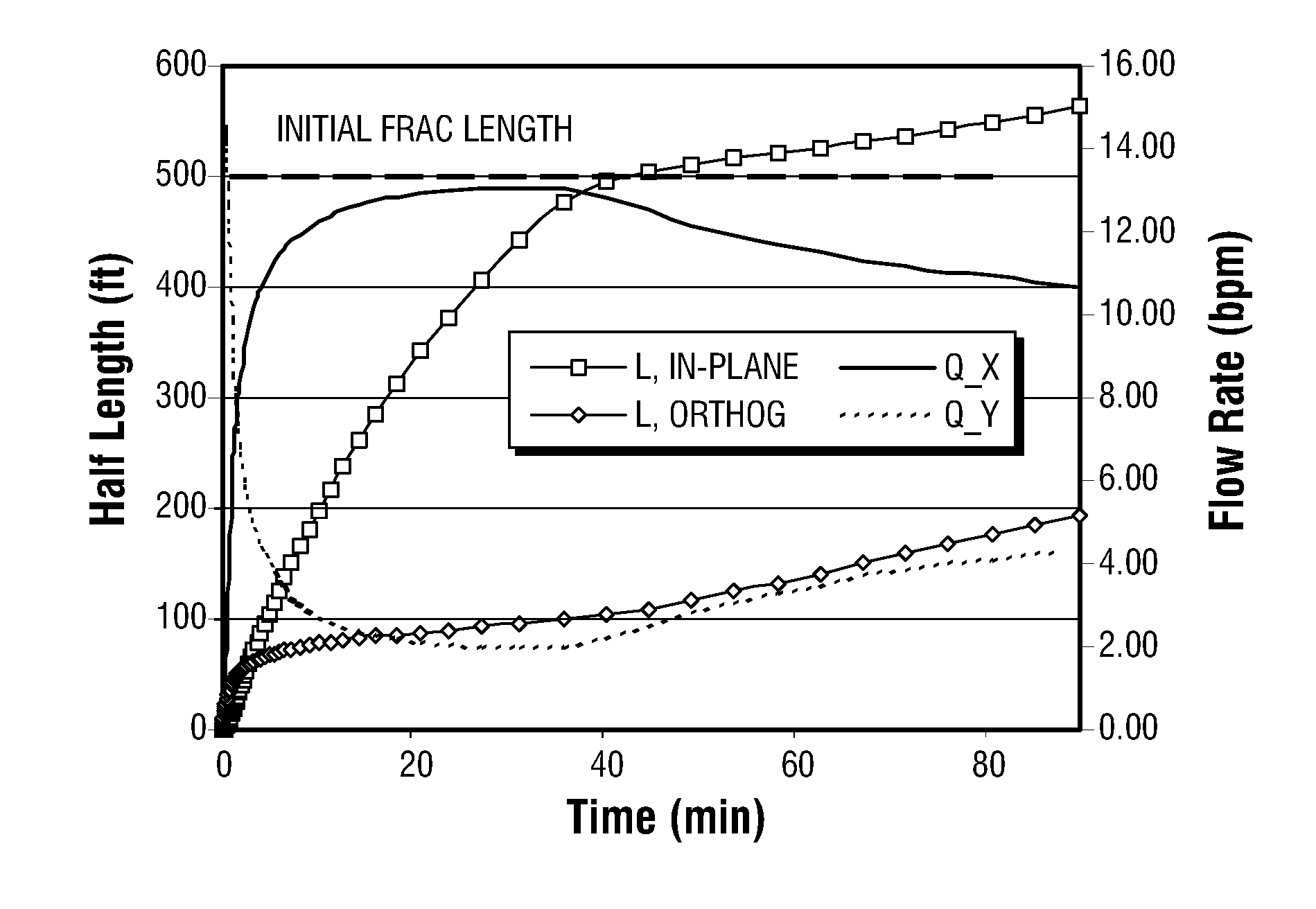
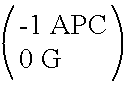Patents
Literature
208 results about "Mathematical equations" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
A mathematical equation is an expression containing at least one variable (=unknown value) and an " equals sign " ( = ) with a mathematical expression on each side of it. The equals sign says that both sides are exactly the same value. An equation can be as simple as x=0, or as hard as 4(3y^99)+76=42÷3x or harder. There are two...
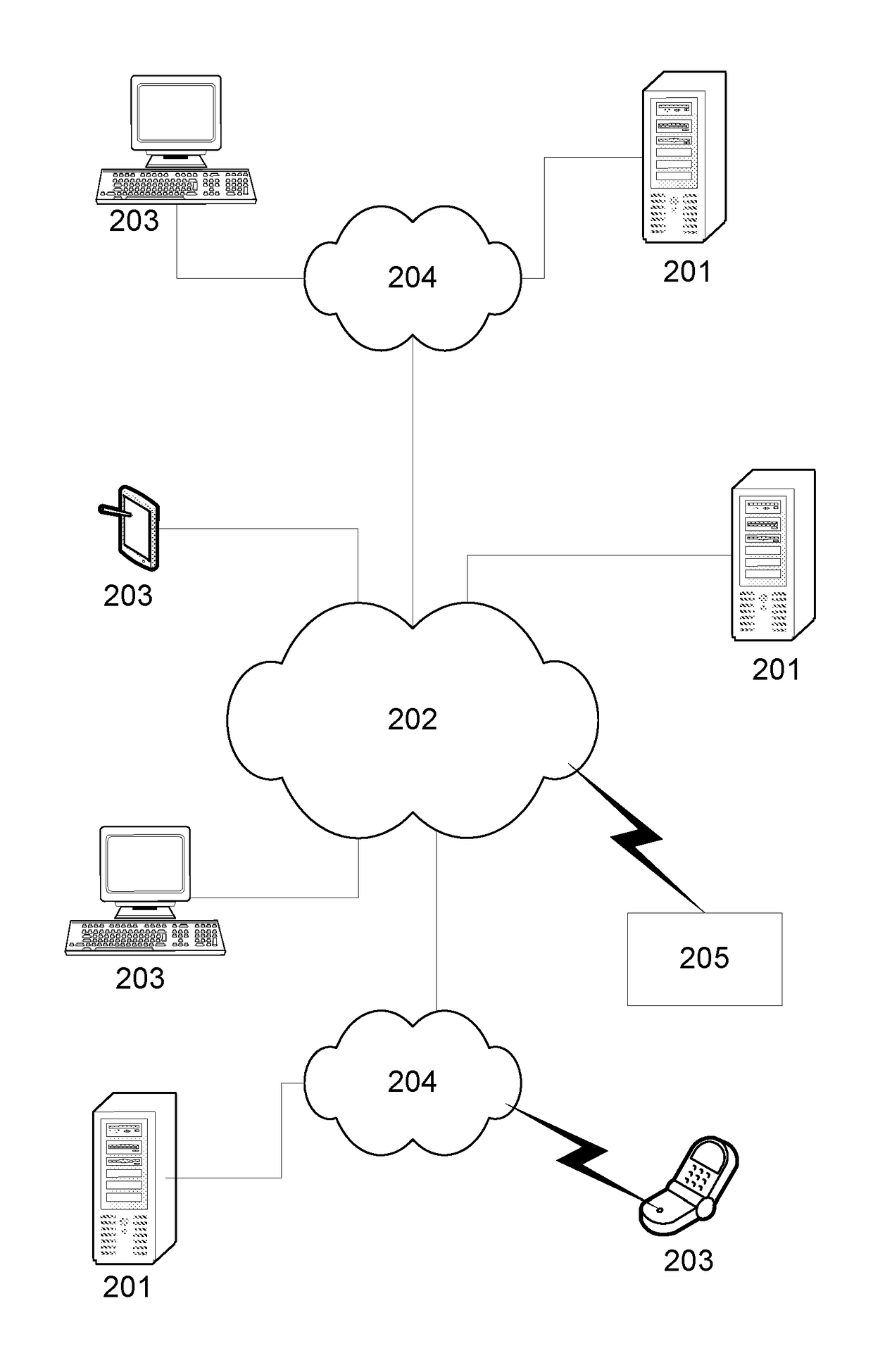
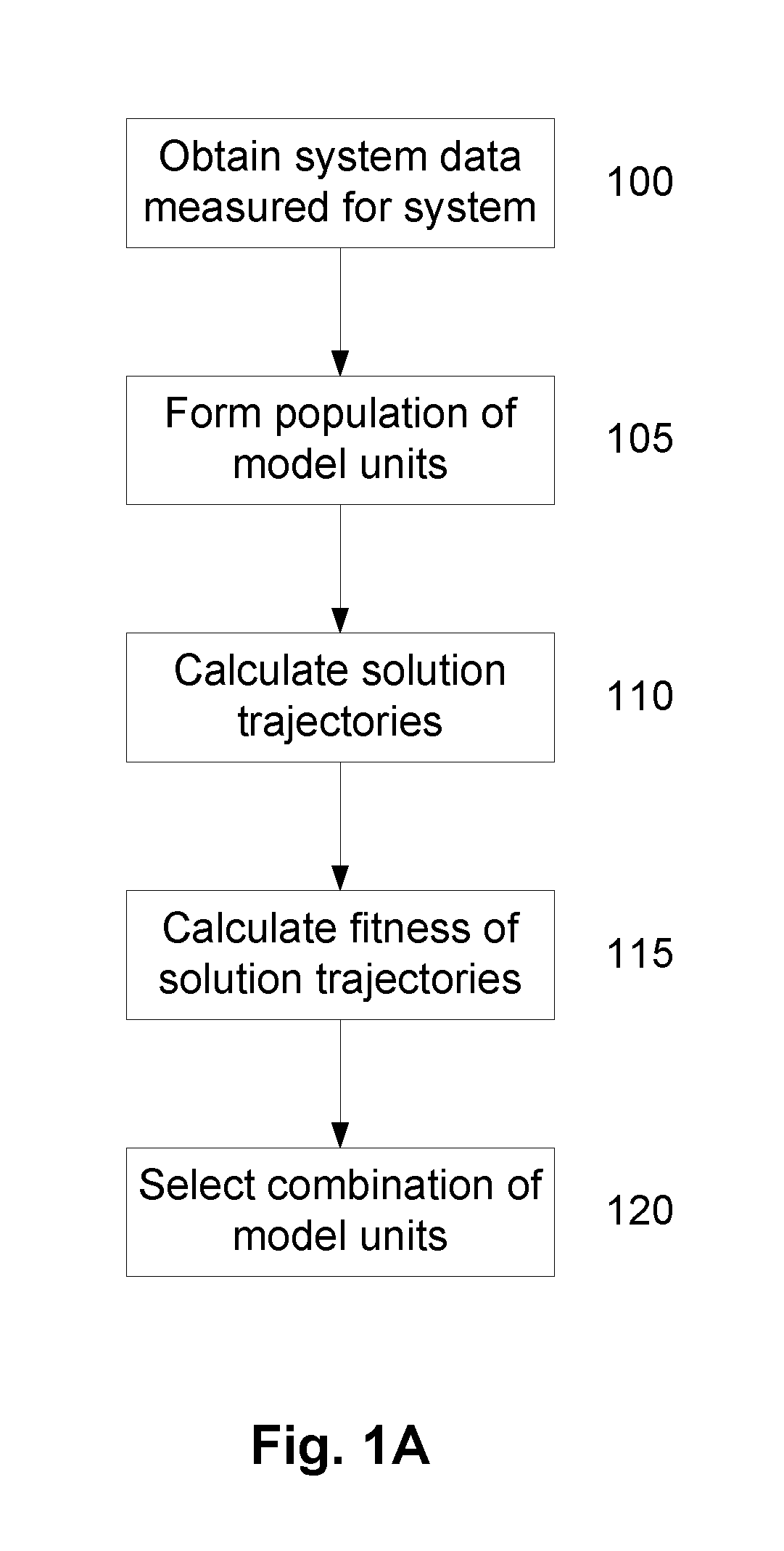
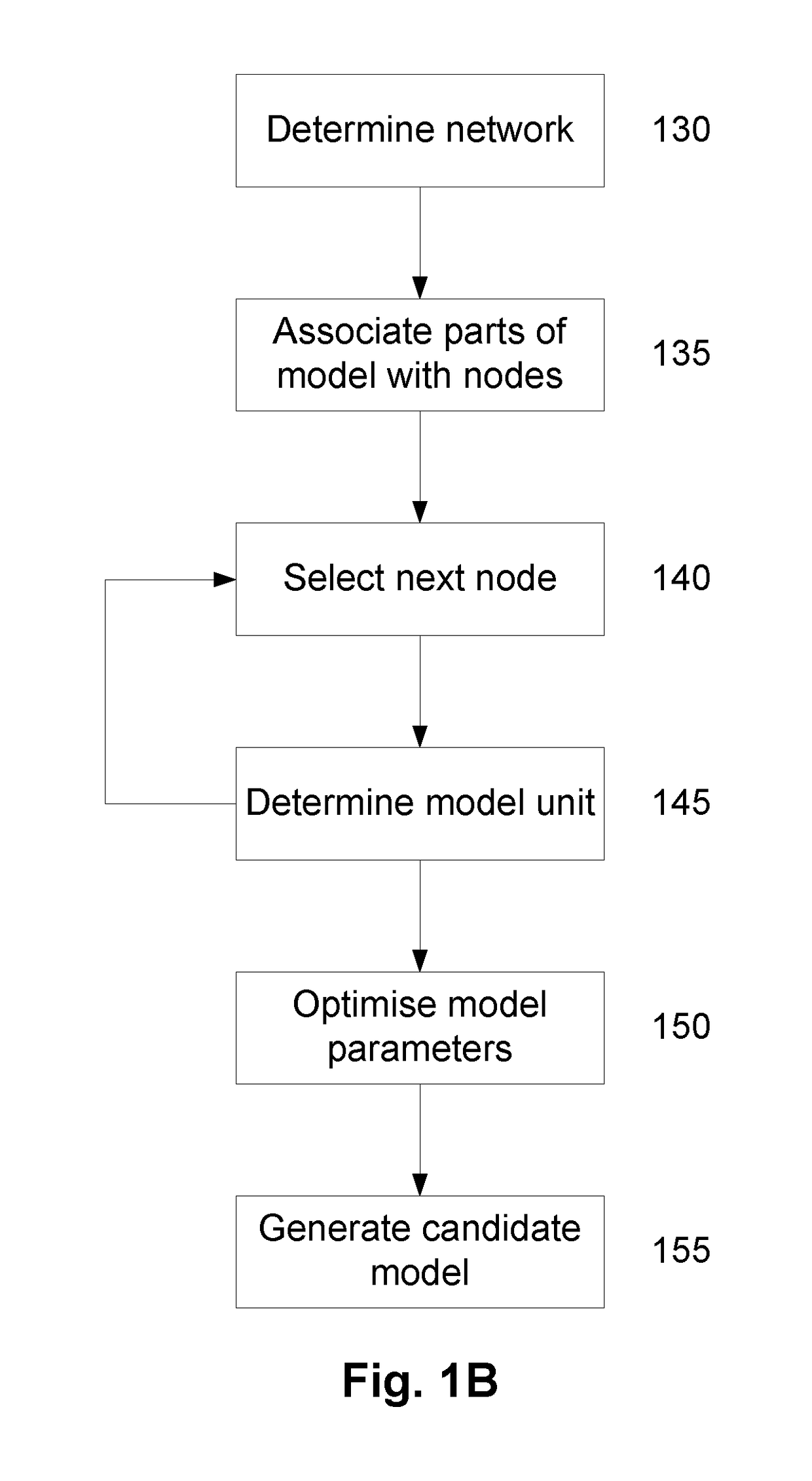
A System and Method for Modelling System Behaviour
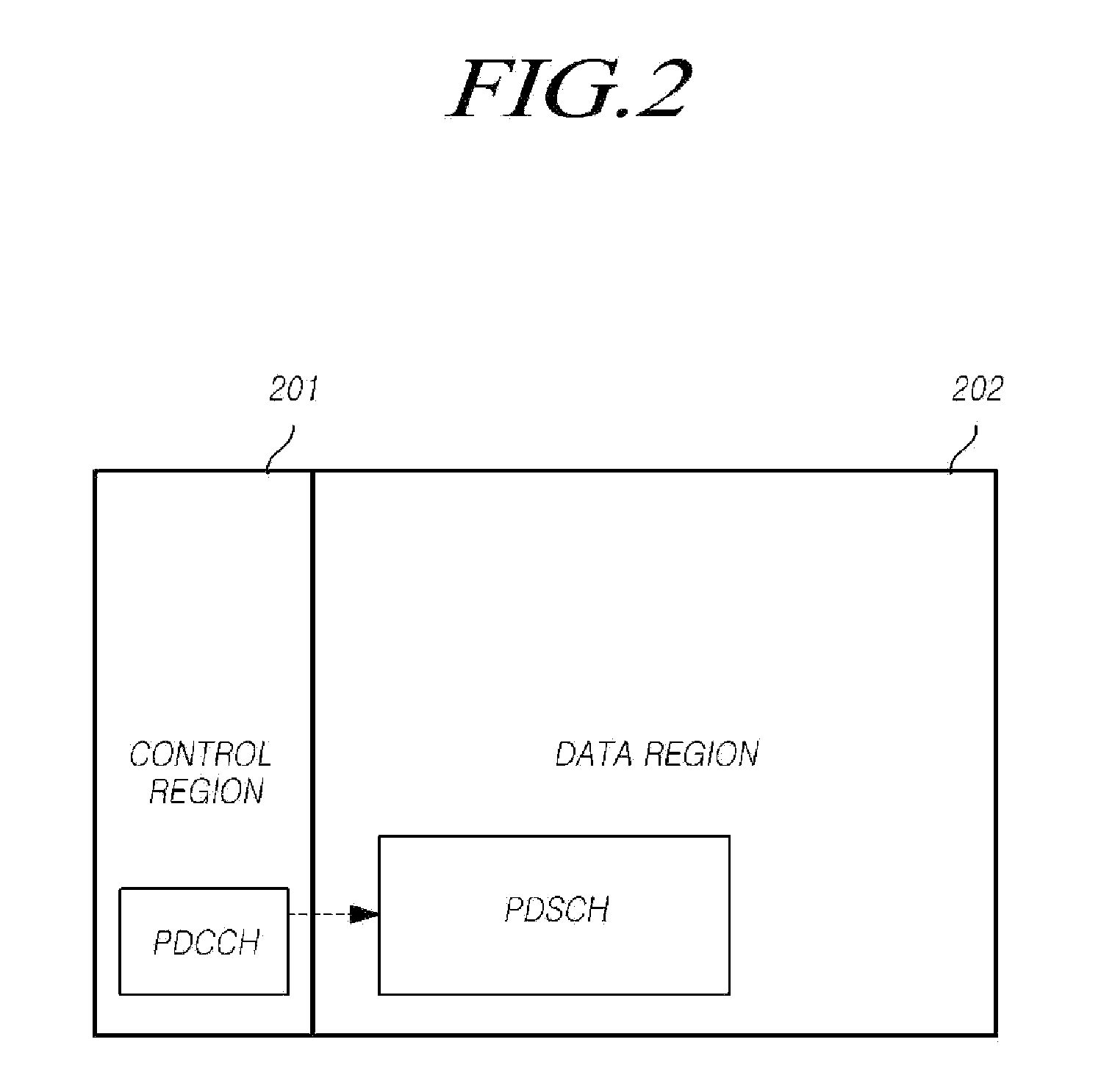
ActiveUS20170147722A1Reduce the impactReduce impactMedical simulationDesign optimisation/simulationCollective modelModel system
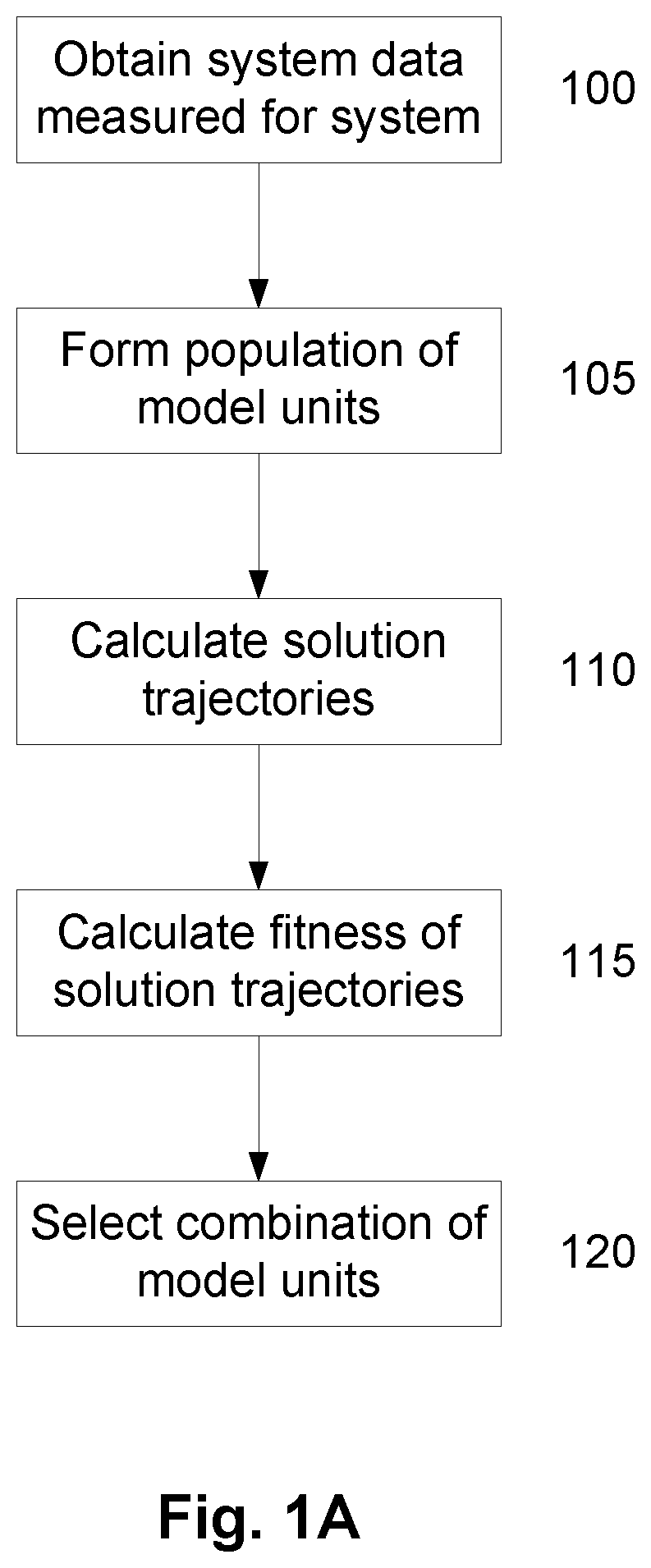
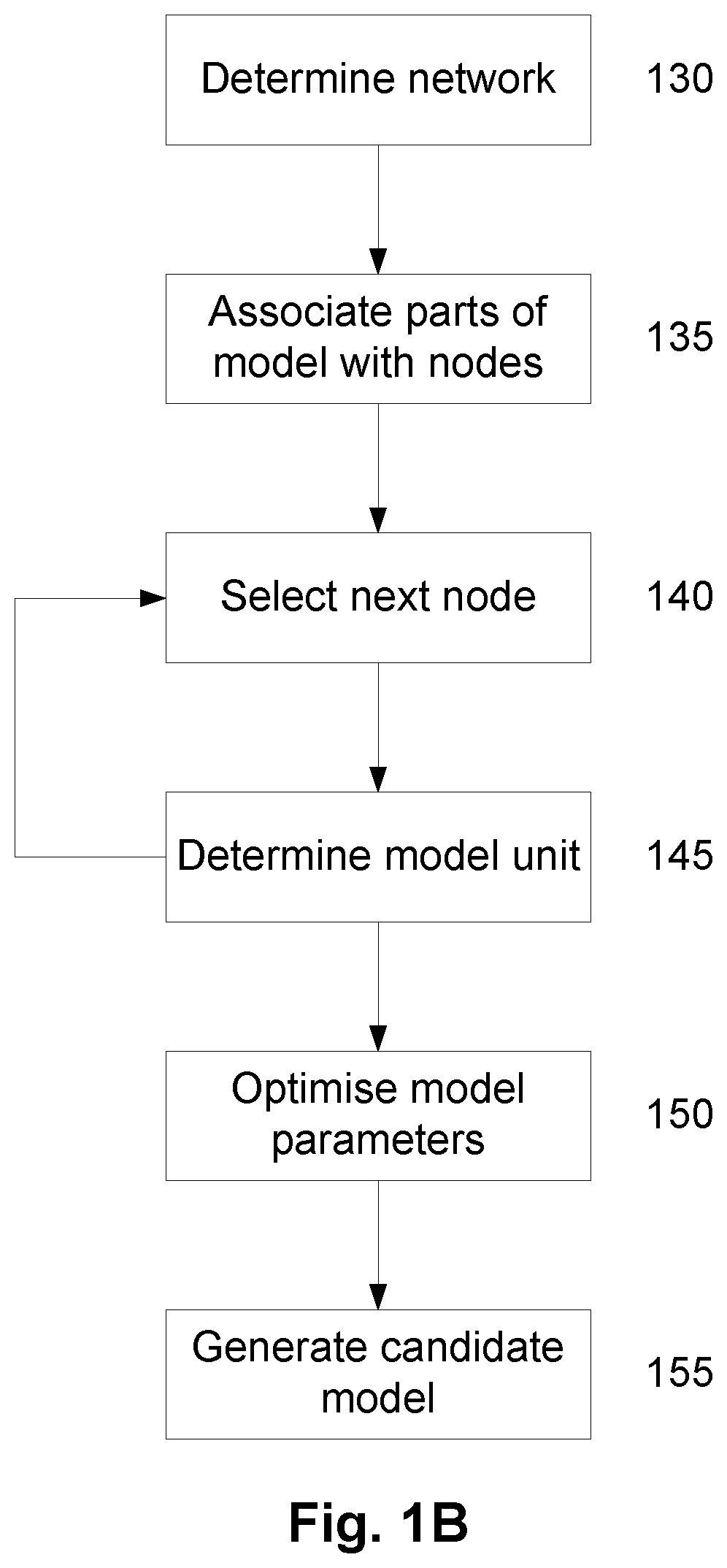
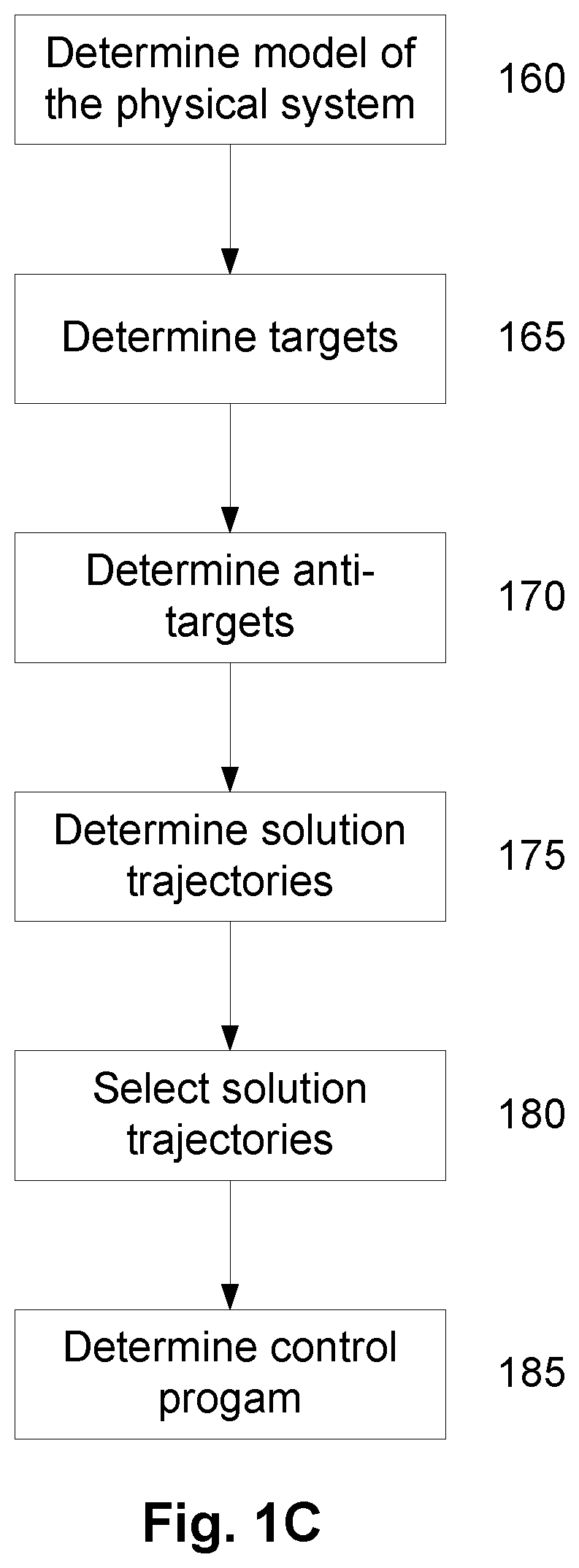
A method of modelling system behaviour of a physical system, the method including, in one or more electronic processing devices obtaining quantified system data measured for the physical system, the quantified system data being at least partially indicative of the system behaviour for at least a time period, forming at least one population of model units, each model unit including model parameters and at least part of a model, the model parameters being at least partially based on the quantified system data, each model including one or more mathematical equations for modelling system behaviour, for each model unit calculating at least one solution trajectory for at least part of the at least one time period; determining a fitness value based at least in part on the at least one solution trajectory; and, selecting a combination of model units using the fitness values of each model unit, the combination of model units representing a collective model that models the system behaviour.
Owner:EVOLVING MACHINE INTELLIGENCE
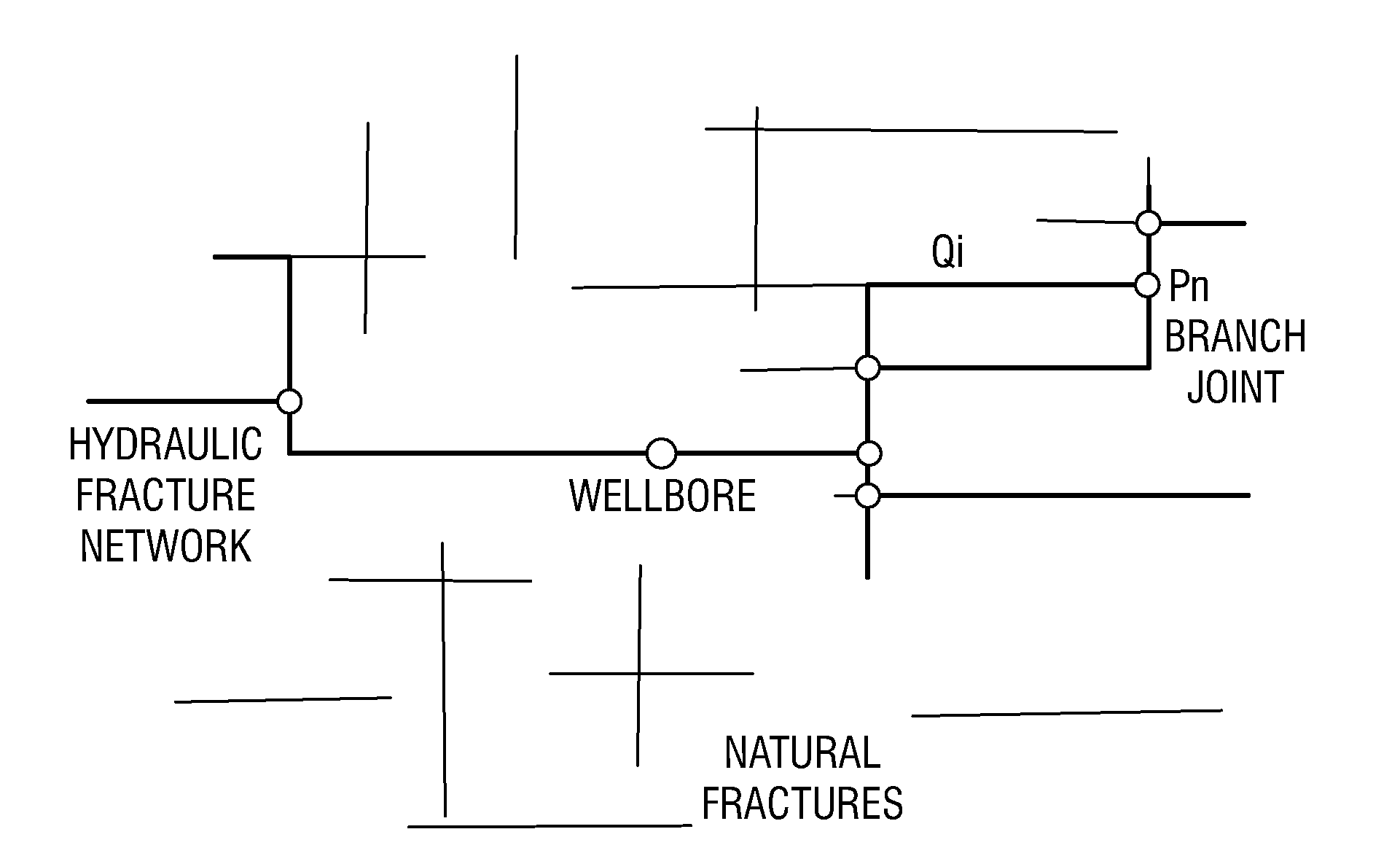
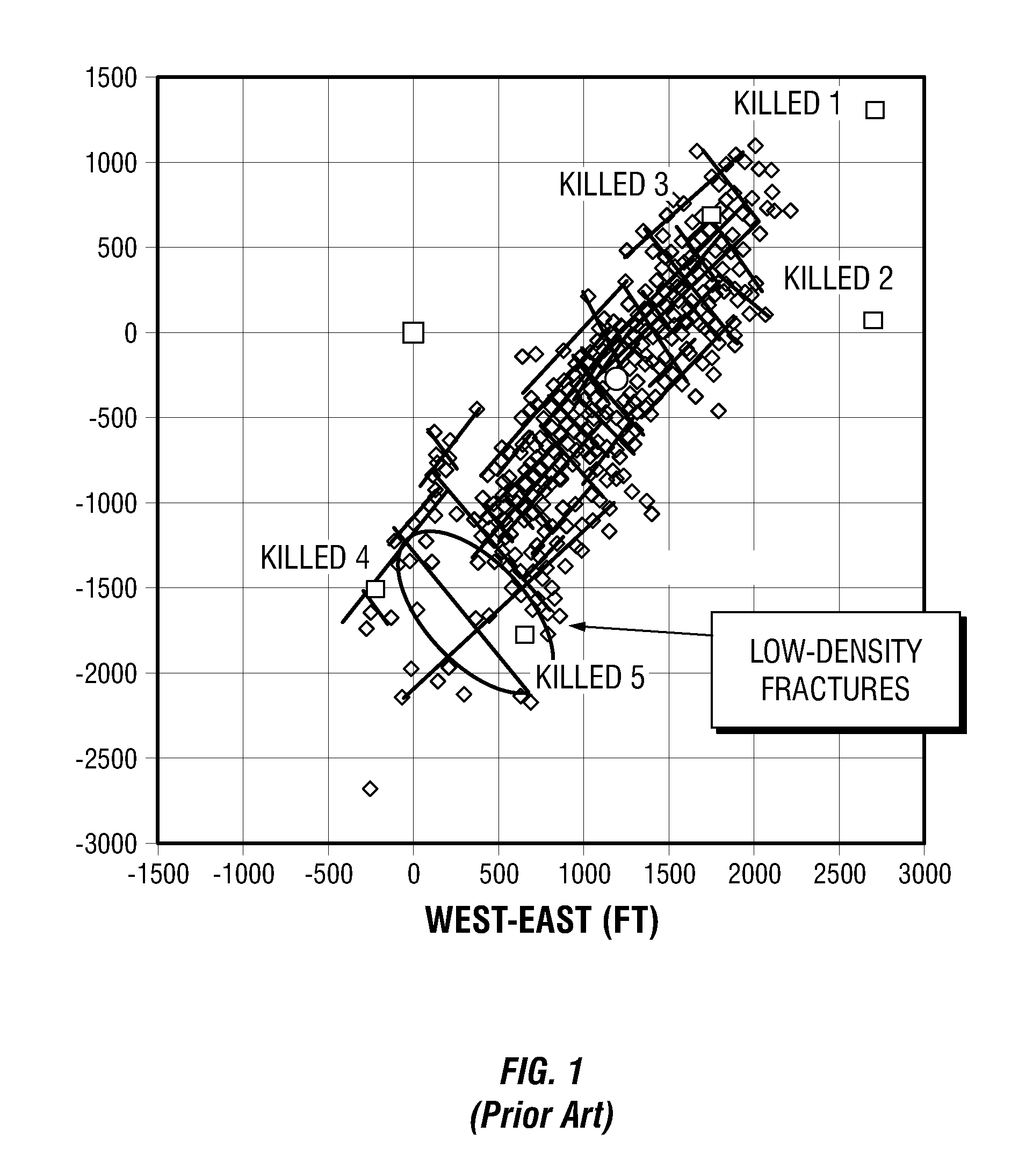
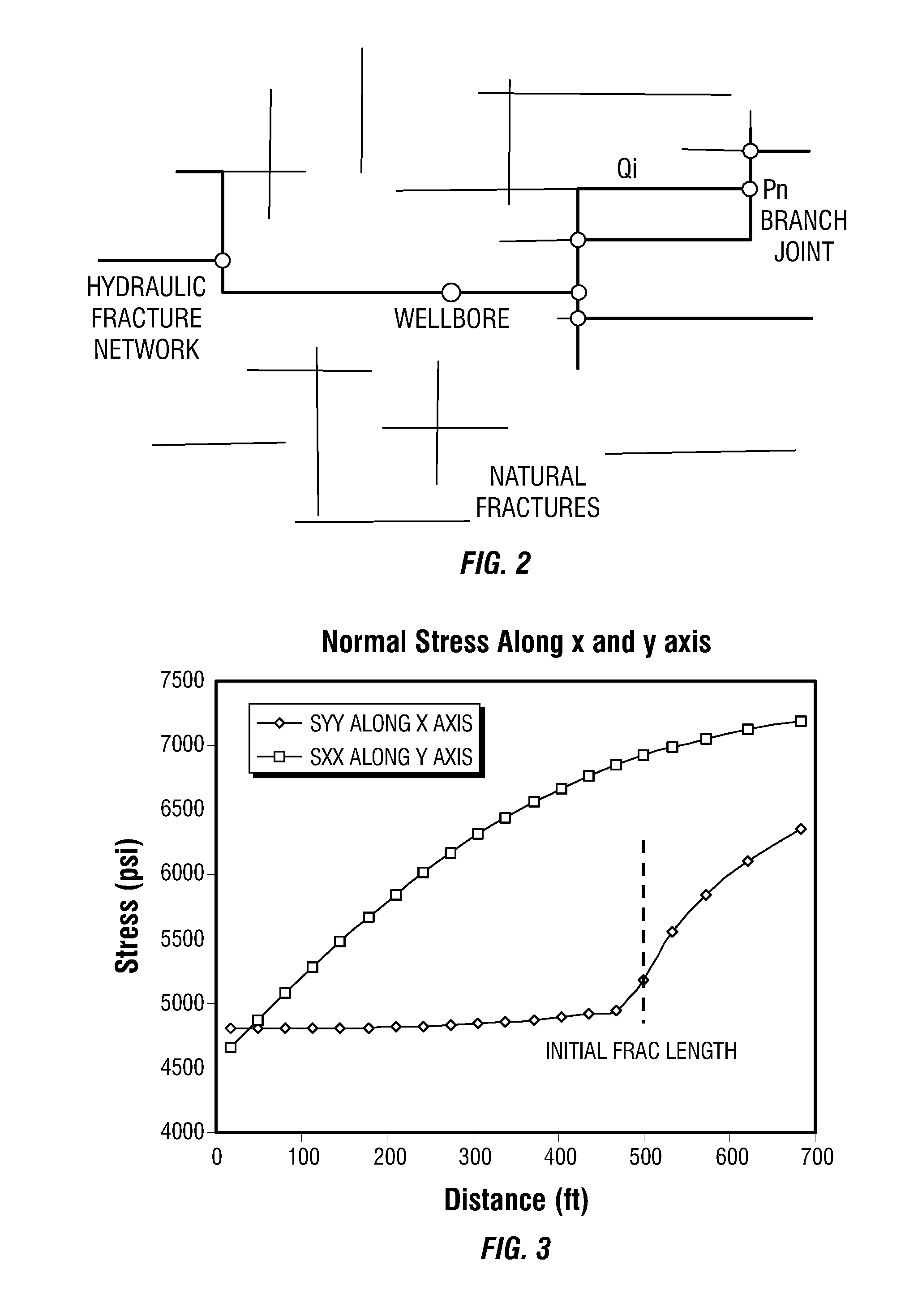
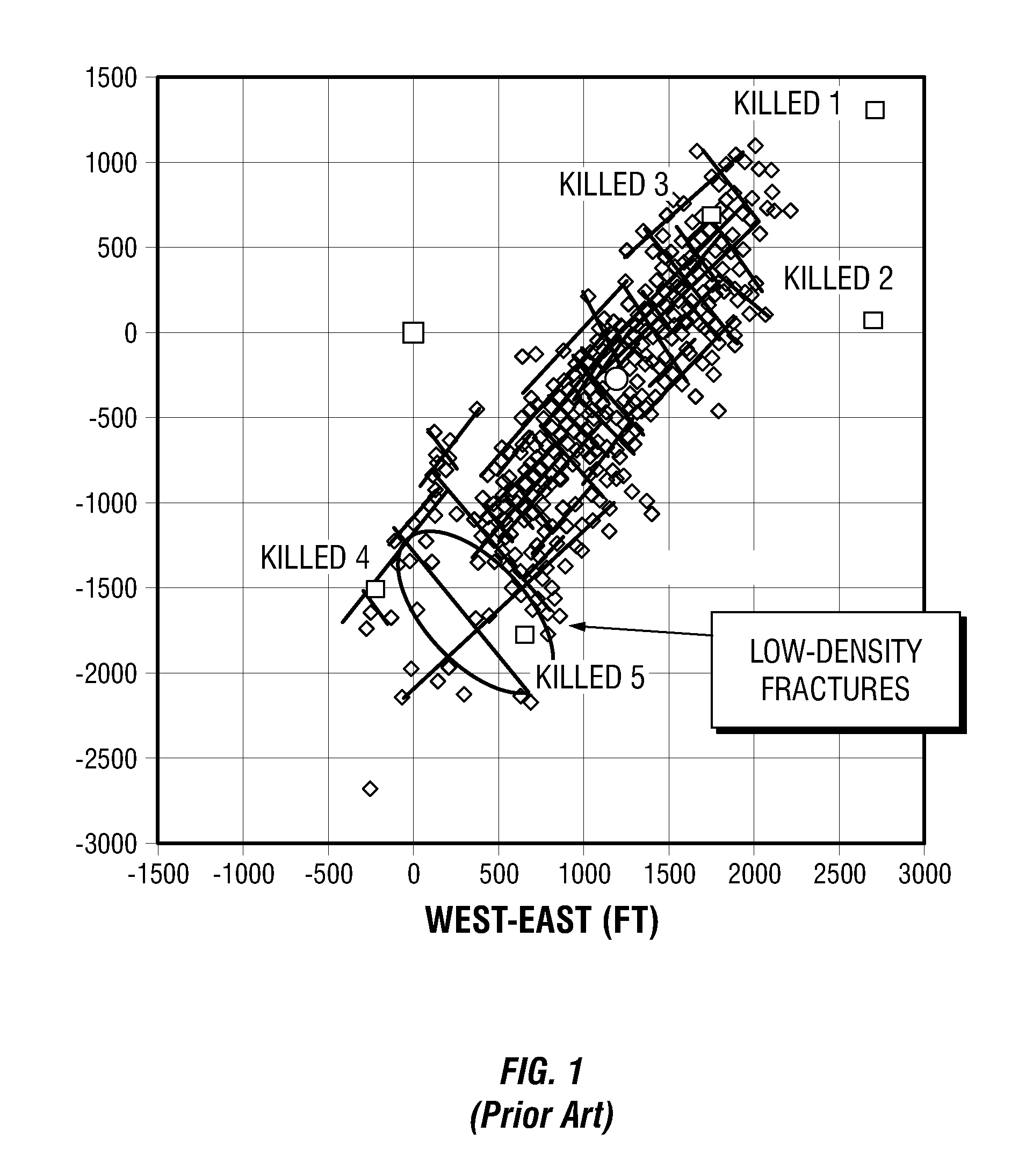
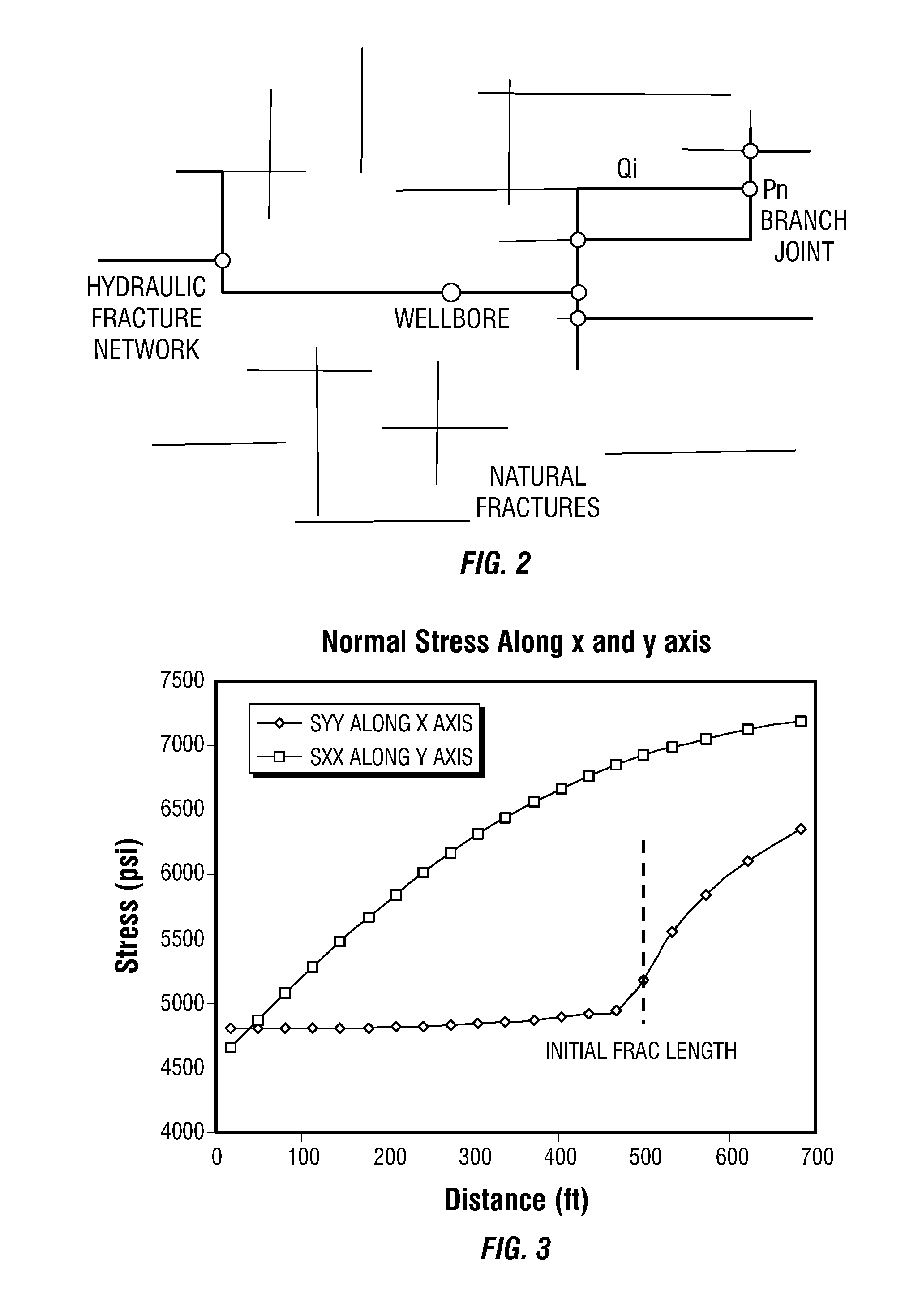
Simulations for Hydraulic Fracturing Treatments and Methods of Fracturing Naturally Fractured Formation
A hydraulic fracture design model that simulates the complex physical process of fracture propagation in the earth driven by the injected fluid through a wellbore. An objective in the model is to adhere with the laws of physics governing the surface deformation of the created fracture subjected to the fluid pressure, the fluid flow in the gap formed by the opposing fracture surfaces, the propagation of the fracture front, the transport of the proppant in the fracture carried by the fluid, and the leakoff of the fracturing fluid into the permeable rock. The models used in accordance with methods of the invention are typically based on the assumptions and the mathematical equations for the conventional 2D or P3D models, and further take into account the network of jointed fracture segments. For each fracture segment, the mathematical equations governing the fracture deformation and fluid flow apply. For each time step, the model predicts the incremental growth of the branch tips and the pressure and flow rate distribution in the system by solving the governing equations and satisfying the boundary conditions at the fracture tips, wellbore and connected branch joints. An iterative technique is used to obtain the solution of this highly nonlinear and complex problem.
Owner:SCHLUMBERGER TECH CORP
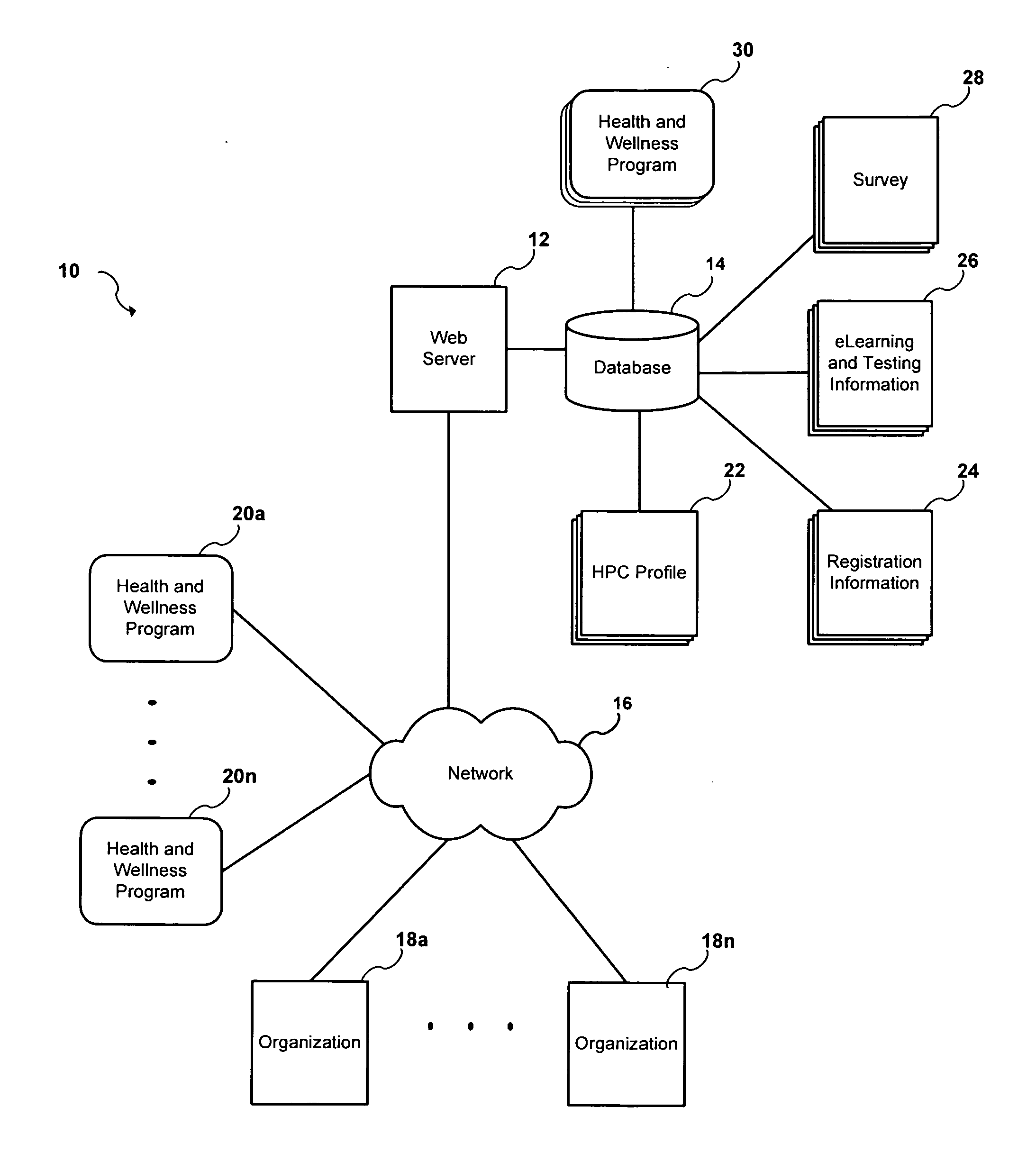
Method and system for managing health and wellness programs
InactiveUS20080177836A1Increase heightGood for healthComputer-assisted medical data acquisitionMultiple digital computer combinationsWeb pageComputer science
A system and method are provided that include a health and wellness system that includes a network configured to provide one or more members of an organization access to the health and wellness system, a web server configured to provide a plurality of web pages over the network, and a plurality of wellness programs. A survey that has a plurality of health and wellness categories is delivered to the one or more members via the plurality of web pages. The one or more members provide a response to the survey that has a plurality of numerical scores for each of the plurality of health and wellness categories. Each of the plurality of wellness programs are configured to improve the health and wellness in at least one of the plurality of health and wellness categories for at least one of the one or more members. Each of the plurality of wellness programs have a predefined numerical equation for each of the plurality of health and wellness categories. One or more of the plurality of wellness programs are identified in a readable format via the plurality of web pages by comparing results from the predefined mathematical equations using the numerical scores as input.
Owner:ORGAL WELLNESS & LEARNING SYST
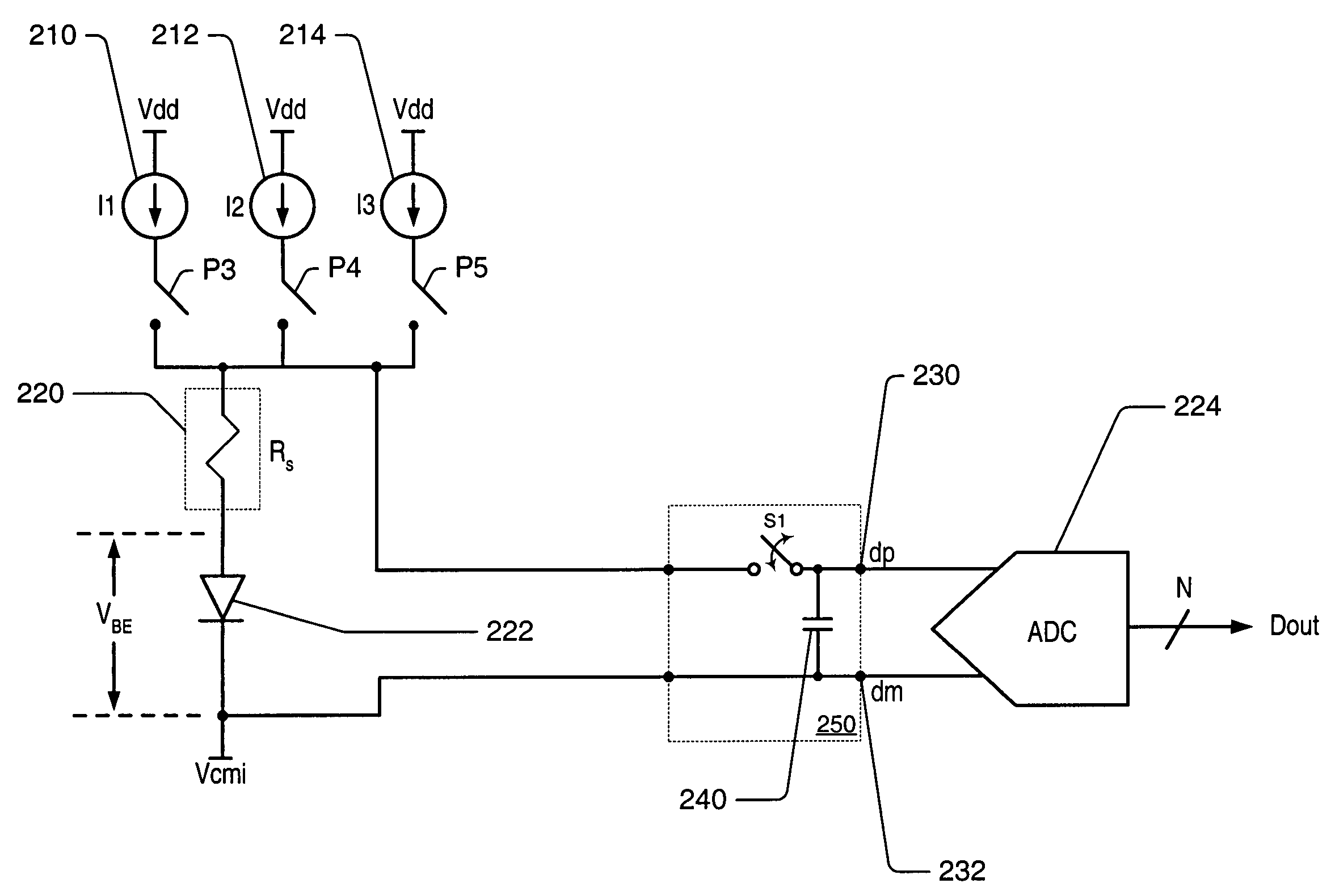
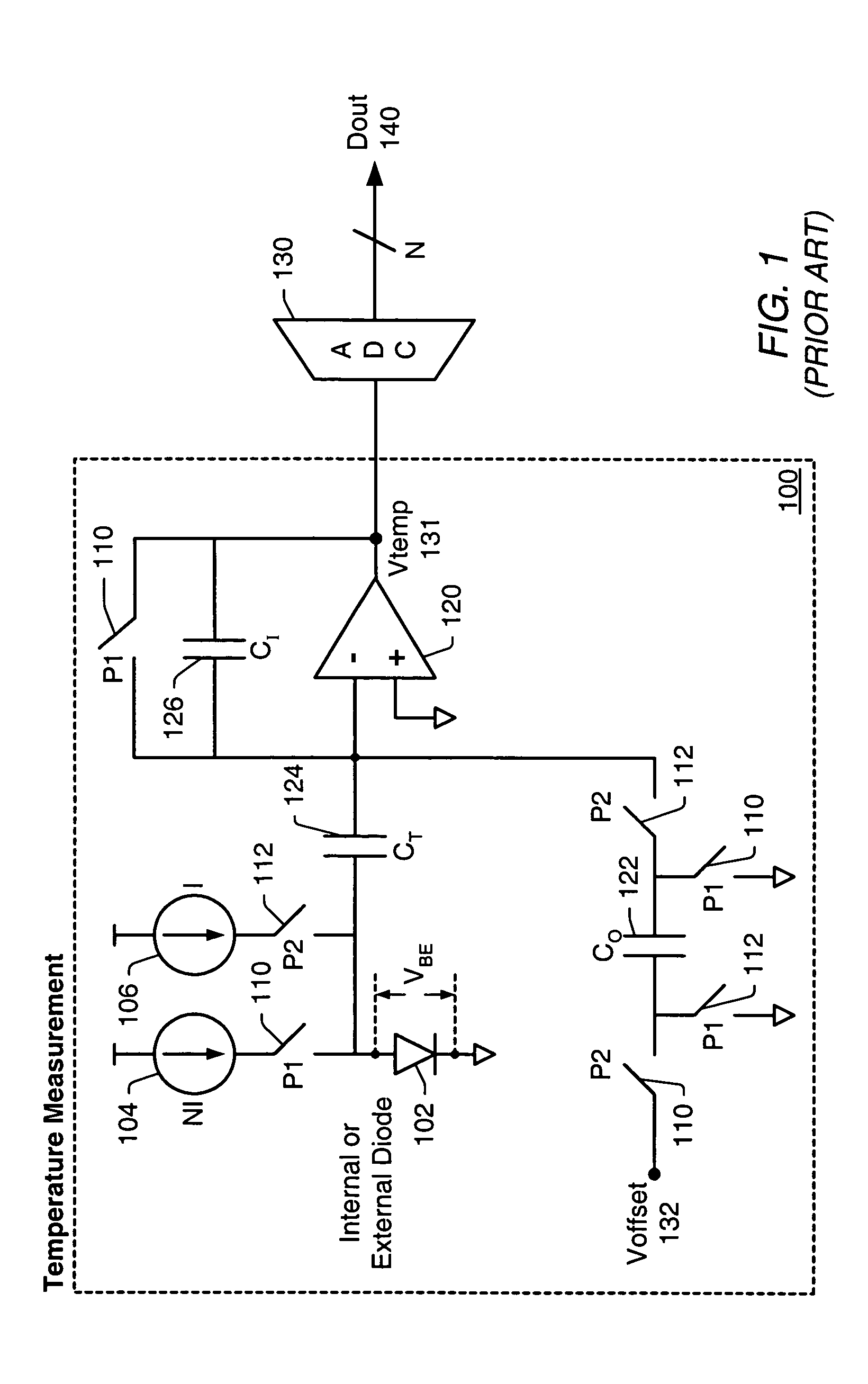
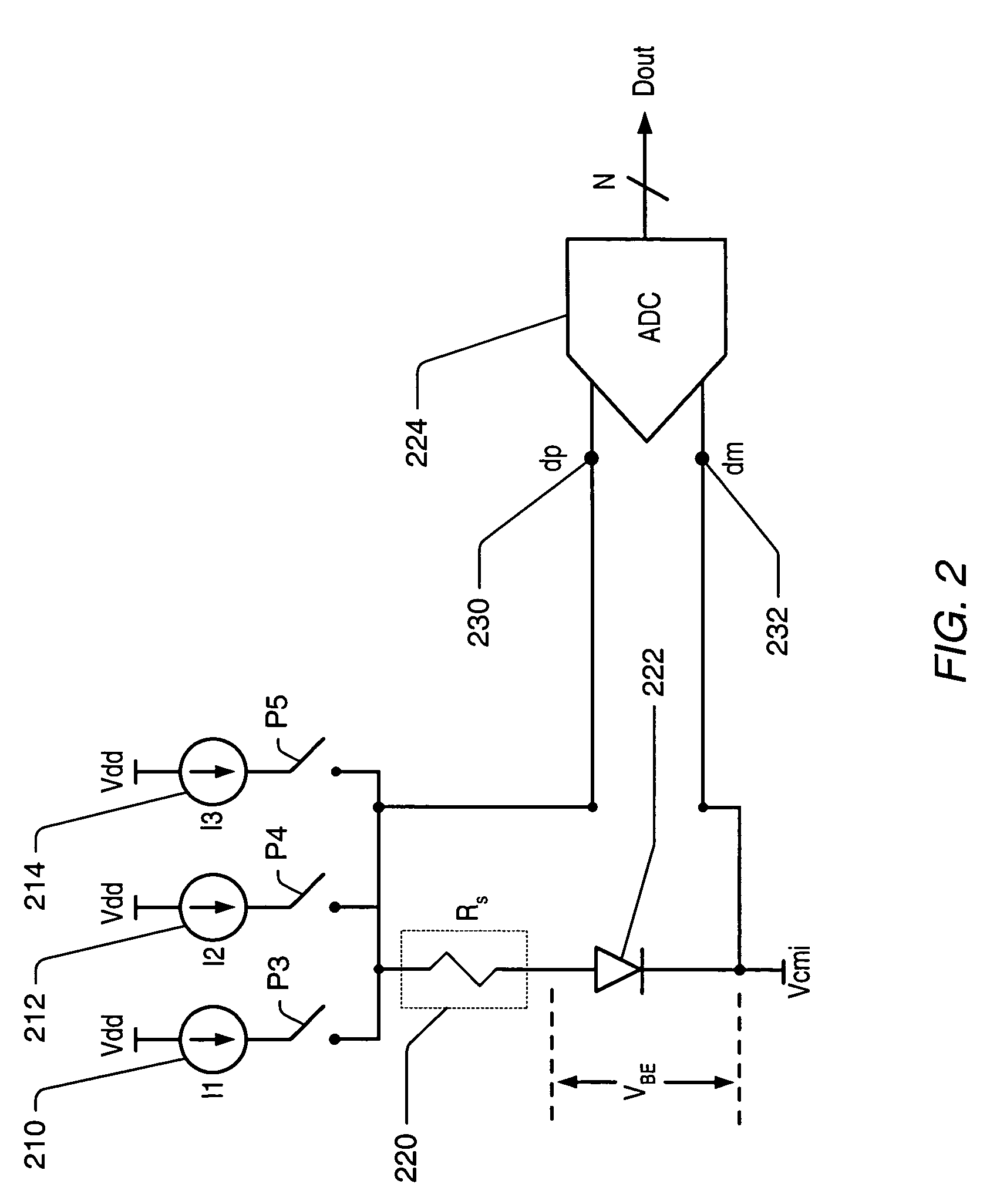
Integrated resistance cancellation in temperature measurement systems
ActiveUS7281846B2Eliminate needThermometers using electric/magnetic elementsUsing electrical meansElectrical resistance and conductanceIntegrator
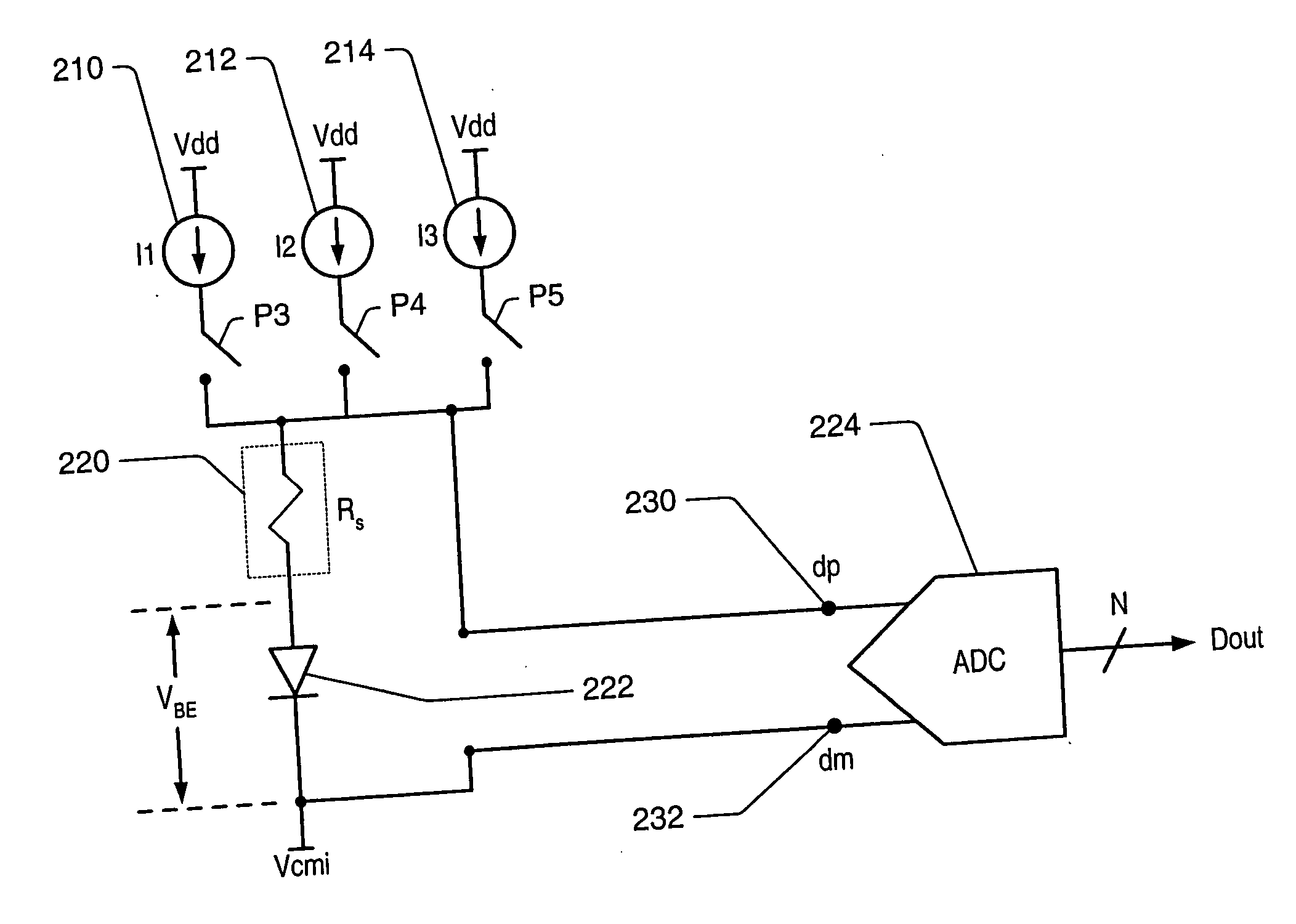
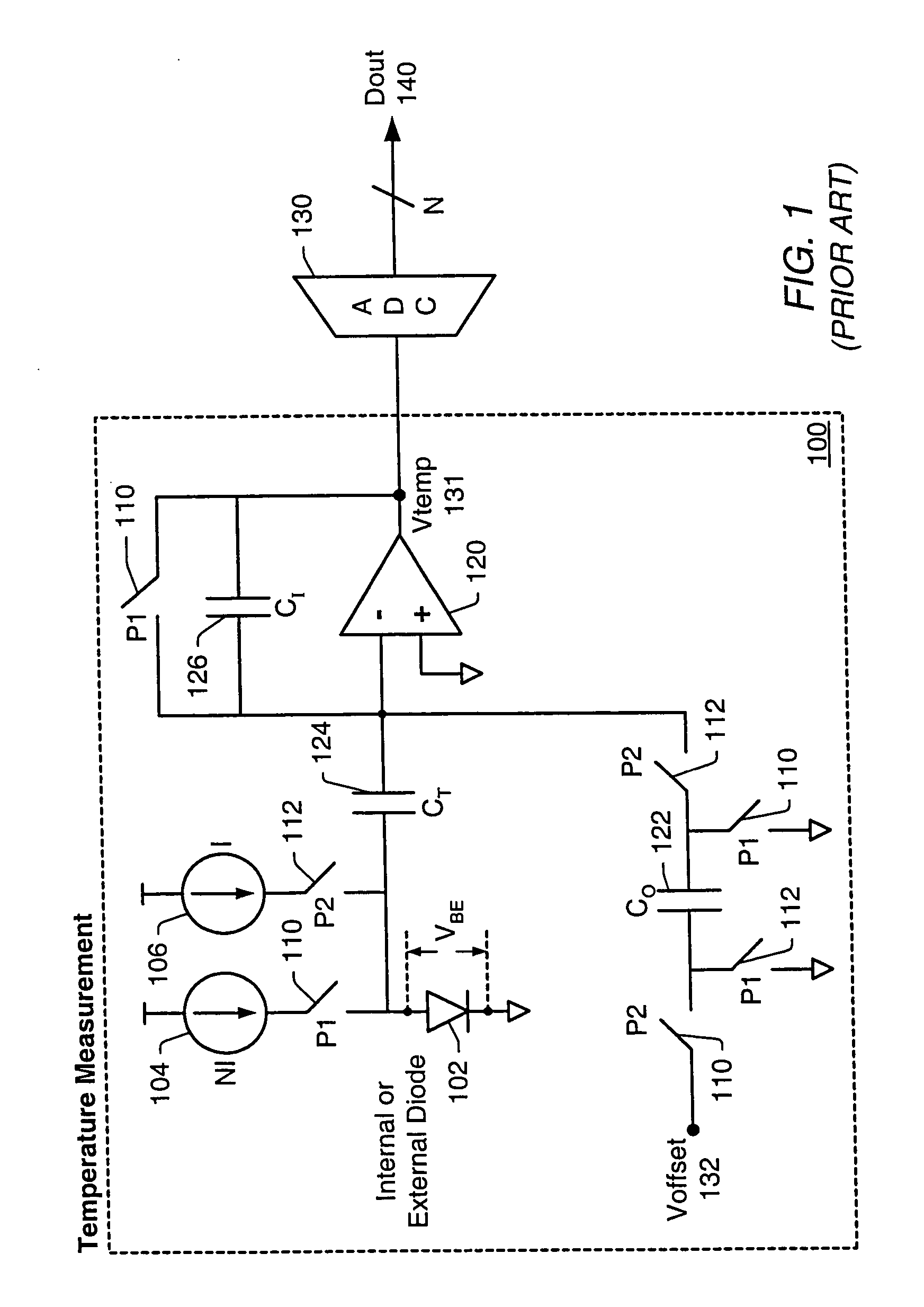
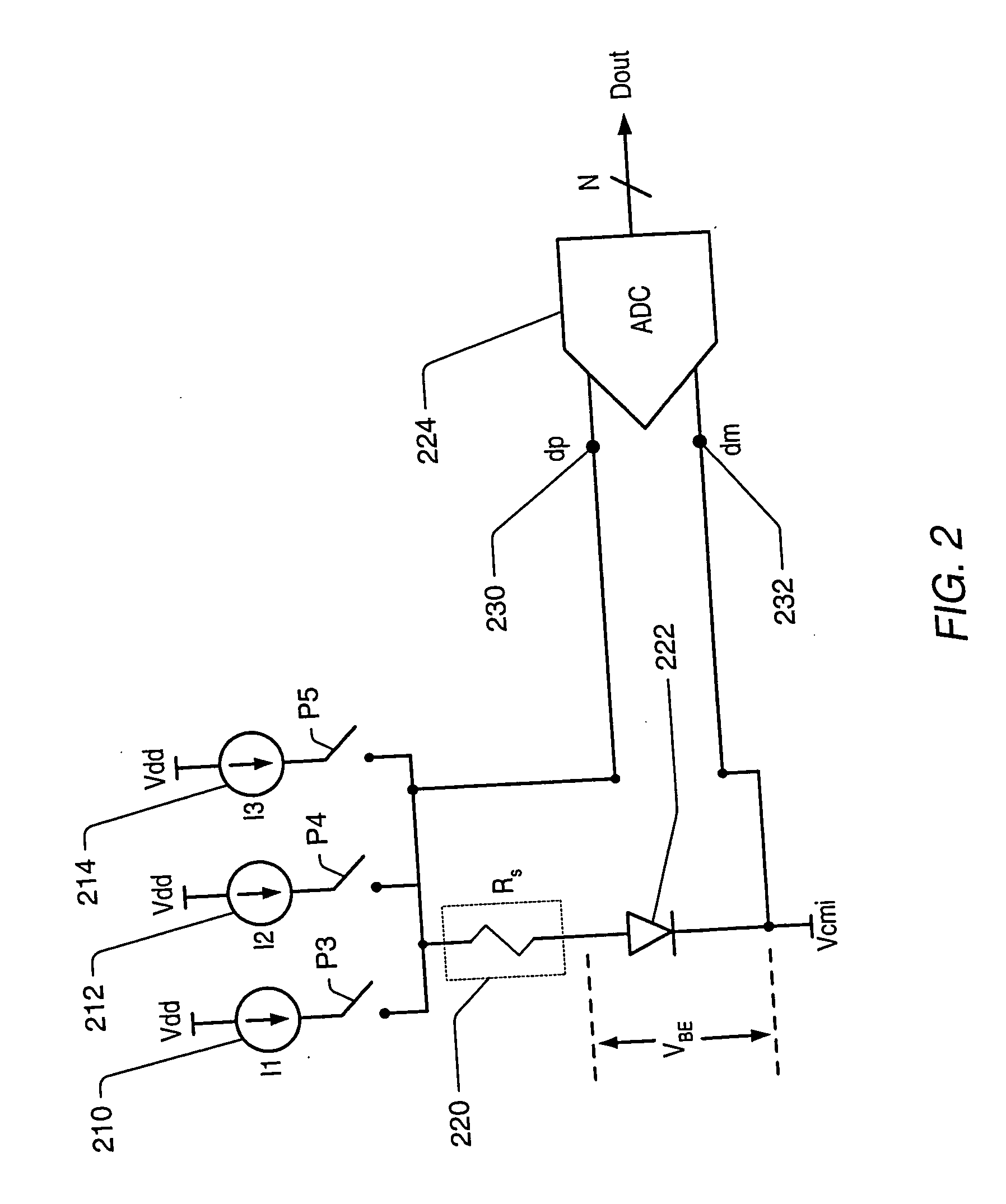
A temperature measurement device may be implemented by coupling a PN-junction, which may be comprised in a diode, to an analog-to-digital converter (ADC) that comprises an integrator. Different currents may be successively applied to the diode, resulting in different VBE values across the diode. The ΔVBE values thus obtained may be successively integrated. Appropriate values for the different currents may be determined based on a set of mathematical equations, each equation relating the VBE value to the temperature of the diode, the current applied to the diode and parasitic series resistance associated with the diode. When the current sources with the appropriate values are sequentially applied to the diode and the resulting diode voltage differences are integrated by the integrator comprised in the ADC, the error in the temperature measurement caused by series resistance is canceled in the ADC, and an accurate temperature reading of the diode is obtained from the output of the ADC.
Owner:MICROCHIP TECH INC
Communications System and Method
InactiveUS20090063427A1Quality improvementProcessingData processing applicationsDigital data processing detailsCommunications systemTheoretical computer science
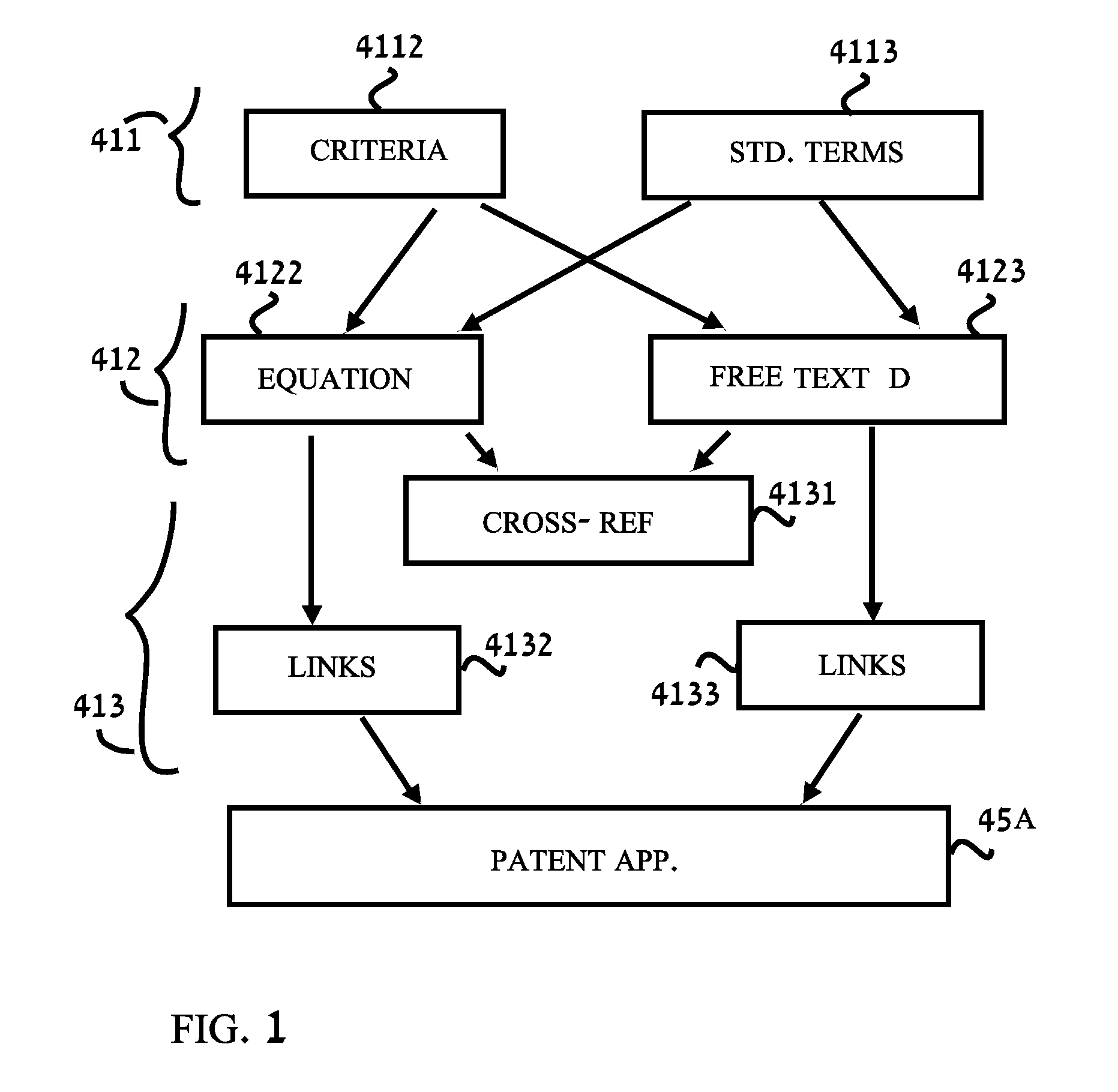
A method for describing an invention comprising: a. defining a vocabulary of standard terms; b. describing the invention in a concise form and using only standard terms for the substantive description. The description may be in concise form uses mathematical equations or mathematical-like statements or tables. The description comprises, for an apparatus invention, the parts of the apparatus and the interrelationships therebetween, and for a method invention the steps of the invention and the order of execution of the steps.
Owner:ZUTA MARC +1
Dual synthetic aperture radar system
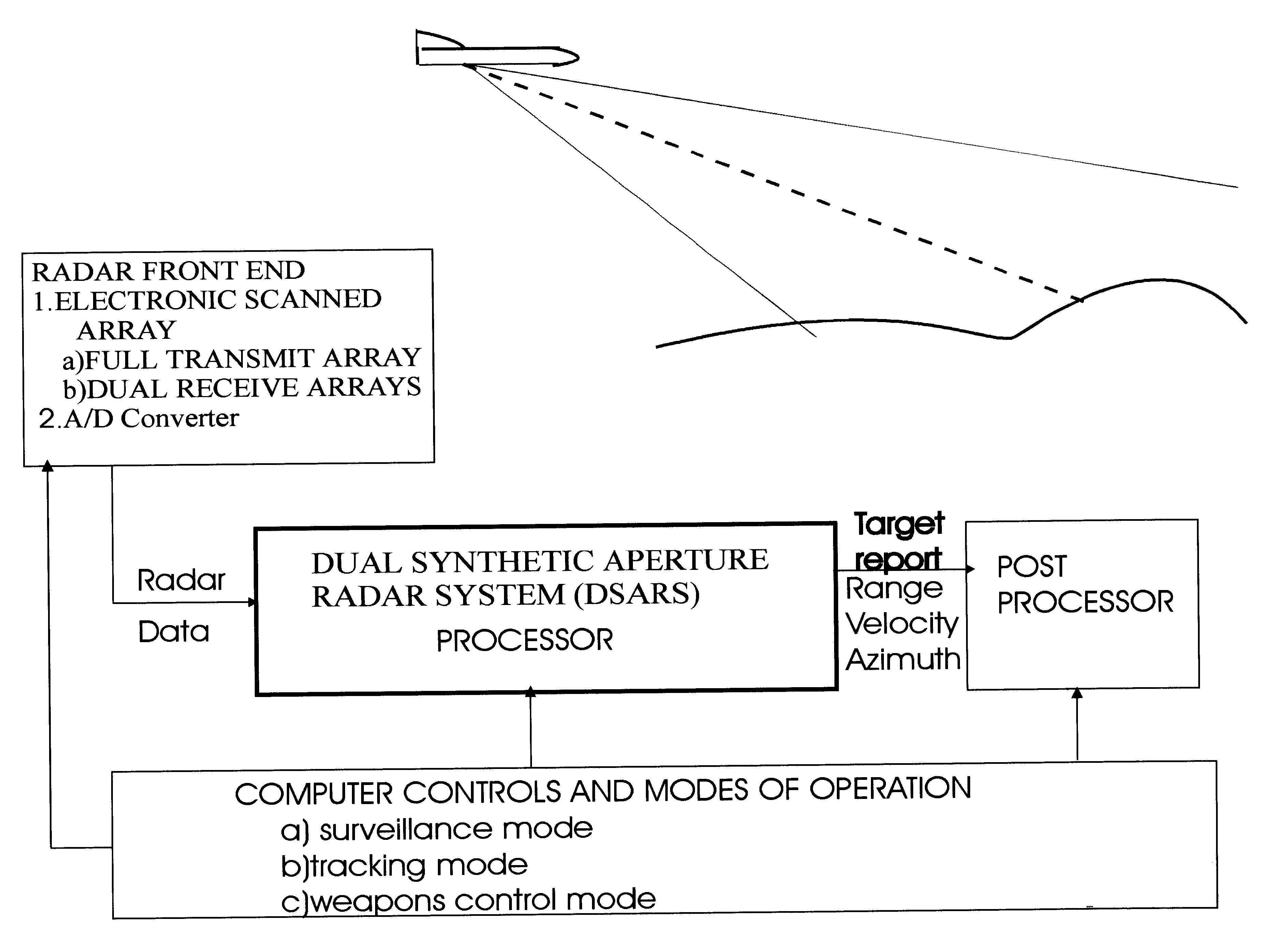
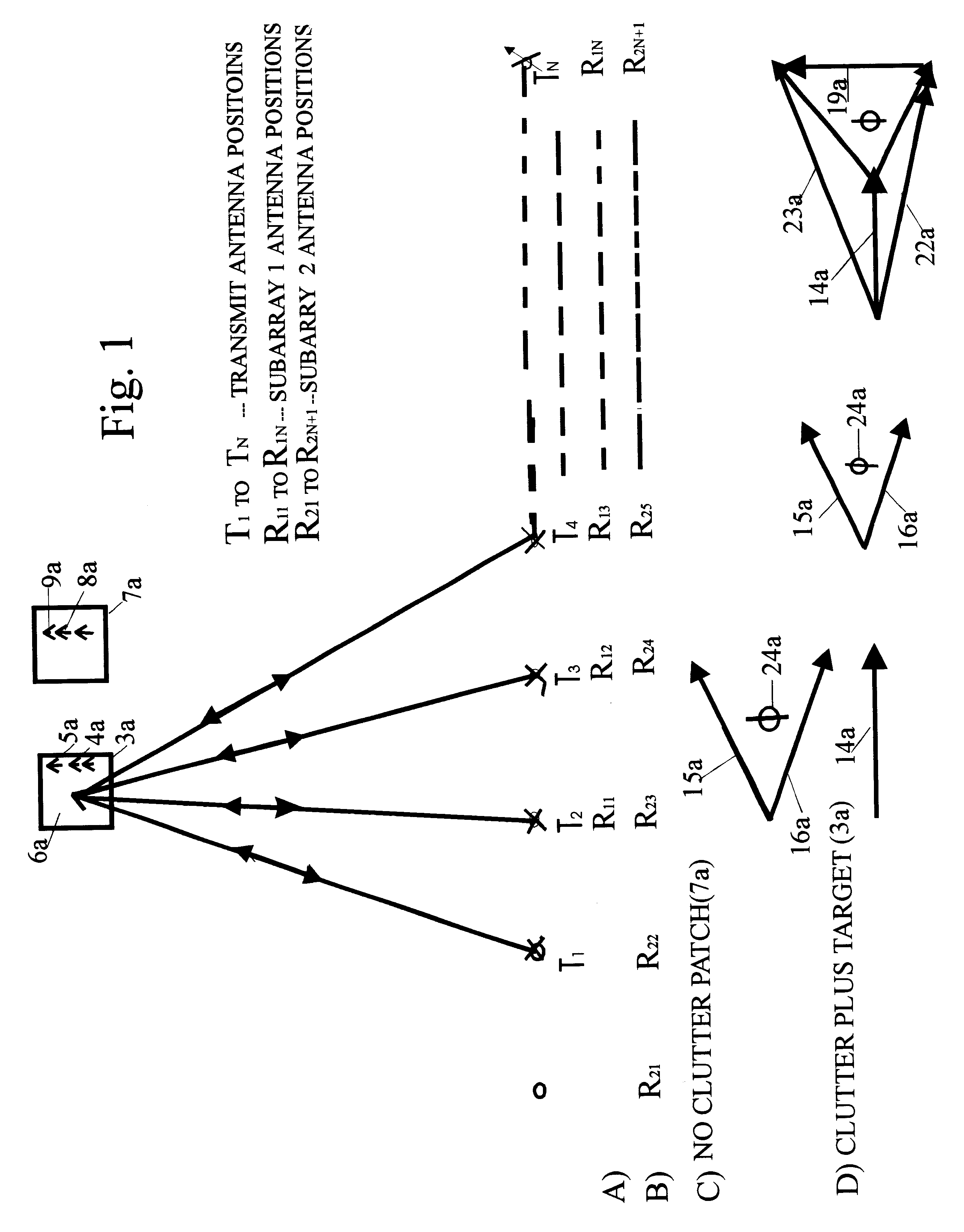
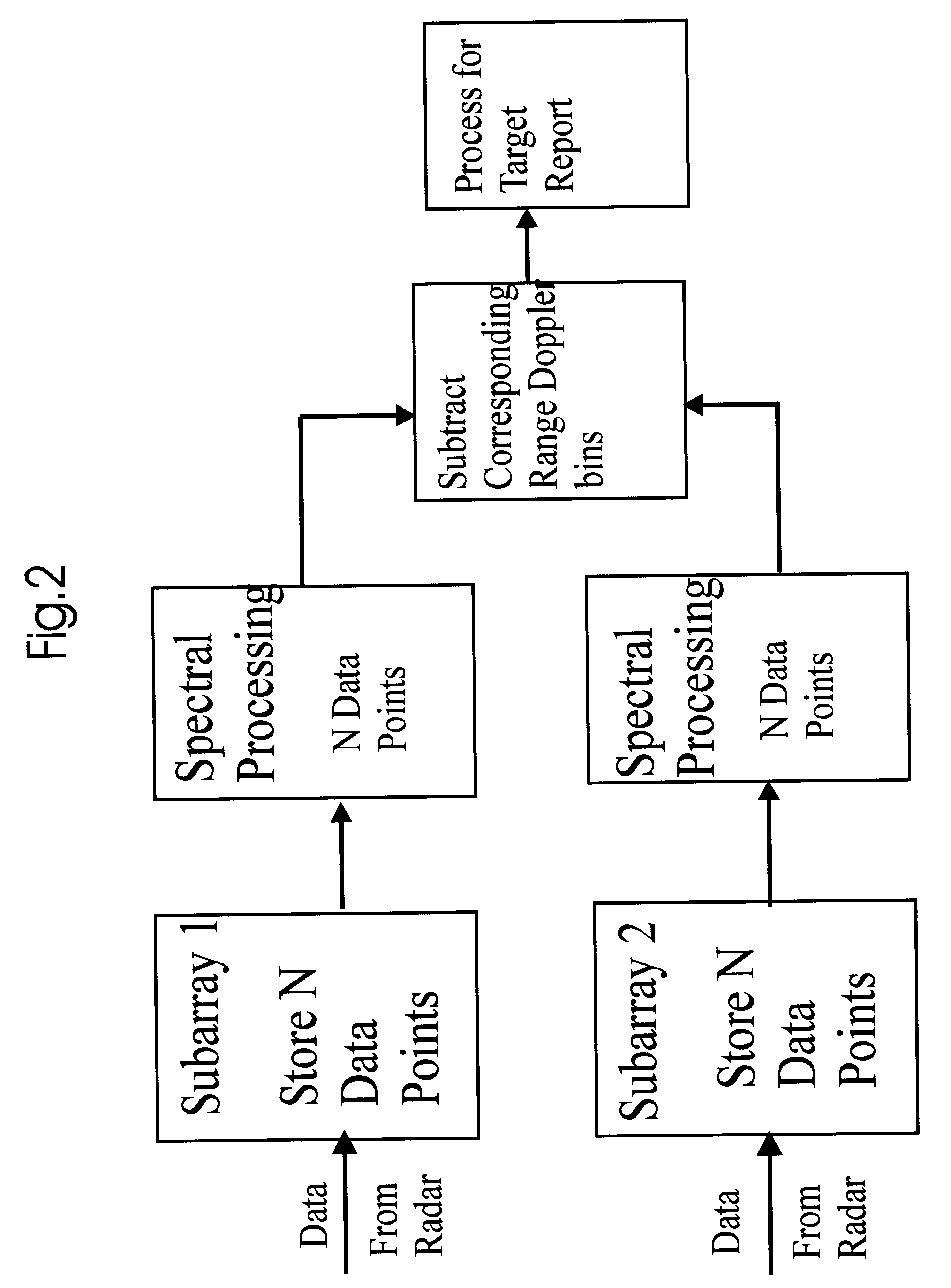
InactiveUS6633253B2Accurate angular positionAccurately radial velocityRadio wave reradiation/reflectionFrequency spectrumControl system
The dual synthetic aperture array system processes returns from the receiving arrays. The two identical receiving arrays employing displaced phase center antenna techniques subtract the corresponding spectrally processed data to cancel clutter. It is further processed that a moving target is detected and its velocity, angular position and range is measured, in or out of the presence of clutter. There are many techniques presented in the disclosure. These techniques are basically independent but are related based on common set of fundamental set of mathematical equations, understanding of radar principles and the implementations involved. These many techniques may be employed singly and / or in combination depending on the application and accuracy required. They are supported by a system that includes, optimization of the number of apertures, pulse repetition frequencies, DPCA techniques to cancel clutter, adaptive techniques to cancel clutter, motion compensation, weighting function for clutter and target, and controlling the system in most optimum fashion to attain the objective of the disclosure.< / PTEXT>
Owner:CATALDO THOMAS J
Dispenser having non-frustro-conical funnel wall
A helix cup for use in a pressurized dispenser. The helix cup has a convergent funnel wall. The funnel wall is not straight and does not satisfy the mathematical equations for surface area or for subtended volume of the frustrum of a cone. Instead, the funnel wall provides a longer flow path than is achieved with straight sidewalls. The longer flow path provides for a tighter particle size distribution at lower pressures than occurs in the prior art.
Owner:PROCTER & GAMBLE CO
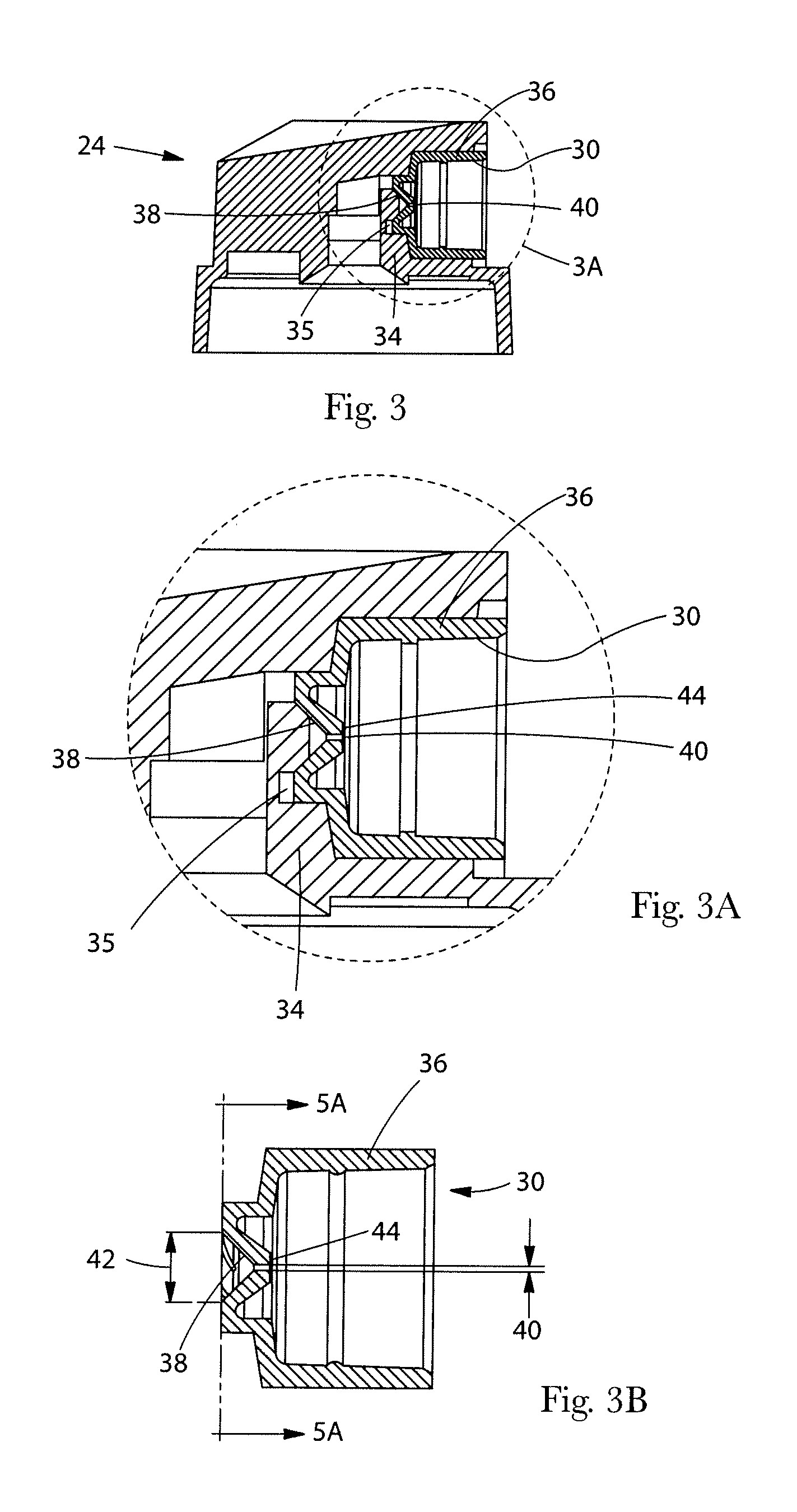
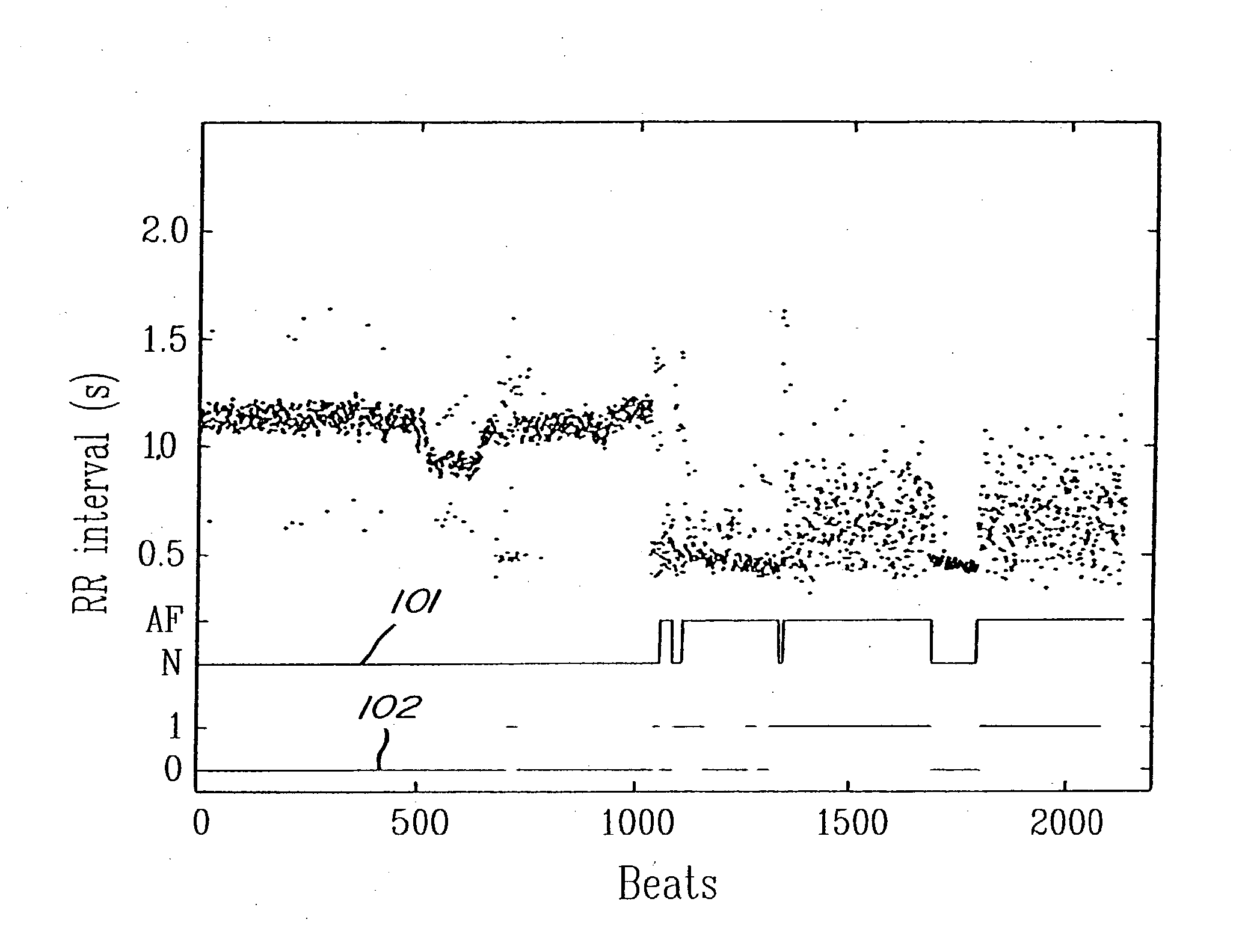
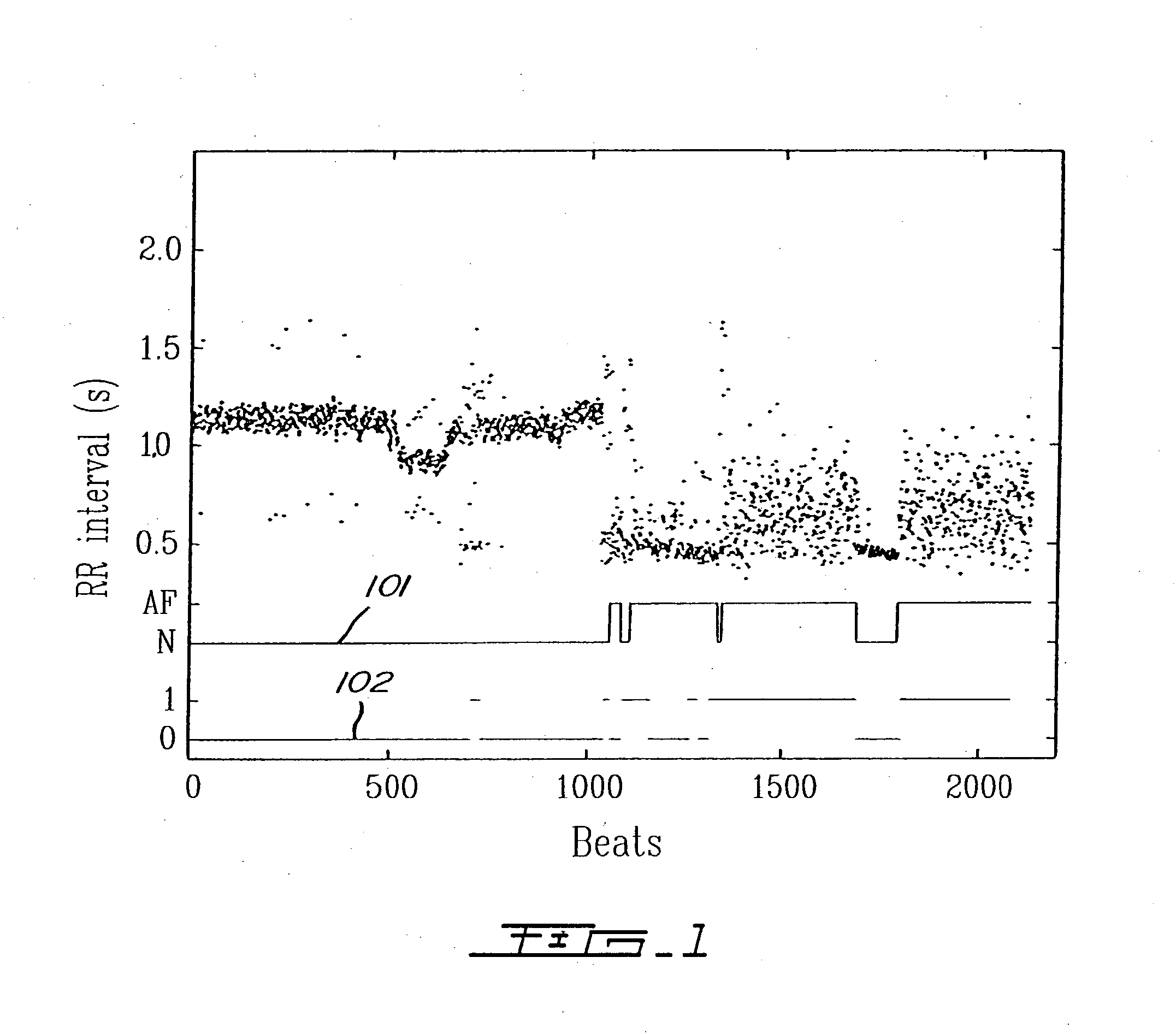
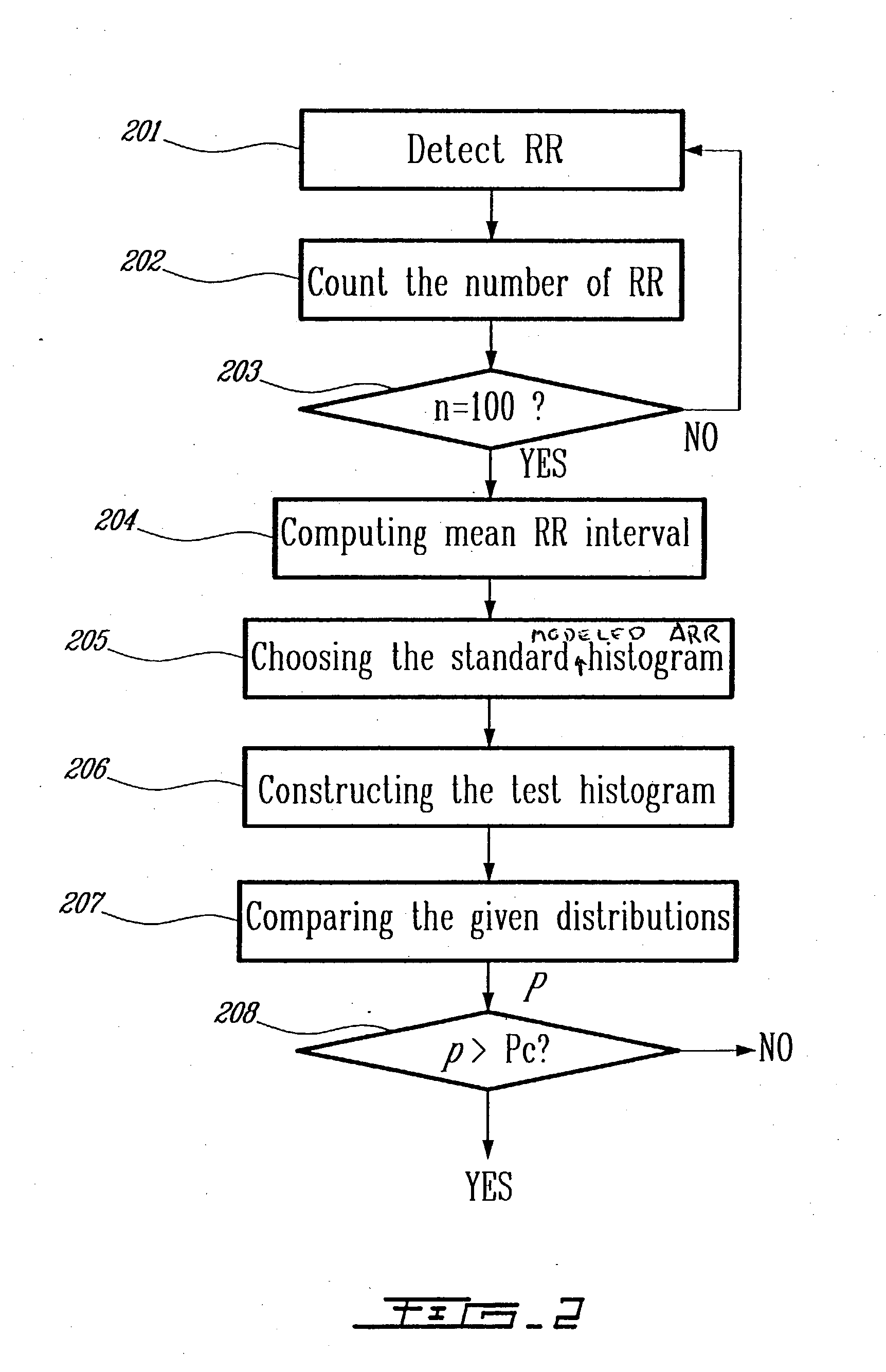
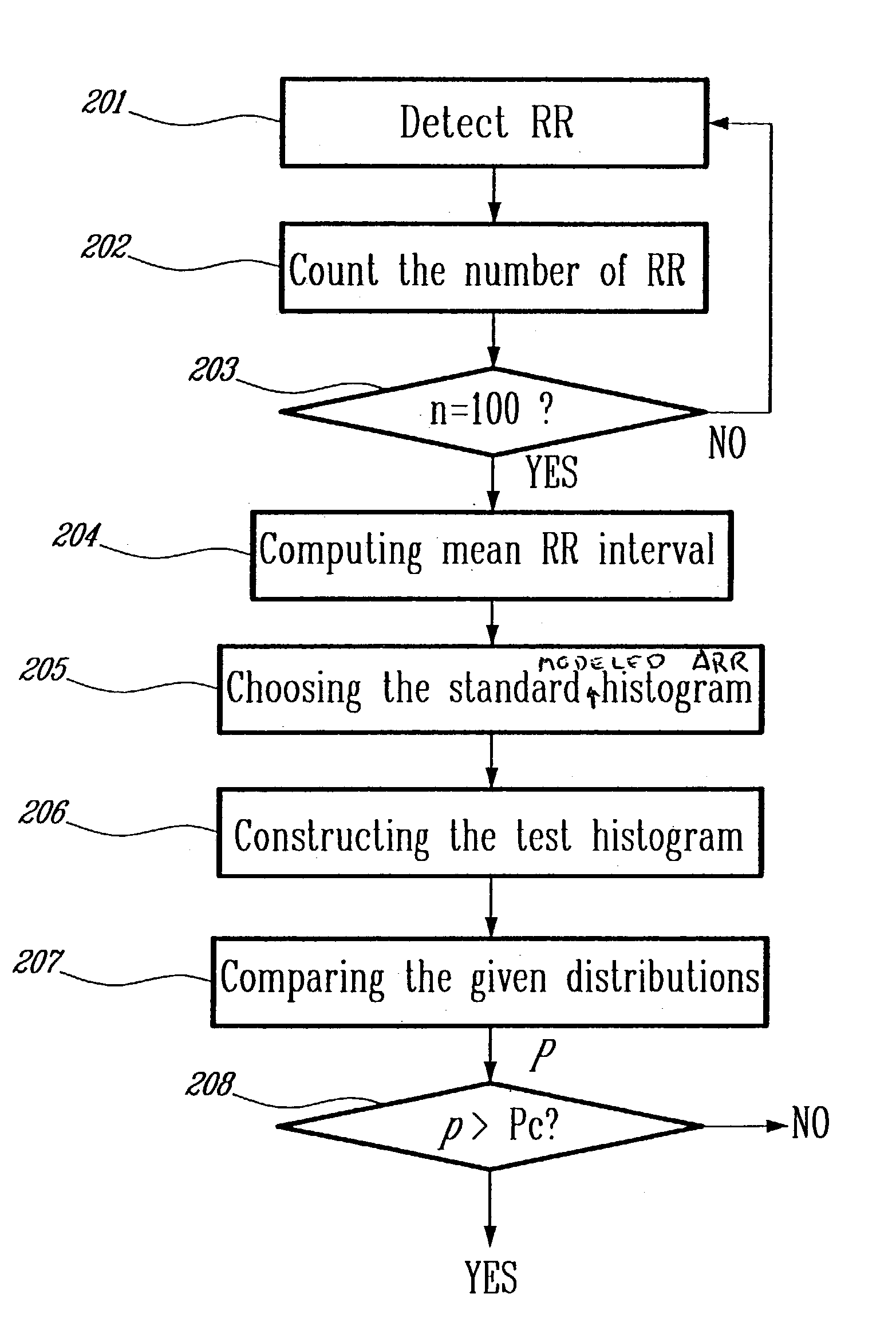
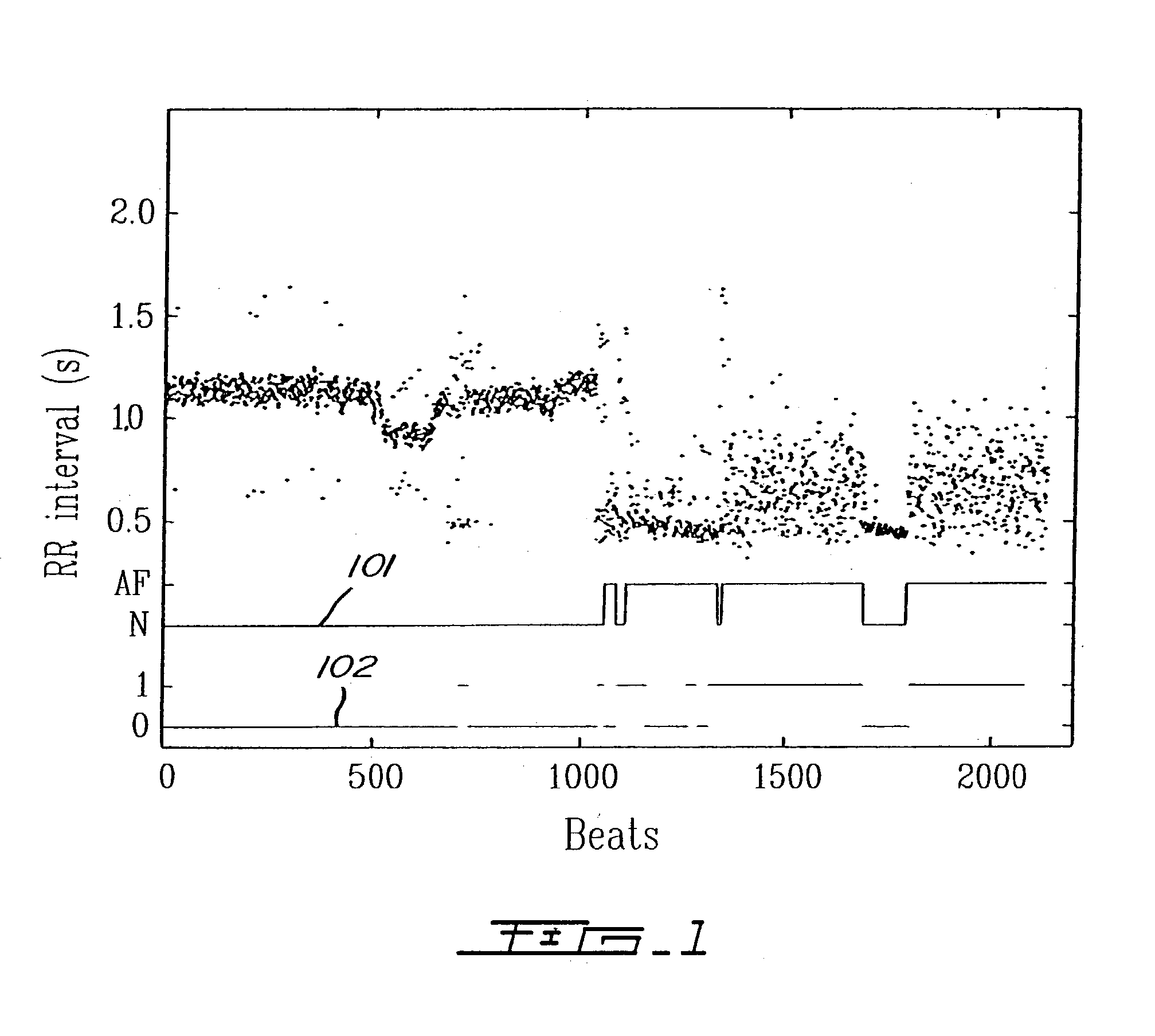
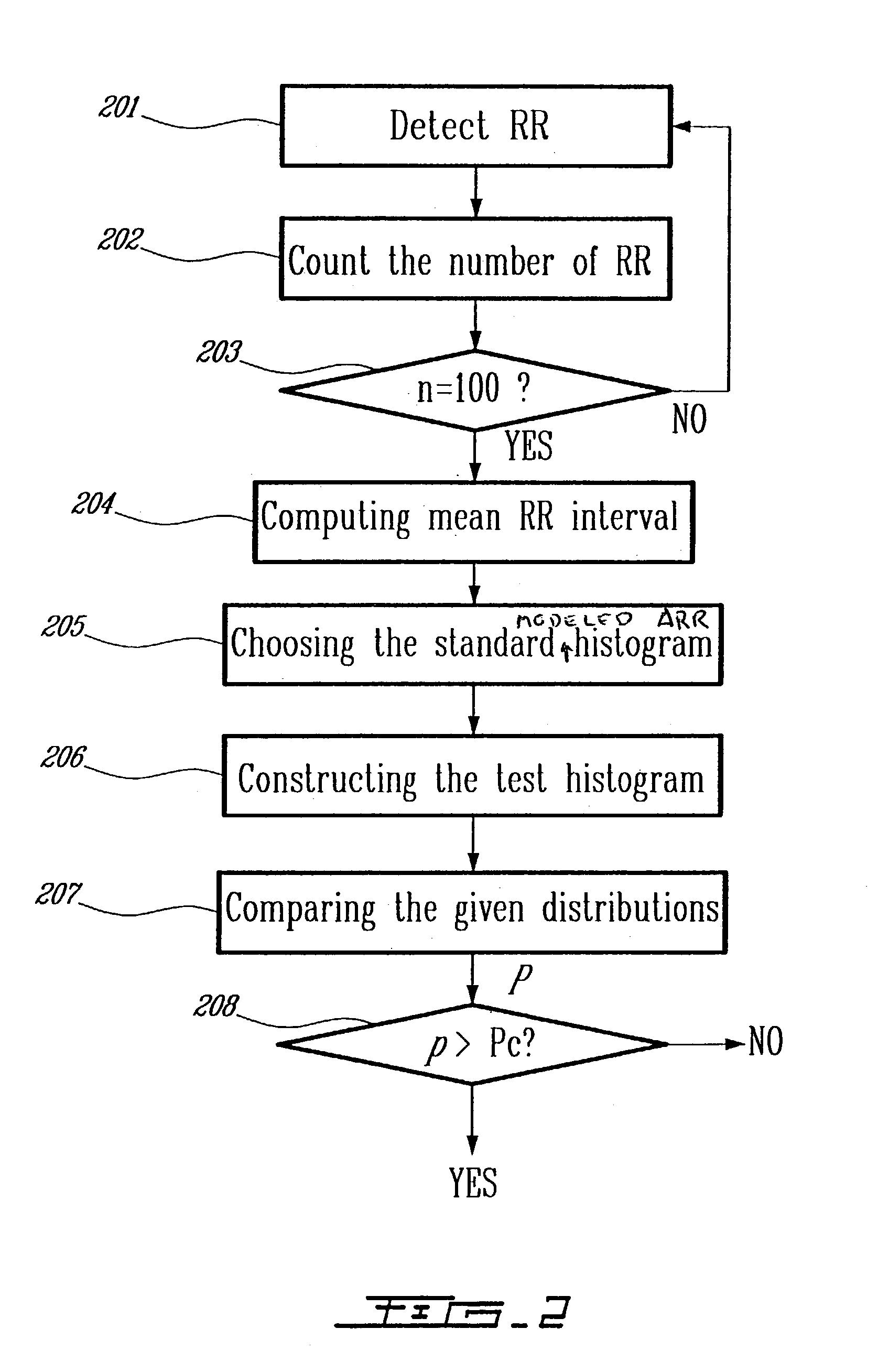
Detection of cardiac arrhythmia using mathematical representation of standard deltaRR probability density histograms
ActiveUS20050004486A1Improve accuracyReduce storage requirementsElectrocardiographySensorsRR intervalMedicine
A method and a system for detecting cardiac arrhythmias that includes detecting RR intervals of the patient wherein each RR interval is an interval between two heart beats, and simulating standard probability density histograms of ΔRRs by means of a suitable probability distribution calculated through at least one mathematical formulae, wherein ΔRR is a difference between two successive RR intervals. Test probability density histograms of ΔRRs of the patient are constructed from the detected RR intervals. Finally, the standard and test histograms are compared to detect whether the patient suffers from cardiac arrhythmia. As non limiting examples, the standard probability density histograms of ΔRRs are modeled by a mathematical equation selected from the group consisting of: the Lorentzian distribution, the Gaussian distribution, the Student's t-distribution, and a probability distribution including a linear combination of the Lorentzian and Gaussian distribution.
Owner:THE ROYAL INSTION FOR THE ADVANCEMENT OF LEARNING MCGILL UNIV A UNIV
Method and system for continuously monitoring cardiac output
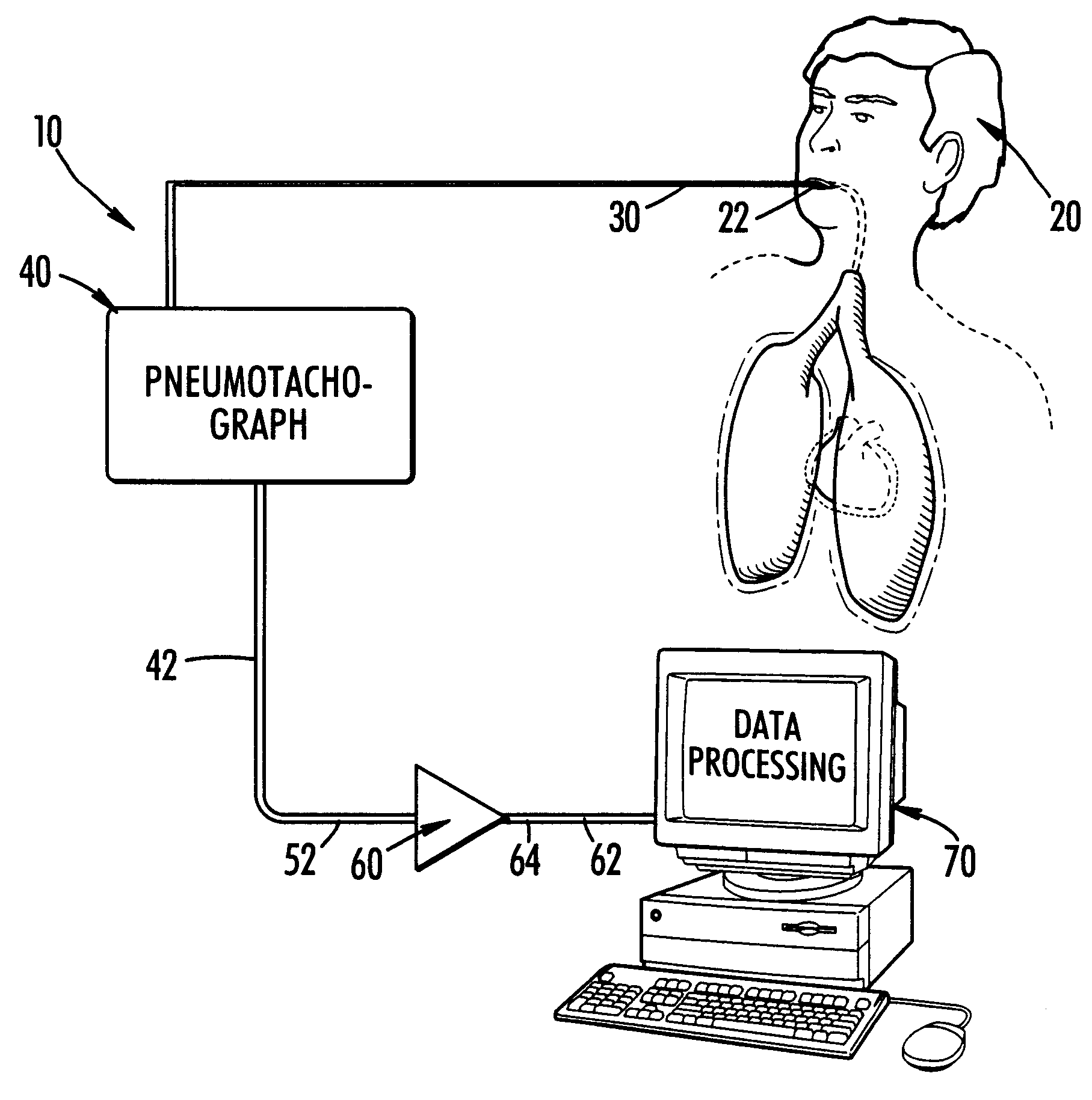
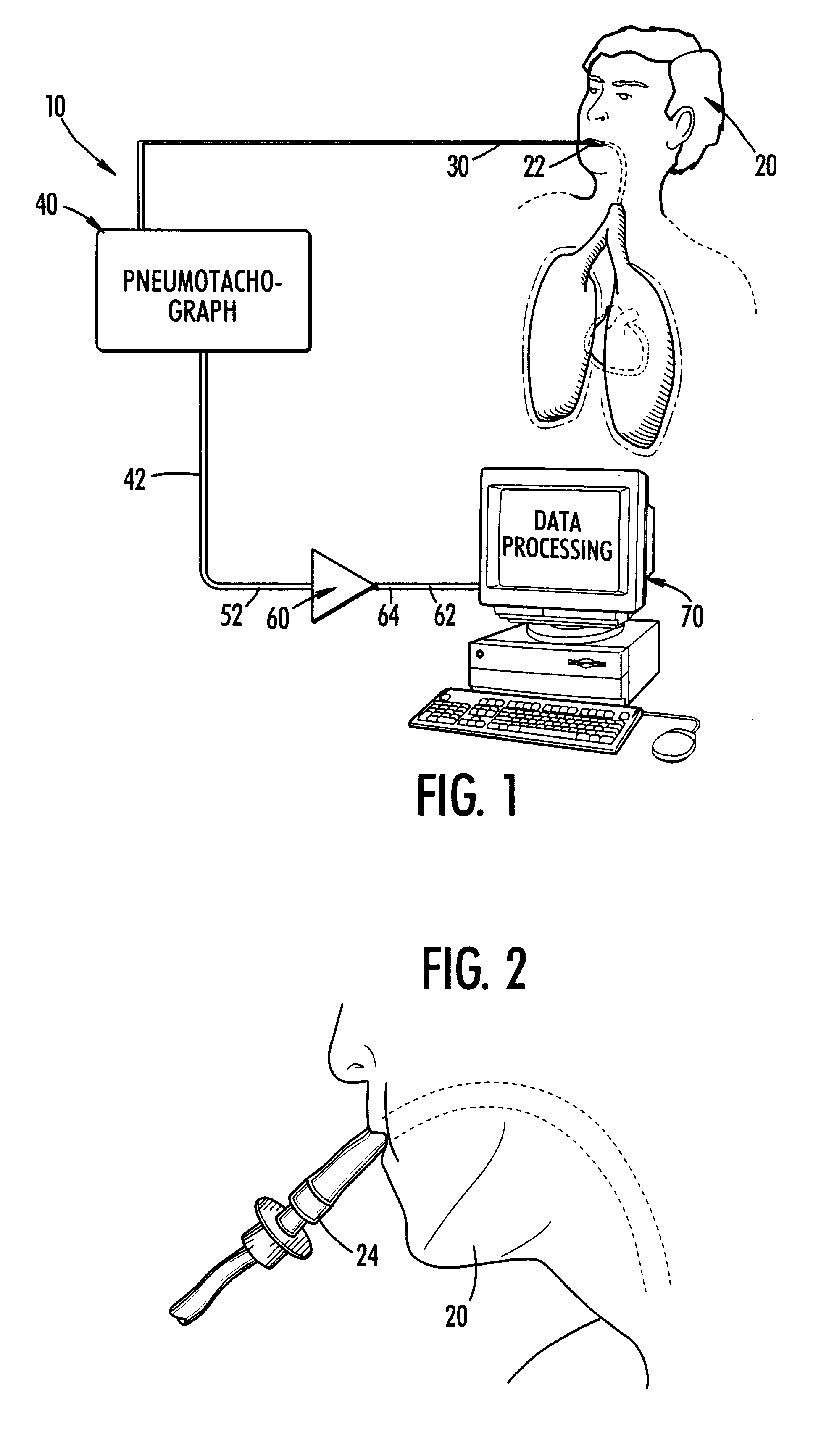
InactiveUS6186956B1Easily moved to patient and transportedLight weightCatheterRespiratory organ evaluationDigital dataAir volume
The present invention is a method and system for continuously monitoring cardiac output. In a preferred embodiment, the method and system of the present invention comprises a pneumotachograph, differential pressure transducer, and a signal amplifier / conditioner interconnected to a programmed digital computer. A patient, preferably, inserts the pneumotachograph in his mouth or, alternatively, the pneumotachograph is connected to a patient's tracheal cannula. As the patient exhales and inhales the differential pressure transducer measures the drop in pressure as air flows through the pneumotachograph thereby producing a weak electrical signal non-linearly proportional to flow. Next, the weak signal is directed through the amplifier / signal conditioner which increases the amplitude and removes some of the noise contained in the transducer output. A digital computer is then utilized to convert the analog time varying electrical signal into a stream of digital data, store it on disk, display it in real time and processes the signal using an experimentally determined correlation factor and mathematical equations relating the fluctuations in air flow with stroke volume to obtain the cardiac output.
Owner:UNIVERSITY OF SOUTH CAROLINA
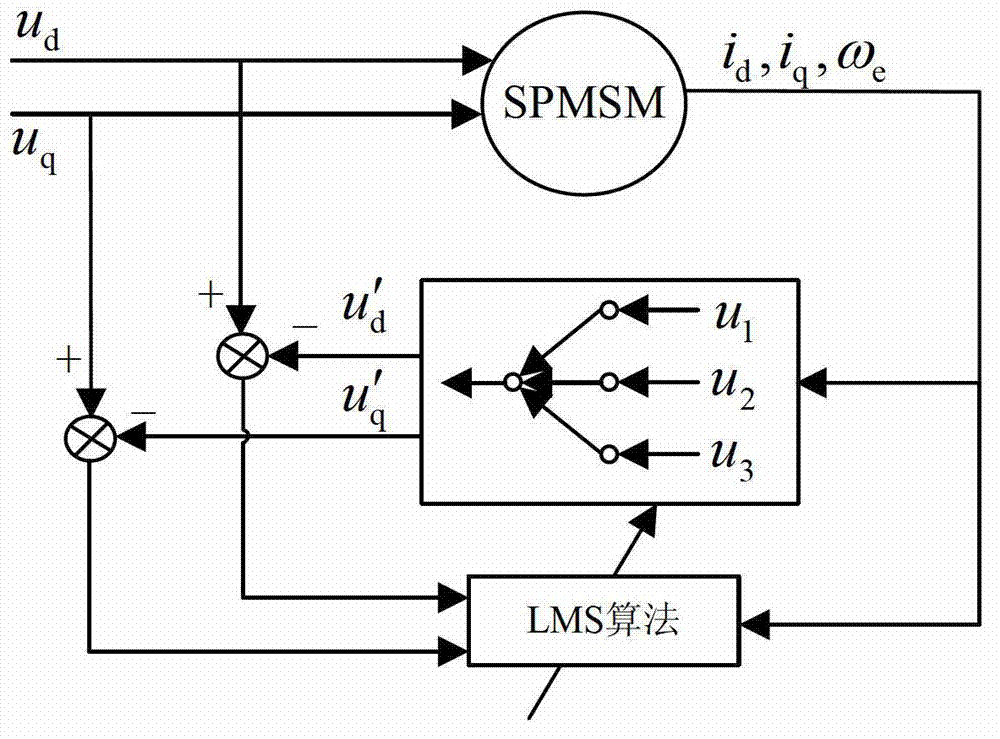
Online decoupling identification method of multiple parameters of PMSM (permanent magnet synchronous motor)
ActiveCN103248306AEliminate false convergenceImprove reliabilityElectronic commutation motor controlAC motor controlPermanent magnet synchronous motorPermanent magnet synchronous generator
The invention relates to the technical field of PMSMs, solves a coupling problem during online identification of multiple parameters of a surface-mounted type PMSM, and achieves online decoupling identification of PMSM inductance, stator resistance and rotor flux linkage. Accordingly, the technical scheme adopted by the invention is that an online decoupling identification method of multiple parameters of the PMSM comprises the steps as follows: 1) identifying and coupling analysis of parameters of the PMSM; 2) a decoupling identification strategy, wherein voltage deviation before and after D shaft current injection is used for increasing the order of a motor mathematical equation, so that the decoupling identification of multiple parameters of the surface-mounted type PMSM inductance, the stator resistance and the rotor flux linkage are achieved; 3) neural network identifier design, wherein according to a parameter online identification problem of the PMSM, online identification is performed on motor parameters by adopting a self-adaptive neural network structure and a weight convergence algorithm based on a least mean square algorithm. The method is mainly applied to the design and manufacture of the PMSM.
Owner:TIANJIN UNIV
Method of nano thin film thickness measurement by auger electron spectroscopy
ActiveUS7582868B2Material analysis using wave/particle radiationUsing wave/particle radiation meansMathematical modelPhysical model
A system and method for measuring the thickness of an ultra-thin multi-layer film on a substrate is disclosed. A physical model of an ultra-thin multilayer structure and Auger electron emission from the nano-multilayer structure is built. A mathematical model for the Auger Electron Spectroscopy (AES) measurement of the multilayer thin film thickness is derived according to the physical model. Auger electron spectroscopy (AES) is first performed on a series of calibration samples. The results are entered into the mathematical model to determine the parameters in the mathematical equation. The parameters may be calibrated by the correlation measurements of the alternate techniques. AES analysis is performed on the ultra-thin multi-layer film structure. The results are entered into the mathematical model and the thickness is calculated.
Owner:SAE MAGNETICS (HK) LTD
Detection of cardiac arrhythmia using mathematical representation of standard DeltaRR probability density histograms
ActiveUS7146206B2Improve accuracyStorage requirements is greatly reducedElectrocardiographySensorsRR intervalMedicine
Owner:THE ROYAL INSTION FOR THE ADVANCEMENT OF LEARNING MCGILL UNIV A UNIV
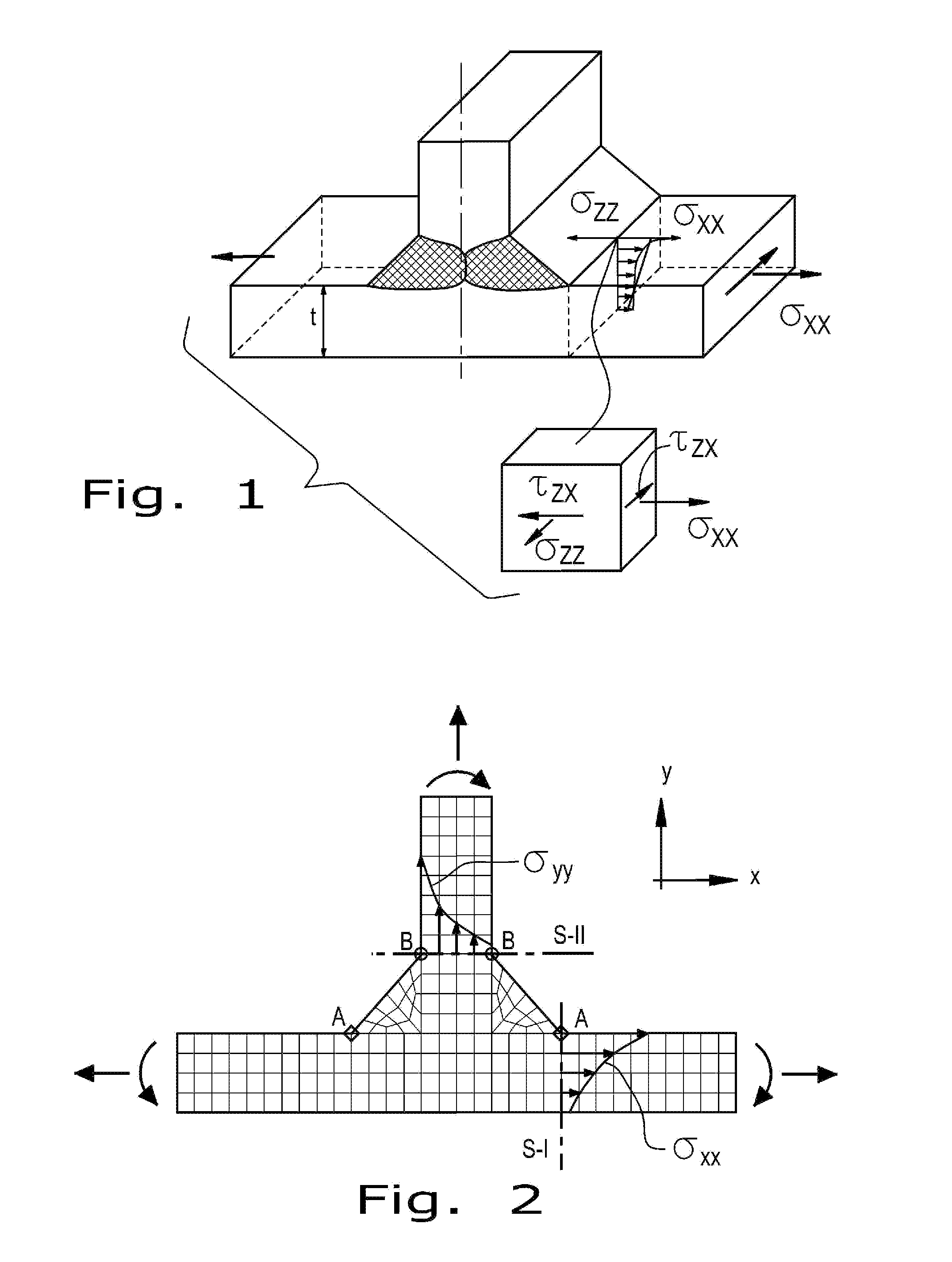
Method for the prediction of fatigue life for structures
A method of determining the fatigue life of a structure includes the steps of:associating a mathematical equation for total strain amplitude with the structure:Δɛ2=σf′E(2Nf)b+ɛf′(2Nf)c,where: Δε / 2=strain amplitude, σf′=fatigue strength coefficient associated with a material of the structure, b=fatigue strength exponent of the material, E=cyclic modulus of elasticity of the material, 2Nf=number of cycles, εf′=fatigue ductility coefficient of the material, and c=fatigue ductility exponent of the material;reducing the fatigue strength exponent (b) such that an elastic portion of a total strain amplitude curve associated with the equation has a reduced slope to account for variable amplitude loading for the structure;generating a total strain amplitude curve, based upon the mathematical equation:Δɛ2=σf′E(2Nf)breduced+ɛf′(2Nf)c,where (breduced) is now the reduced fatigue strength exponent; anddetermining a fatigue life of the structure, based on the total strain amplitude curve with the reduced fatigue strength exponent.
Owner:DEERE & CO
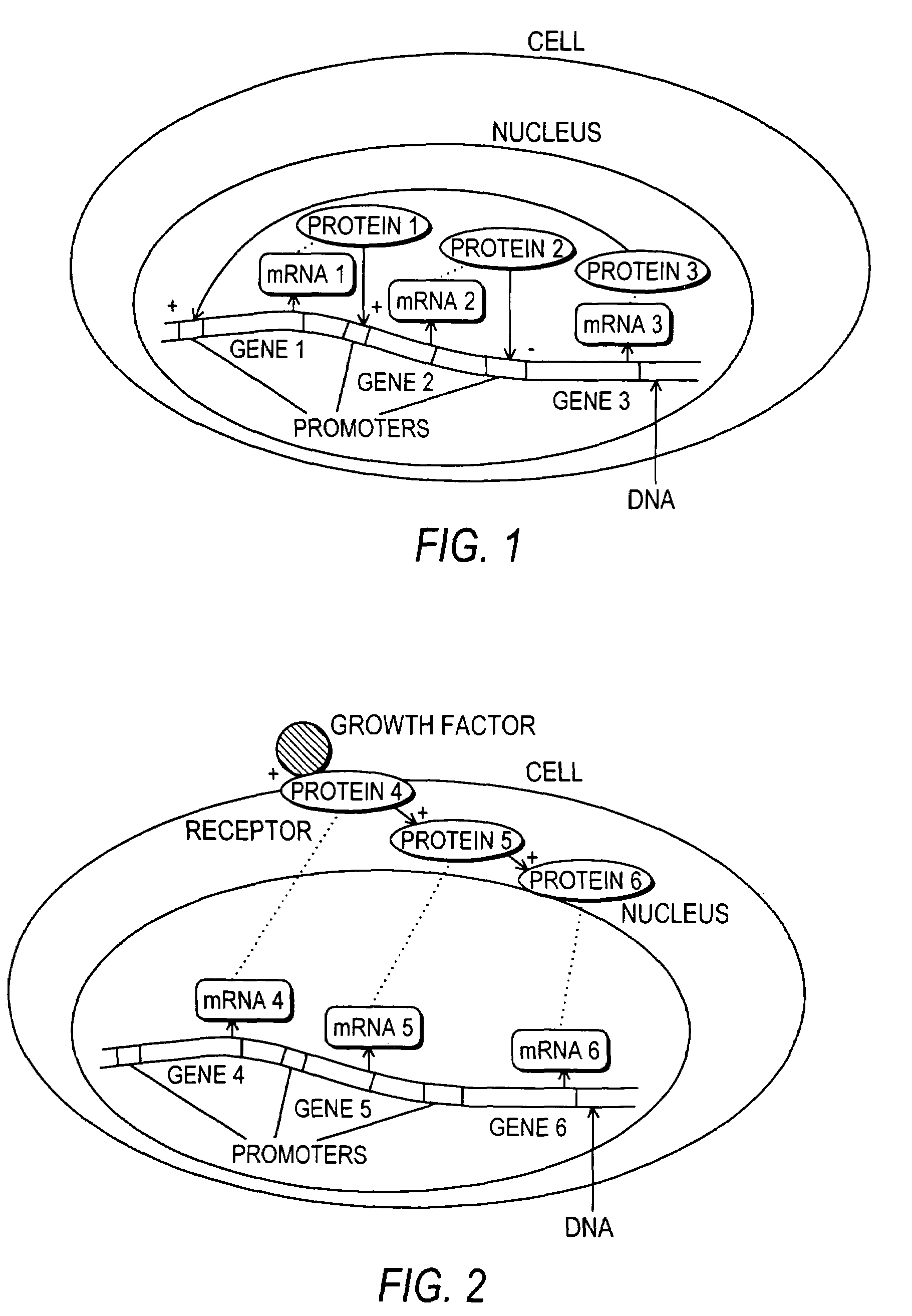
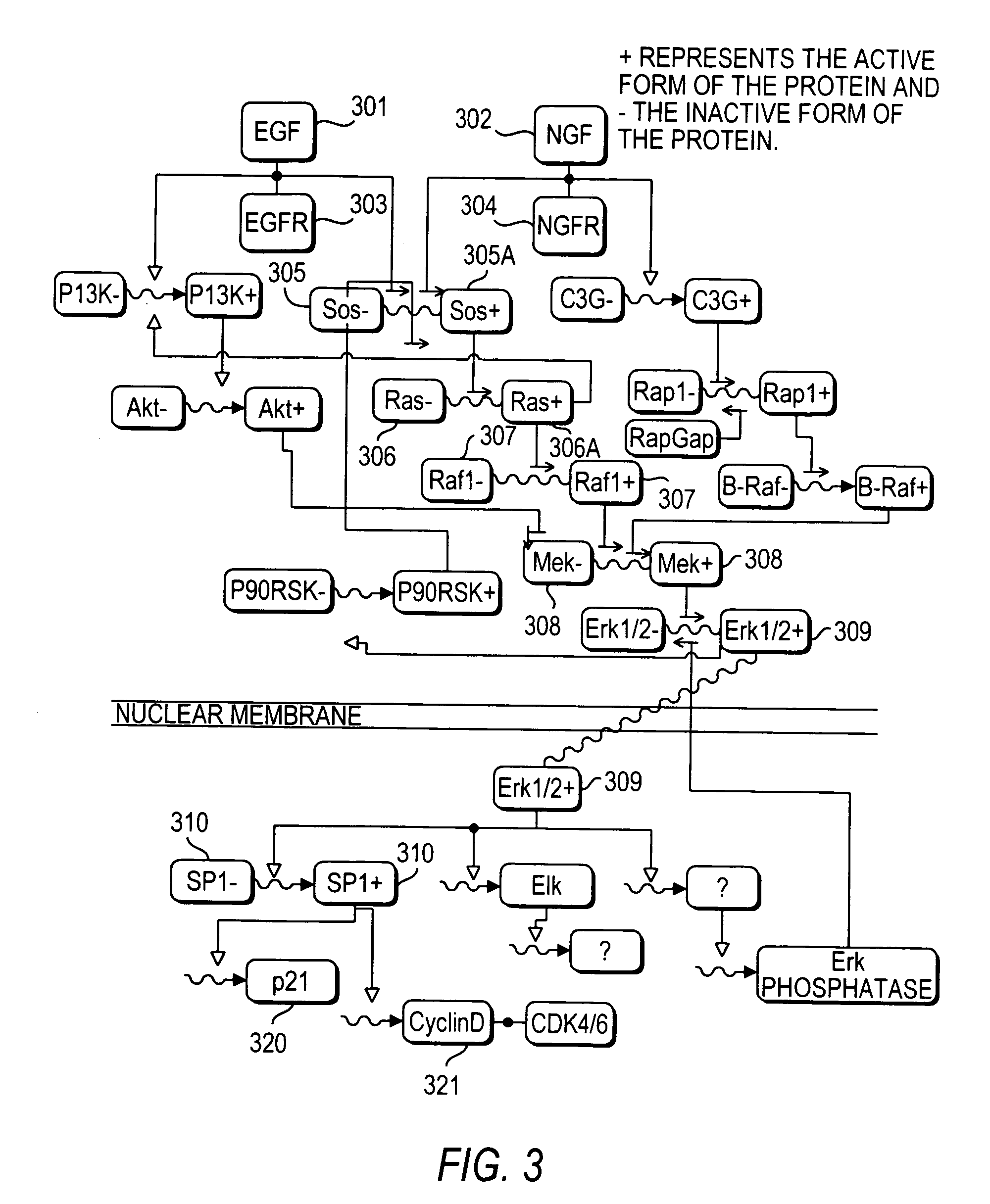
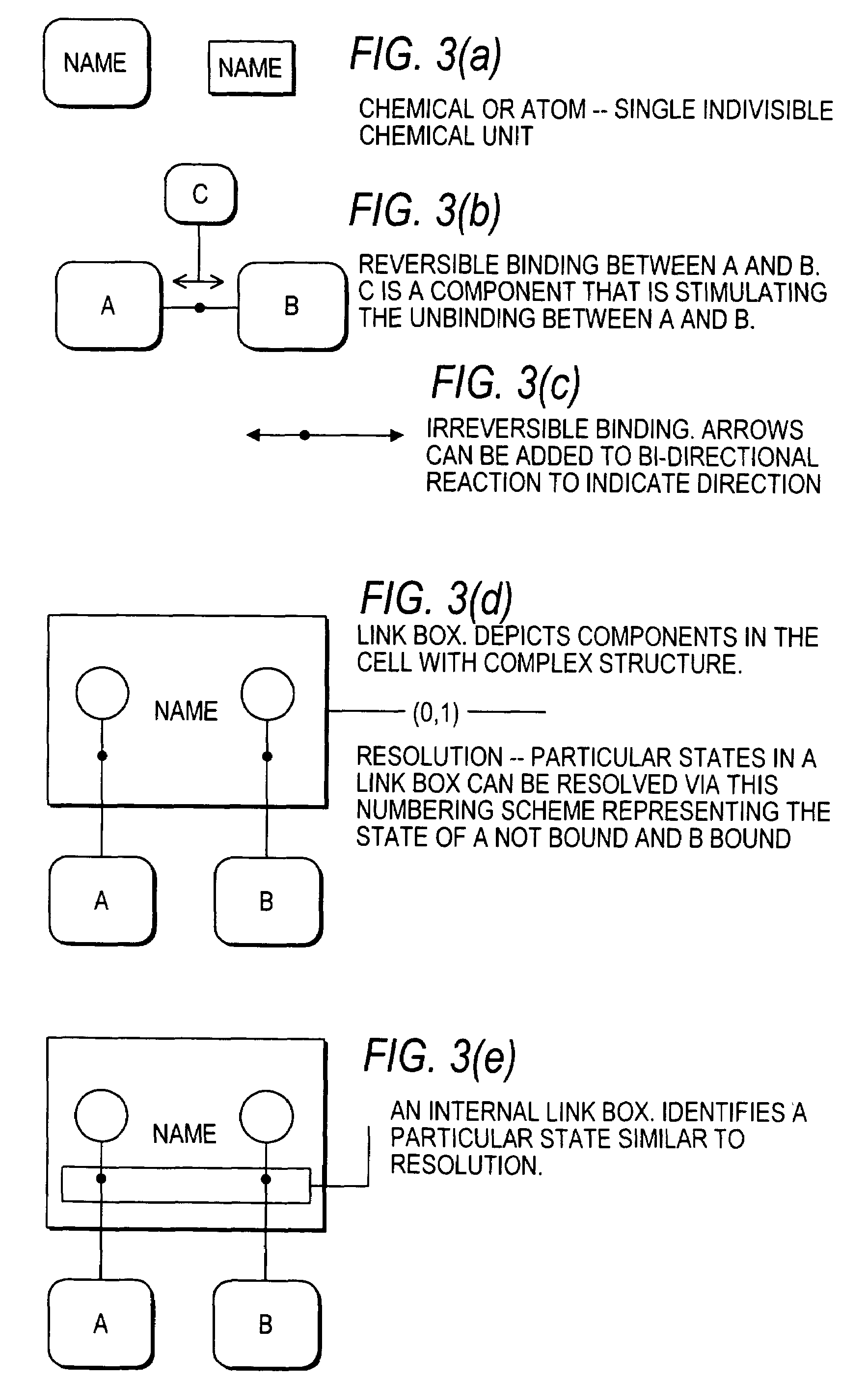
Methods and systems for the identification of components of mammalian biochemical networks as targets for therapeutic agents
InactiveUS7415359B2Low costAvoid delayData processing applicationsAnalogue computers for chemical processesCell stateAnalysis method
Systems and methods are presented for cell simulation and cell state prediction. For example, a cellular biochemical network intrinsic to a phenotype of a cell can be simulated by specifying its components and their interrelationships. The various interrelationships can be represented with one or more mathematical equations which can be solved to simulate a first state of the cell. The simulated network can then be perturbed, and the equations representing the perturbed network can be solved to simulate a second state of the cell which can then be compared to the first state, identifying the effect of such perturbation on the network, and thereby identifying one or more components as targets. Alternatively, components of a cell can be identified as targets for interaction with therapeutic agents based upon an analytical approach, in which a stable phenotype of a cell is specified and correlated to the state of the cell and the role of that cellular state to its operation. A cellular biochemical network believed intrinsic to that phenotype can then be specified, mathematically represented, and perturbed, and the equations representing the perturbed network solved, thereby identifying one or more components as targets.
Owner:CORNELL RES FOUNDATION INC +1
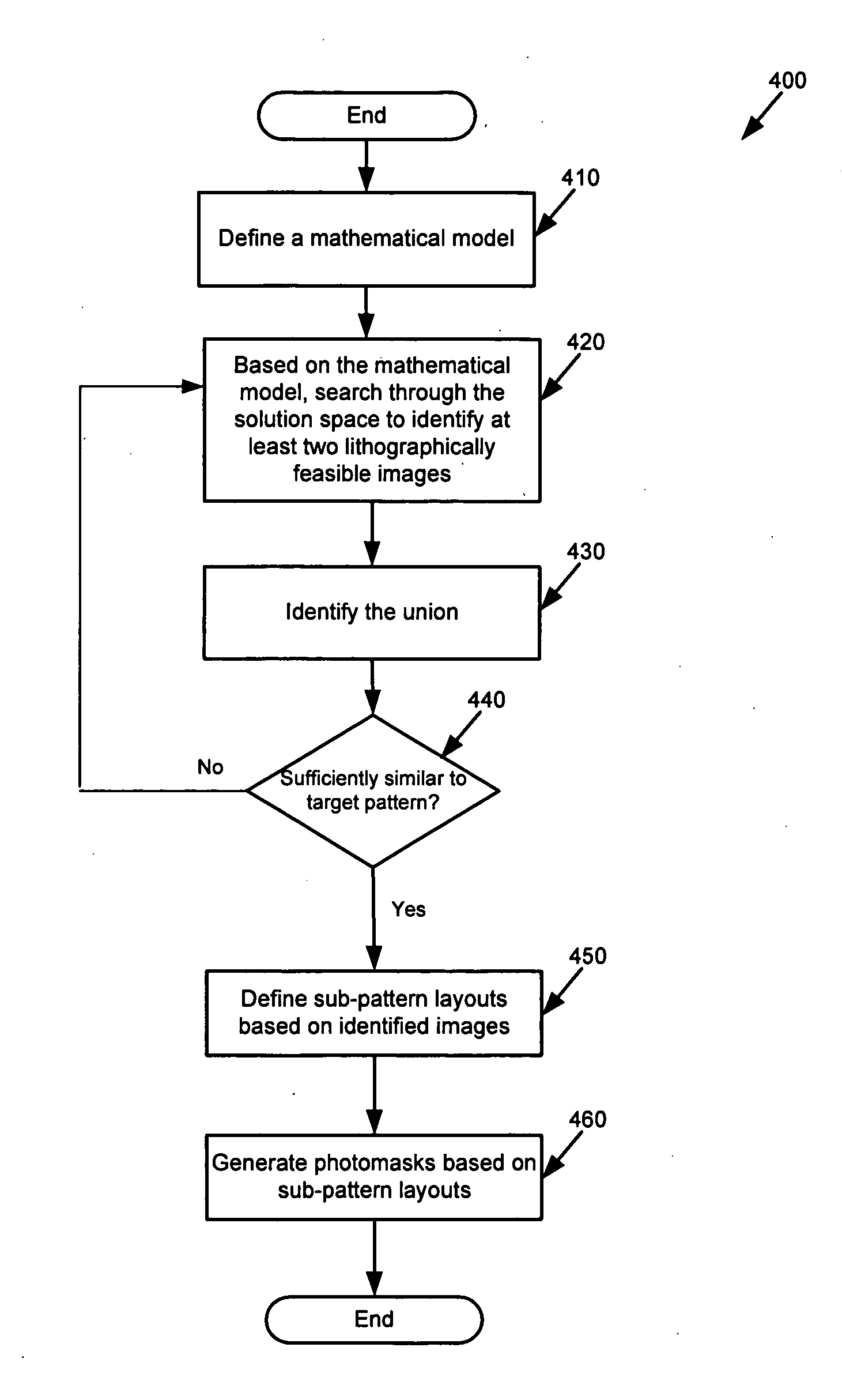
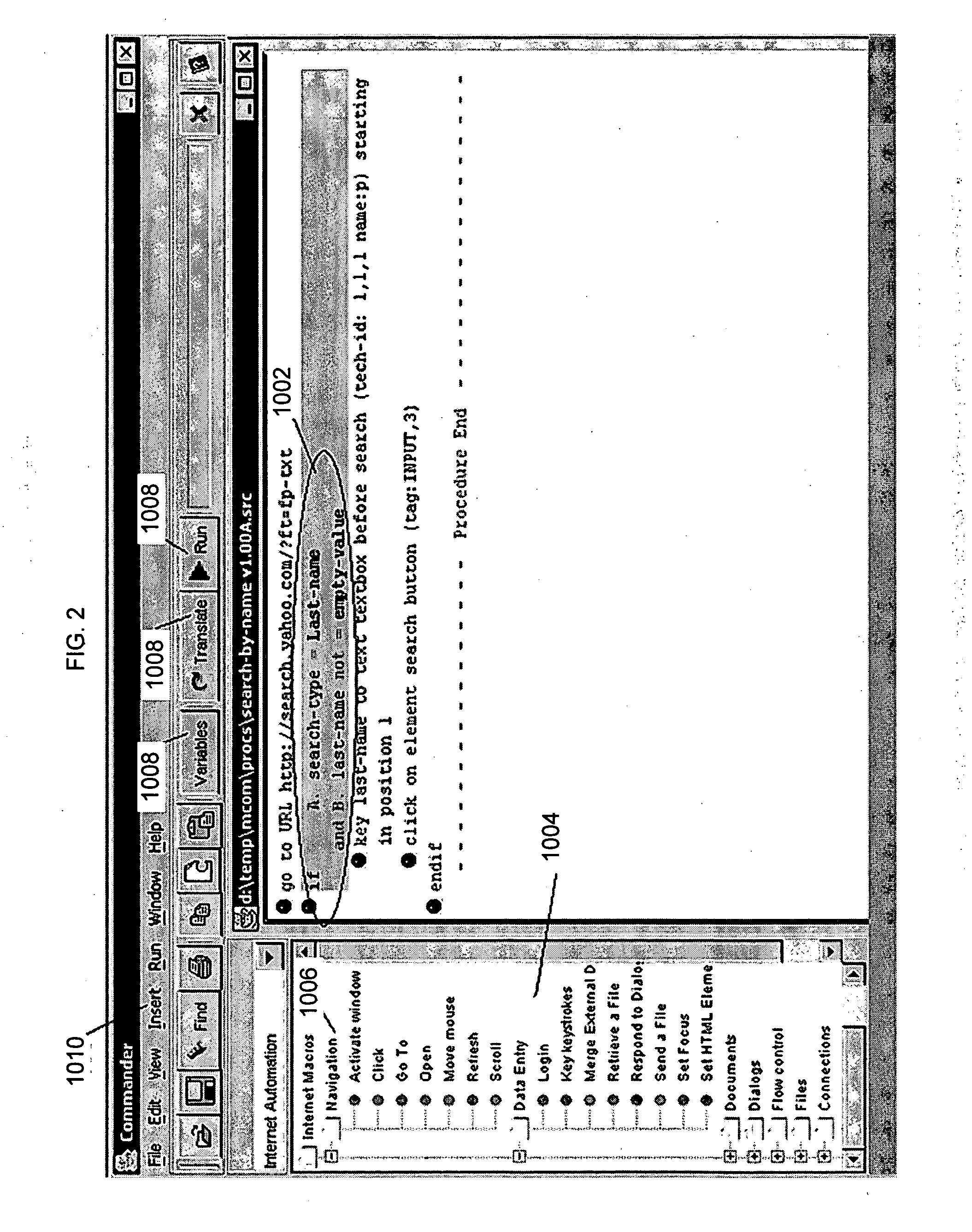
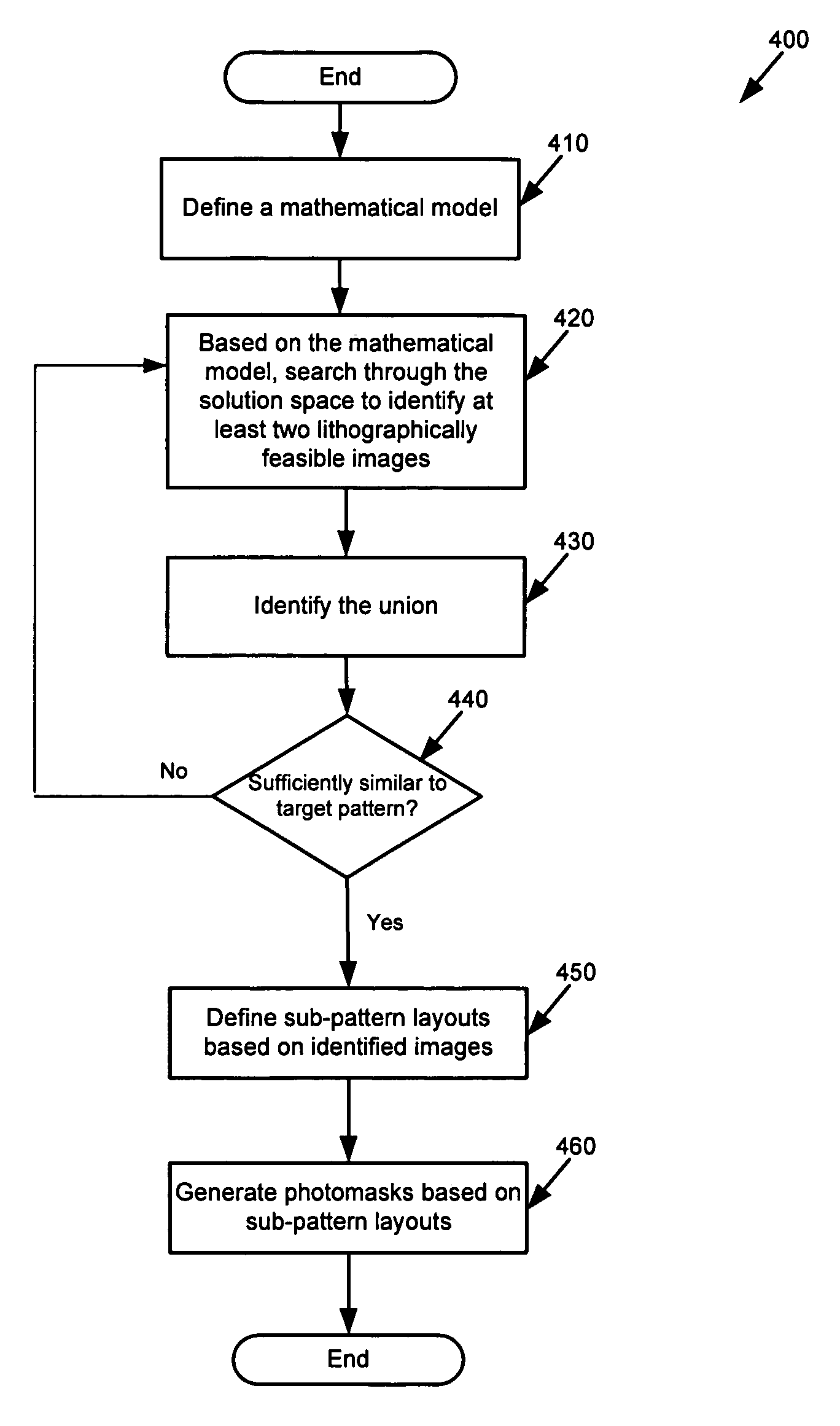
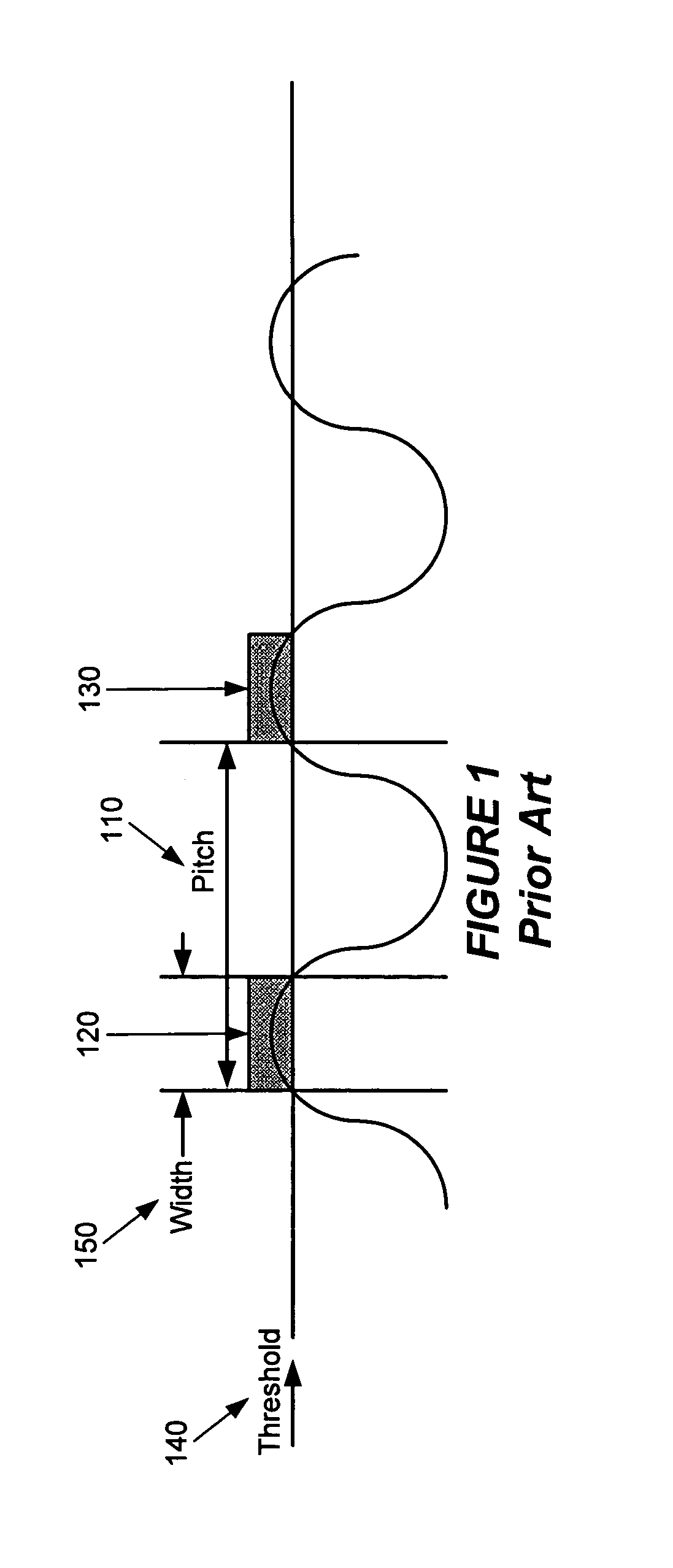
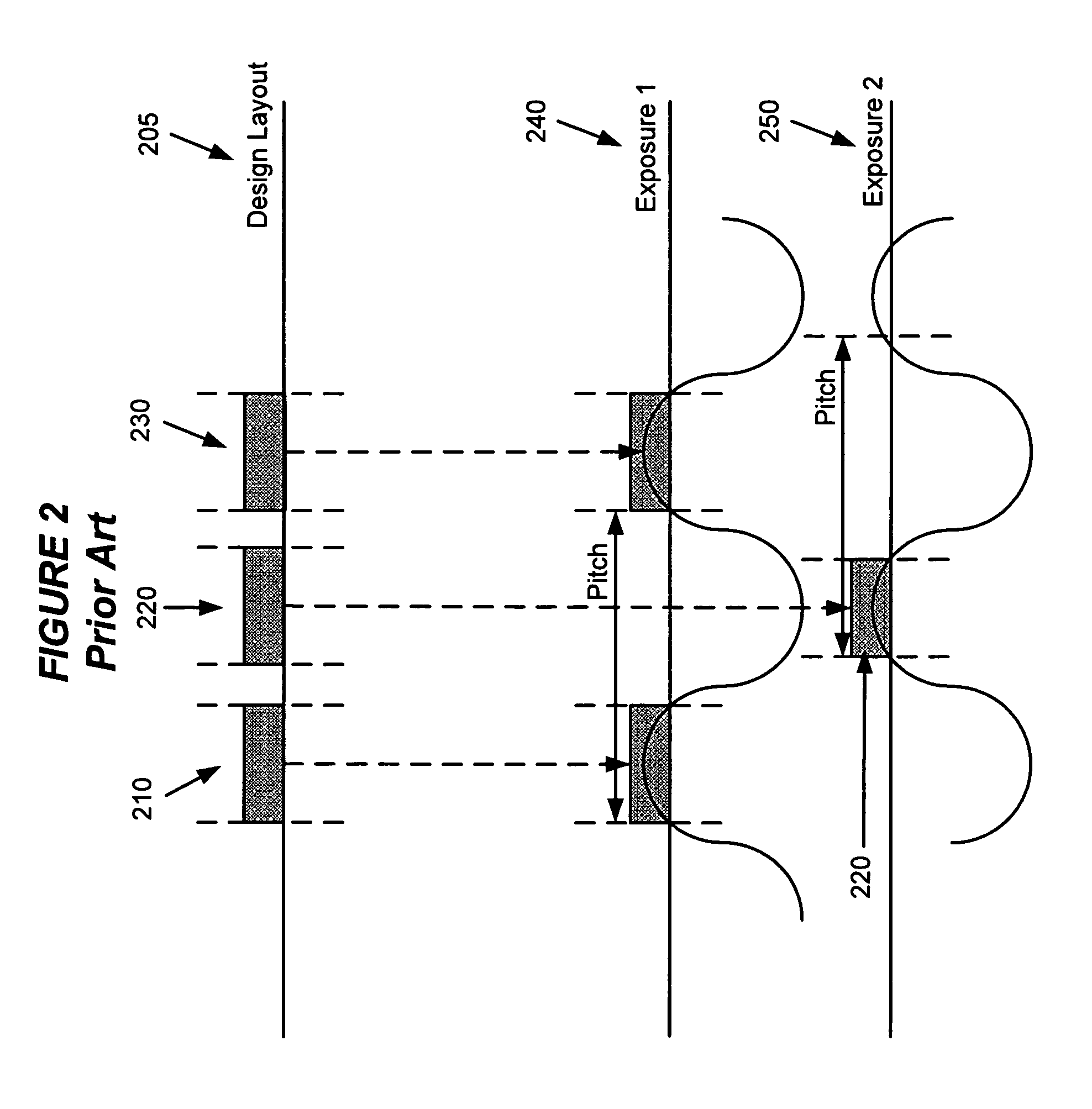
System and method for model based multi-patterning optimization
ActiveUS20100037200A1Photomechanical apparatusOriginals for photomechanical treatmentLithography processComputational physics
Some embodiments provide a method for optimally decomposing patterns within particular spatial regions of interest on a particular layer of a design layout for a multi-exposure photolithographic process. Specifically, some embodiments model the spatial region using a mathematical equation in terms of two or more intensities. Some embodiments then optimize the model across a set of feasible intensities. The optimization yields a set of intensities such that the union of the patterns created / printed from each exposure intensity most closely approximates the patterns within the particular regions. Based on the set of intensities, some embodiments then determine a decomposition solution for the patterns that satisfies design constraints of a multi-exposure photolithographic printing process. In this manner, some embodiments achieve an optimal photolithographic printing of the particular regions of interest without performing geometric rule based decomposition.
Owner:CADENCE DESIGN SYST INC
Method of nano thin film thickness measurement by auger electron spectroscopy
ActiveUS20060138326A1Material analysis using wave/particle radiationUsing wave/particle radiation meansMock upsMathematical model
A system and method for measuring the thickness of an ultra-thin multi-layer film on a substrate is disclosed. A physical model of an ultra-thin multilayer structure and Auger electron emission from the nano-multilayer structure is built. A mathematical model for the Auger Electron Spectroscopy (AES) measurement of the multilayer thin film thickness is derived according to the physical model. Auger electron spectroscopy (AES) is first performed on a series of calibration samples. The results are entered into the mathematical model to determine the parameters in the mathematical equation. The parameters may be calibrated by the correlation measurements of the alternate techniques. AES analysis is performed on the ultra-thin multi-layer film structure. The results are entered into the mathematical model and the thickness is calculated.
Owner:SAE MAGNETICS (HK) LTD
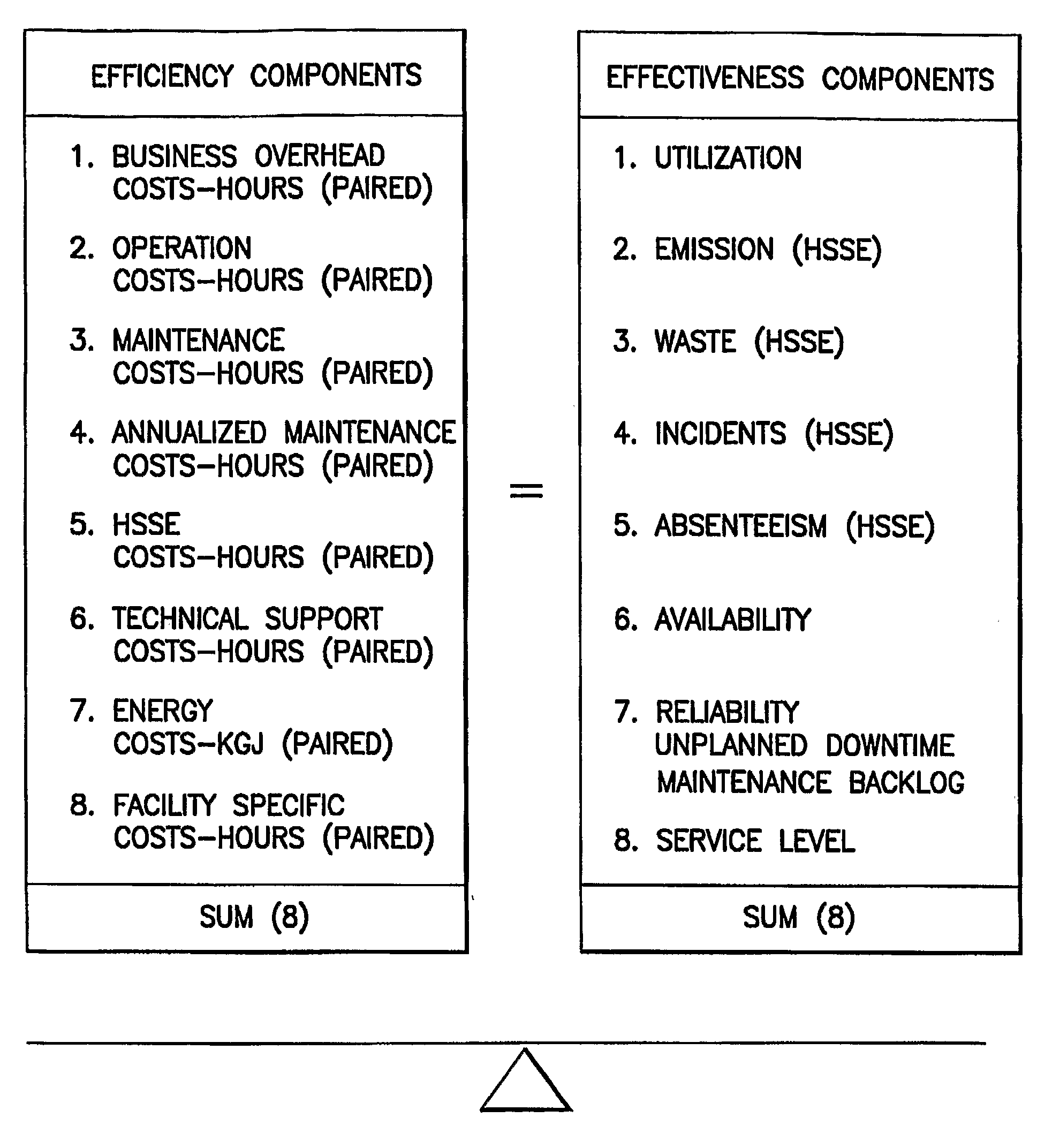
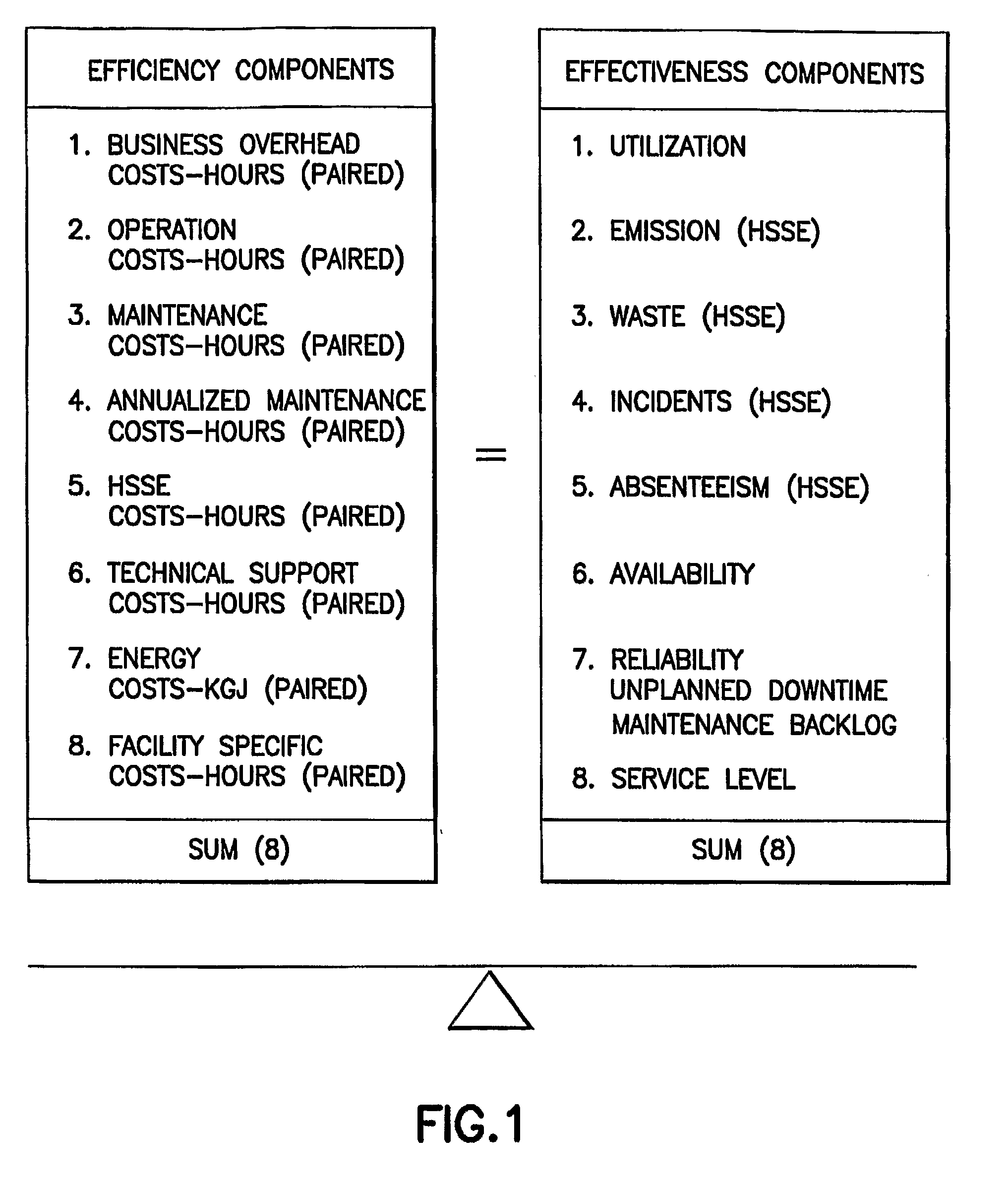
Method For Measuring The Overall Operational Performance Of Hydrocarbon Facilities
InactiveUS20080262898A1Inform balanceEasy to monitorData processing applicationsOil and natural gasPetroleum
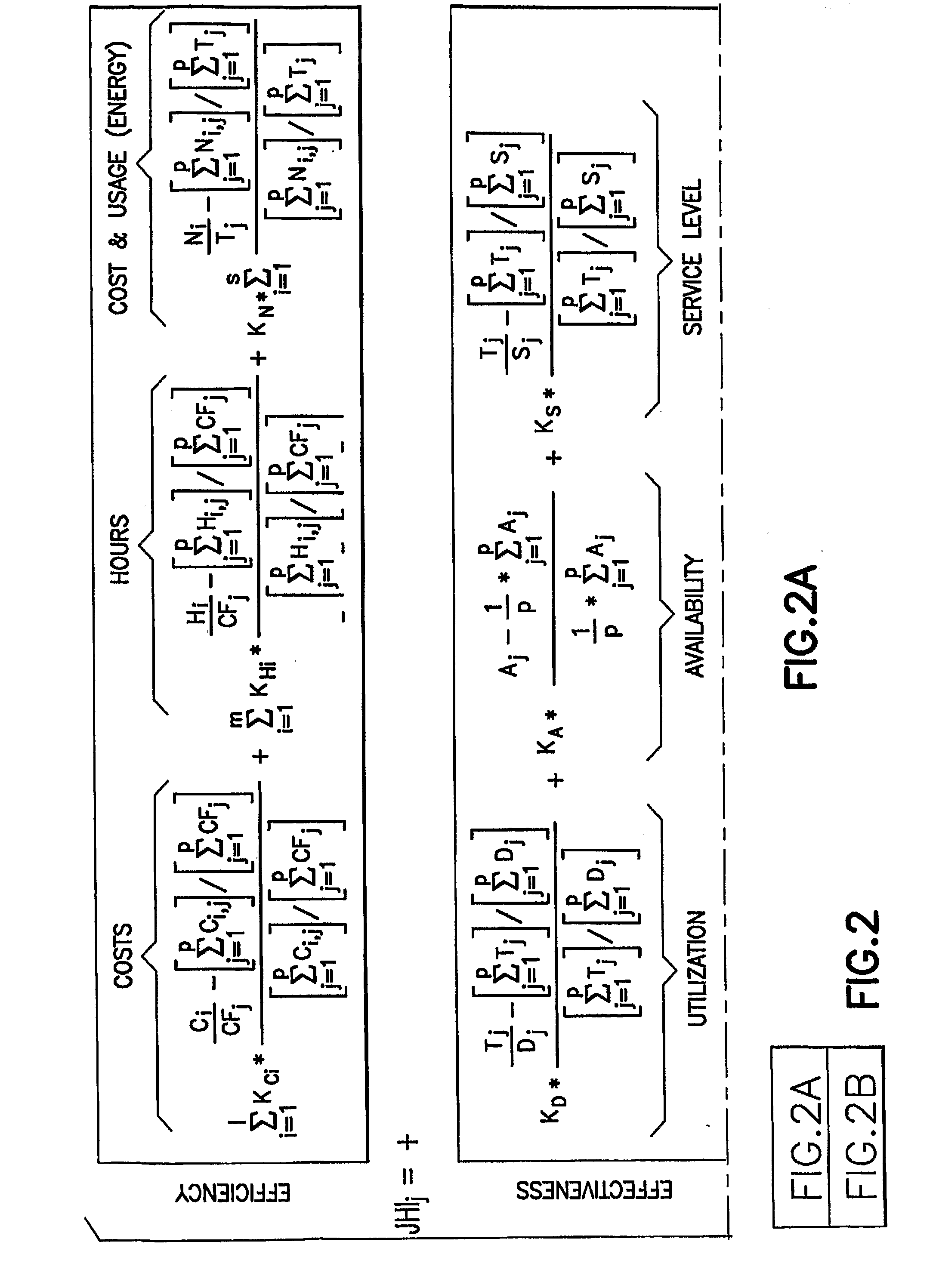
A method, named Juran Hydrocarbons Index (JHI), for measuring the overall competitiveness of an oil and gas facility (i.e. platform, pipeline, marine terminal, processing terminal, refinery, or LNG plant) that incorporates operational efficiency and effectiveness. The method describes a plurality of performance metrics that reflect measurable properties of an oil & gas organization. It further calculates the percentage deviation of these metrics relative to the corresponding industry or peer group average and assins weights of importance in the form of gap cofficients. Finally, it integrates all the individual performances into a mathematical equation to provide a numerical value (an index) of overall hydrocarbons competitiveness.
Owner:JURAN INST
Integrated resistance cancellation in temperature measurement systems
ActiveUS20060193370A1Eliminate needThermometer detailsThermometers using material expansion/contactionElectrical resistance and conductanceIntegrator
A temperature measurement device may be implemented by coupling a PN-junction, which may be comprised in a diode, to an analog-to-digital converter (ADC) that comprises an integrator. Different currents may be successively applied to the diode, resulting in different VBE values across the diode. The ΔVBE values thus obtained may be successively integrated. Appropriate values for the different currents may be determined based on a set of mathematical equations, each equation relating the VBE value to the temperature of the diode, the current applied to the diode and parasitic series resistance associated with the diode. When the current sources with the appropriate values are sequentially applied to the diode and the resulting diode voltage differences are integrated by the integrator comprised in the ADC, the error in the temperature measurement caused by series resistance is canceled in the ADC, and an accurate temperature reading of the diode is obtained from the output of the ADC.
Owner:MICROCHIP TECH INC
Method for implementing flexible combined overload control for aircraft in large airspace
InactiveCN102645933AOptimal Control StrategySimple overload controlSimulator controlAttitude controlAccelerometerFlight vehicle
The invention discloses a method for implementing flexible combined overload control for an aircraft in a large airspace. A combined measured value of an inertial navigation linear accelerometer and an angular speed meter is adopted as a control value, the technical difficulty in realizing overload control of a non-minimum phase system is solved by means of reconstructing a mathematical equation, asymptotic stability of an attach angle and a sideslip angle in a whole flying journey is completely realized, and the method is applicable to flexible small-space non-planar flying control for supersonic aircrafts with large-space variable trajectories. The method for implementing combined overload control has the advantages of intuitive structure, simple design and capability of completely meeting engineering application requirements.
Owner:NAVAL AERONAUTICAL & ASTRONAUTICAL UNIV PLA
Simulations for hydraulic fracturing treatments and methods of fracturing naturally fractured formation
A hydraulic fracture design model that simulates the complex physical process of fracture propagation in the earth driven by the injected fluid through a wellbore. An objective in the model is to adhere with the laws of physics governing the surface deformation of the created fracture subjected to the fluid pressure, the fluid flow in the gap formed by the opposing fracture surfaces, the propagation of the fracture front, the transport of the proppant in the fracture carried by the fluid, and the leakoff of the fracturing fluid into the permeable rock. The models used in accordance with methods of the invention are typically based on the assumptions and the mathematical equations for the conventional 2D or P3D models, and further take into account the network of jointed fracture segments. For each fracture segment, the mathematical equations governing the fracture deformation and fluid flow apply. For each time step, the model predicts the incremental growth of the branch tips and the pressure and flow rate distribution in the system by solving the governing equations and satisfying the boundary conditions at the fracture tips, wellbore and connected branch joints. An iterative technique is used to obtain the solution of this highly nonlinear and complex problem.
Owner:SCHLUMBERGER TECH CORP
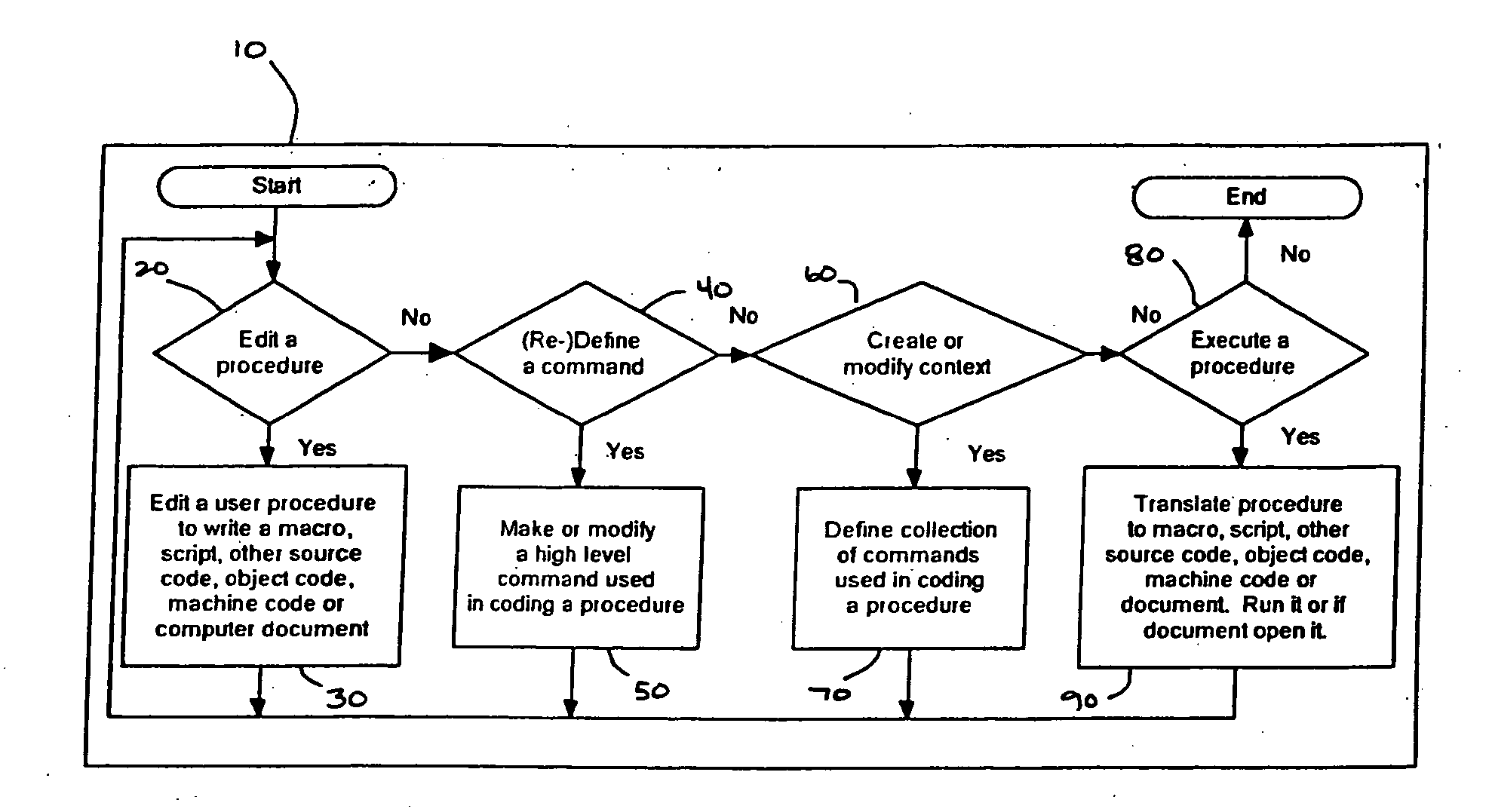
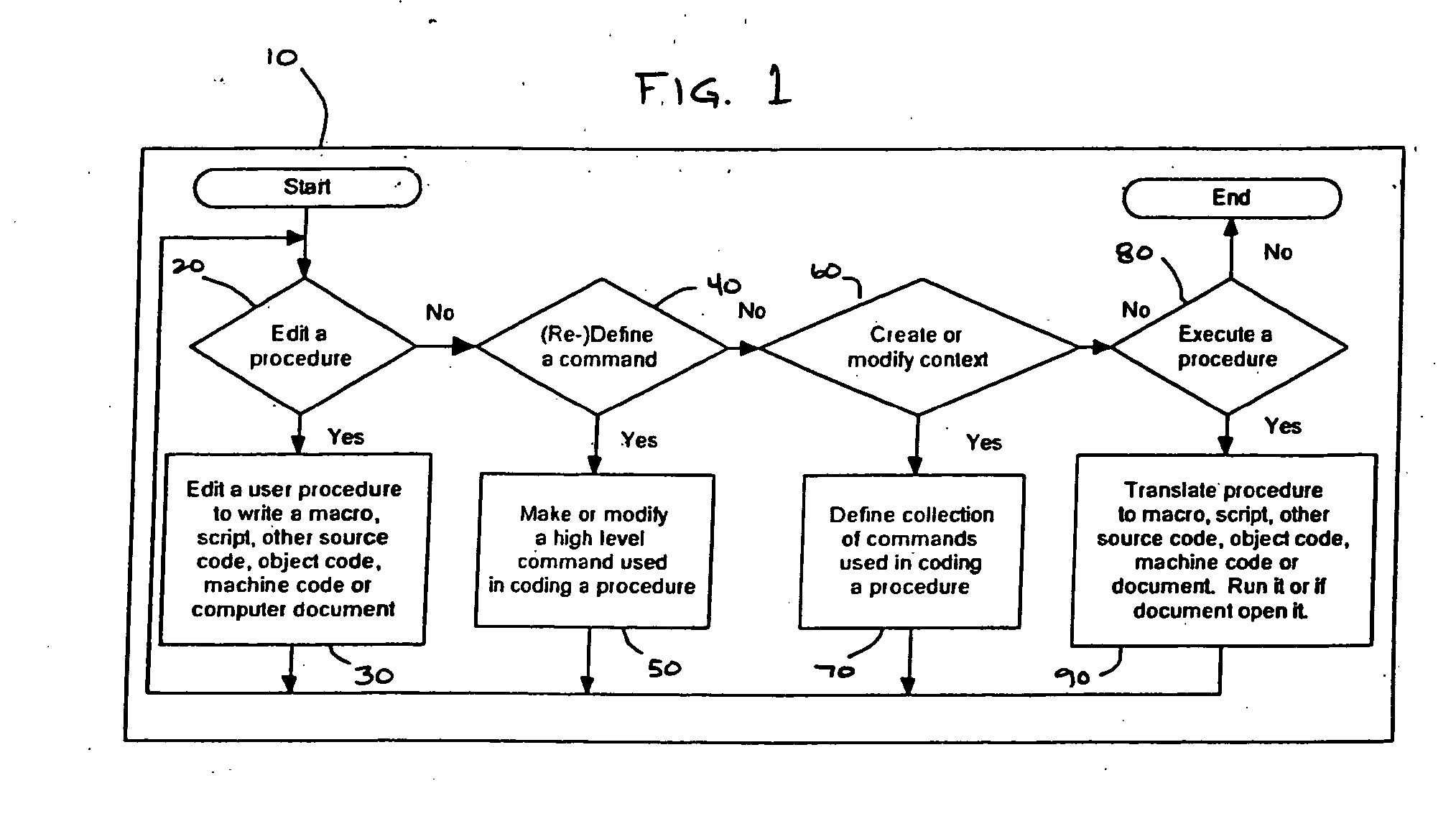
Computer source code generator
InactiveUS20070005342A1Simple processSimplify the coding processIntelligent editorsSpecific program execution arrangementsMinimal knowledgeGoal programming
A programming editor and creator allows users with minimal knowledge of a computer language syntax to generate computer source code using a structured rather than freeform process. A predefined command sentence is provided representing at least a portion of a programming command, and includes words, which may be defined as constant or enterable words. Enterable words may be required or optional, and correspond to entry components. Each word may be defined according to word types, including list words, variable list words, variable words, mathematical words, condition words or Boolean words. Enterable words may include repeatable words which may be inserted multiple times within a command to create conditional statements, mathematical equations and string concatenations. Input values are provided for the entry components. The predefined command sentence with the input values is converted to a completed command sentence corresponding to the programming command, which is translated to a target programming language.
Owner:ORTSCHEID RONALD
System and method for model based multi-patterning optimization
ActiveUS8069423B2Photomechanical apparatusOriginals for photomechanical treatmentDecompositionAlgorithm
Some embodiments provide a method for optimally decomposing patterns within particular spatial regions of interest on a particular layer of a design layout for a multi-exposure photolithographic process. Specifically, some embodiments model the spatial region using a mathematical equation in terms of two or more intensities. Some embodiments then optimize the model across a set of feasible intensities. The optimization yields a set of intensities such that the union of the patterns created / printed from each exposure intensity most closely approximates the patterns within the particular regions. Based on the set of intensities, some embodiments then determine a decomposition solution for the patterns that satisfies design constraints of a multi-exposure photolithographic printing process. In this manner, some embodiments achieve an optimal photolithographic printing of the particular regions of interest without performing geometric rule based decomposition.
Owner:CADENCE DESIGN SYST INC
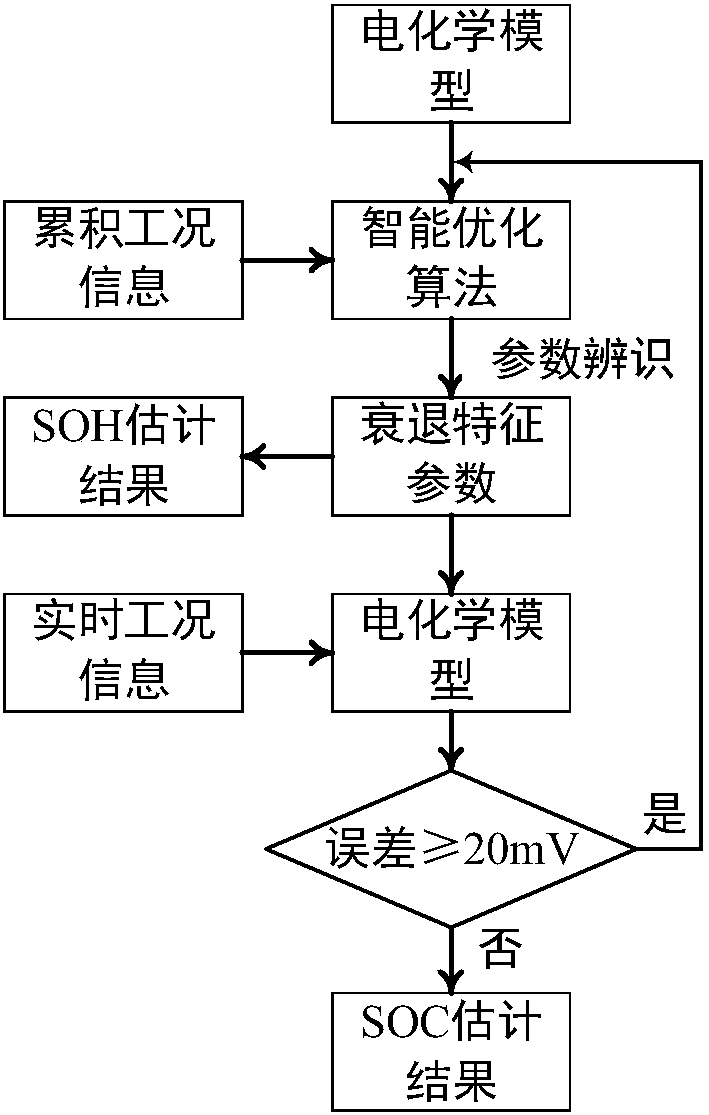
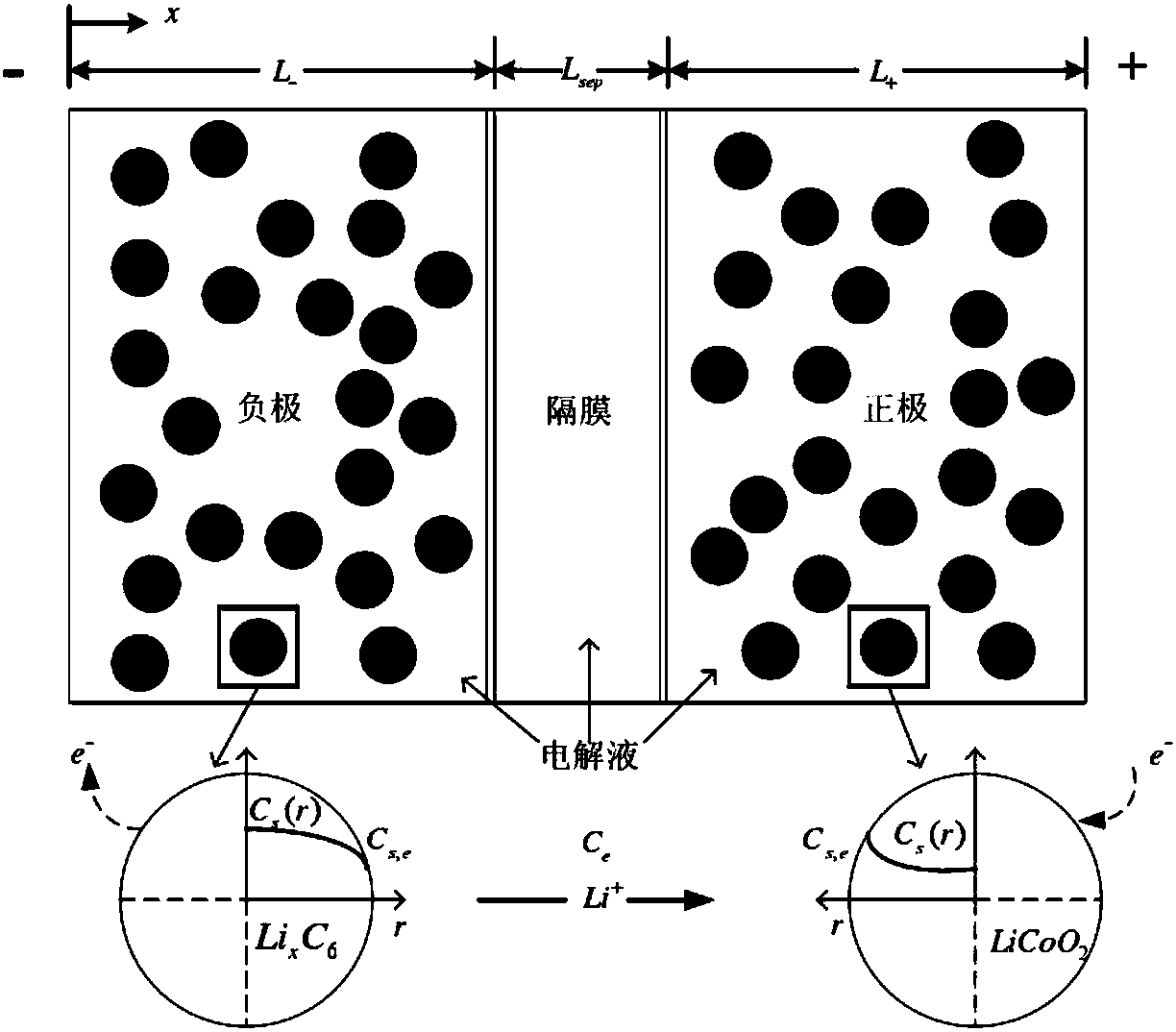
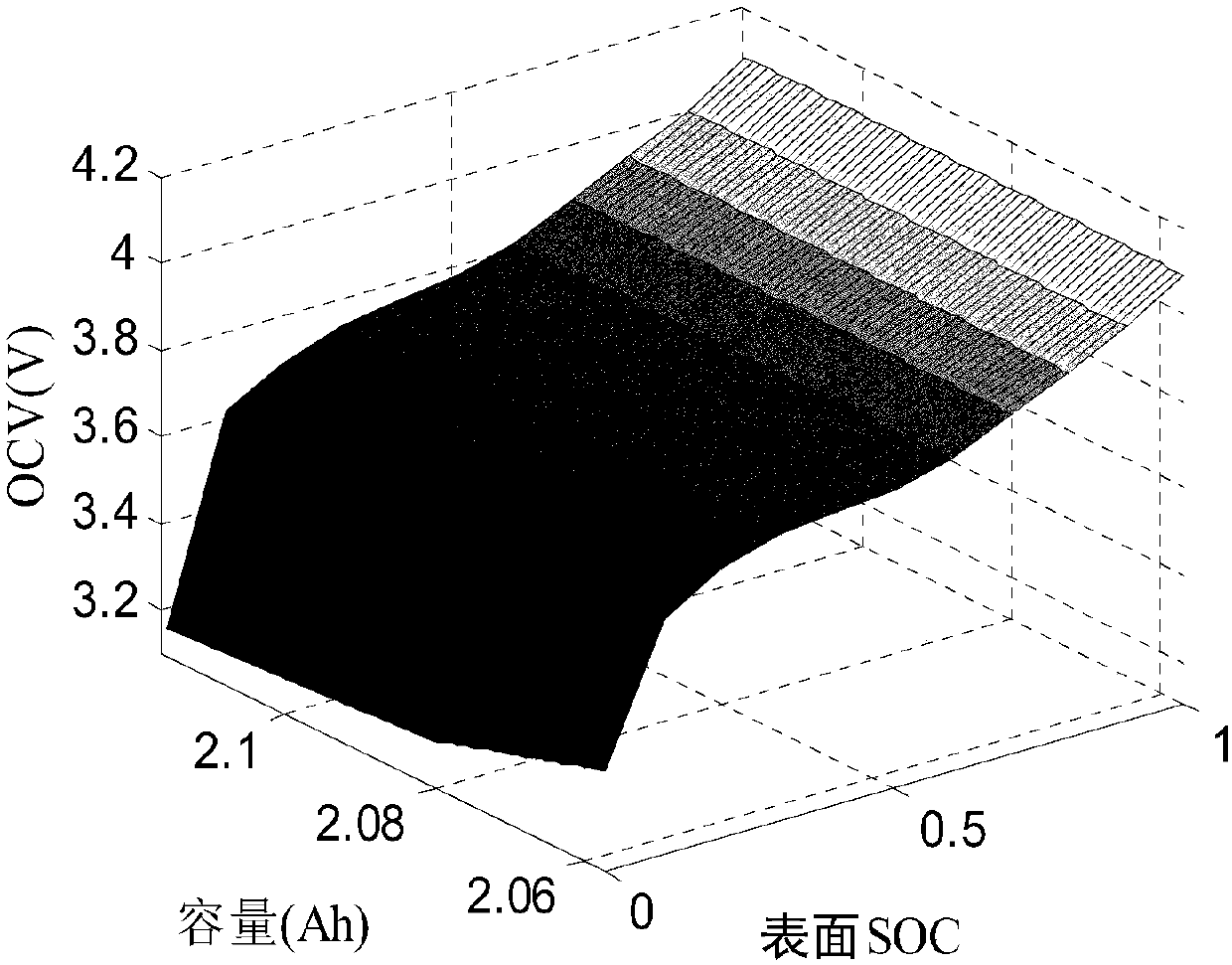
Joint estimation method of state of charge and state of health of power battery system based on electrochemical model
ActiveCN107066722AImprove reliabilityReduce computing timeDesign optimisation/simulationSpecial data processing applicationsState of healthElectrical battery
The invention relates to a joint estimation method of a state of charge (SOC) and a state of health (SOH) of a power battery system based on an electrochemical model. The method comprises the steps of establishing a pseudo two-dimensional electrochemical model correlation mathematical equation based on an electrochemical correlation theory, performing dimensionality reduction processing and calculation solution on a complicated partial differential equation by a finite analysis method and a numerical calculation method, allowing an open circuit potential of an electrode to be equivalent to open circuit voltage of a battery, establishing a capacity-surface SOC-EOCV three-dimensional response surface, extracting and establishing a degradation path graph of a battery performance degradation characteristic parameter by an intelligent optimization algorithm based on battery ageing test data, and finally realizing estimation of the SOH of the power battery based on the degradation path graph and realizing estimation of the SOC of the power battery based on concentration distribution and change rules of lithium ions in the battery.
Owner:BEIJING INSTITUTE OF TECHNOLOGYGY
Method of measuring the flux of a soil gas
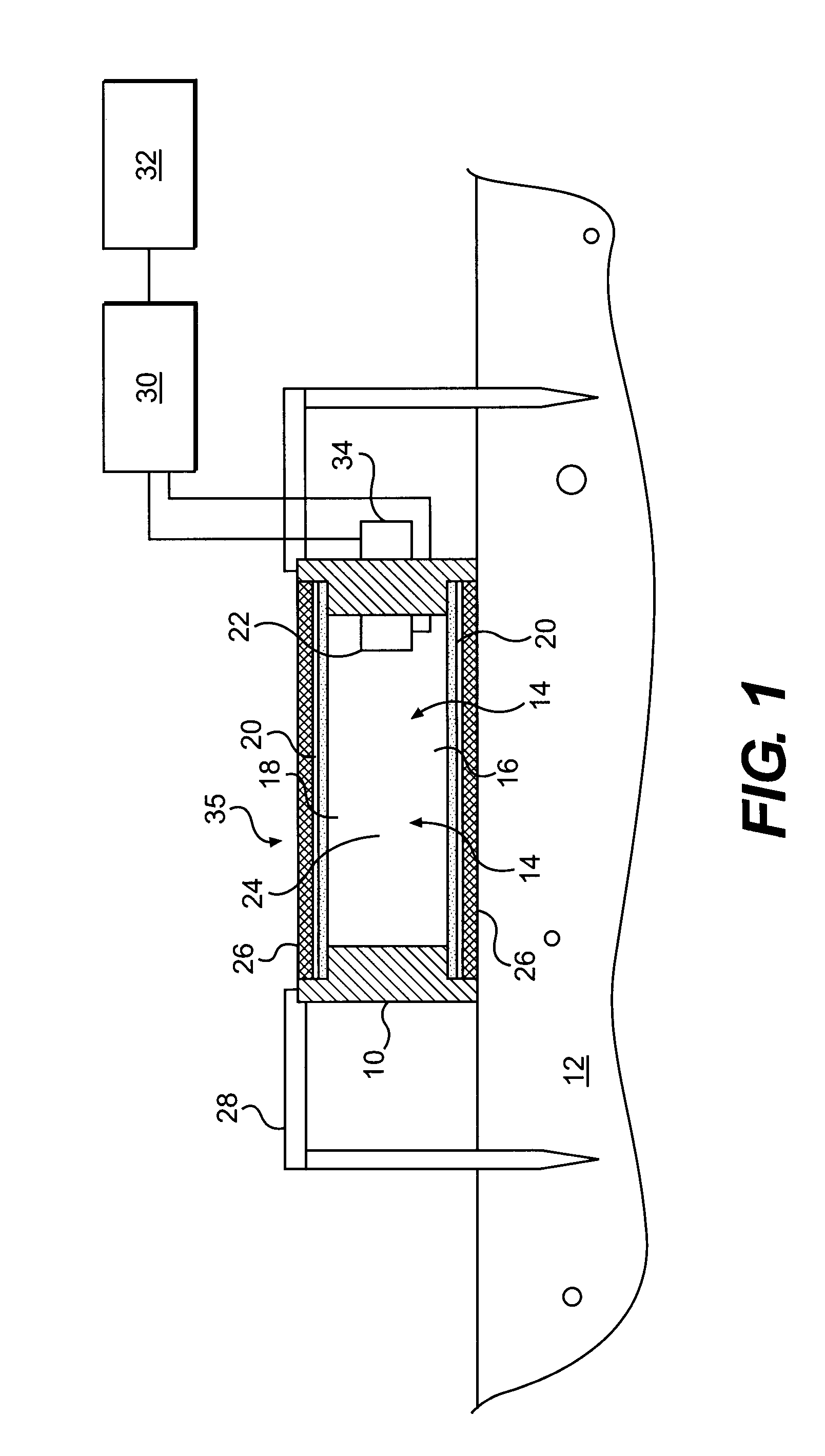
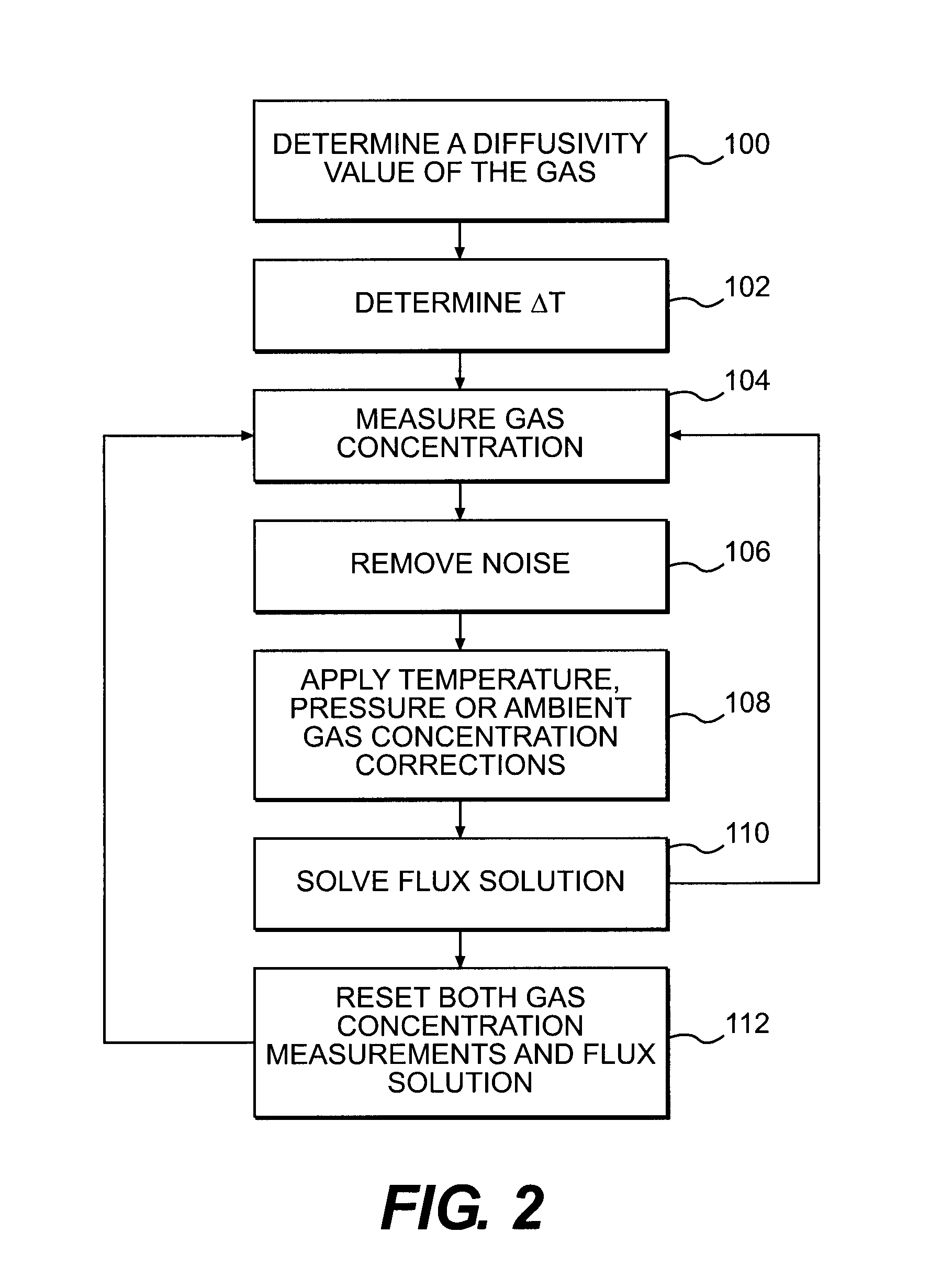
ActiveUS20120035850A1Easy to practiceEasy to deploySamplingProspecting/detection of underground/near-surface gasesSoil gasGas flux
A method for determining a flux of a gas contained in a medium through a boundary of the medium comprises 1) measuring at least twice with a probe a concentration C of the gas over a time interval Δt and 2) determining the flux of the gas using the following mathematical equation: (I) where F is the gas flux, D is the diffusivity value and ΔC is a variation in the gas concentration during the time interval Δt. The probe is placed proximate the boundary. The probe has a gas inlet, a cavity, a gas concentration sensor and a membrane. Each element is in fluid communication with each other so that the gas flows from the gas inlet through the membrane and contacts the gas concentration sensor.F=(D∂C(z,t)∂z)z=0=DΔCπΔt(I)
Owner:EOSENSE INC
Transmitter, method and terminal for allocating resources, and method of receiving resource allocation information
ActiveUS20140307692A1Inter user/terminal allocationDistributed allocationCommunications systemResource block
The present invention relates to resource allocation in a wireless communication system. Specifically, the method comprises: an encoding step in which a resource block or a resource block group generates a resource indication value (RIV) which is a transform of the length (L) and the offset (j) of a contiguous resource allocation region; and a transmission step in which the resource allocation information is transmitted to a terminal, wherein the resource allocation information is calculated by means of a prescribed mathematical equation.
Owner:PANTECH CORP
System and method for modelling system behaviour
ActiveUS10565329B2Reduce impactMedical simulationDesign optimisation/simulationCollective modelAlgorithm
A method of modelling system behaviour of a physical system, the method including, in one or more electronic processing devices obtaining quantified system data measured for the physical system, the quantified system data being at least partially indicative of the system behaviour for at least a time period, forming at least one population of model units, each model unit including model parameters and at least part of a model, the model parameters being at least partially based on the quantified system data, each model including one or more mathematical equations for modelling system behaviour, for each model unit calculating at least one solution trajectory for at least part of the at least one time period; determining a fitness value based at least in part on the at least one solution trajectory; and, selecting a combination of model units using the fitness values of each model unit, the combination of model units representing a collective model that models the system behaviour.
Owner:EVOLVING MACHINE INTELLIGENCE
Boost circuit modeling approach and use thereof based on finite state machine
InactiveCN101393578AEffective modelingReduce volumeSpecial data processing applicationsCircuit modelingVirtual finite-state machine
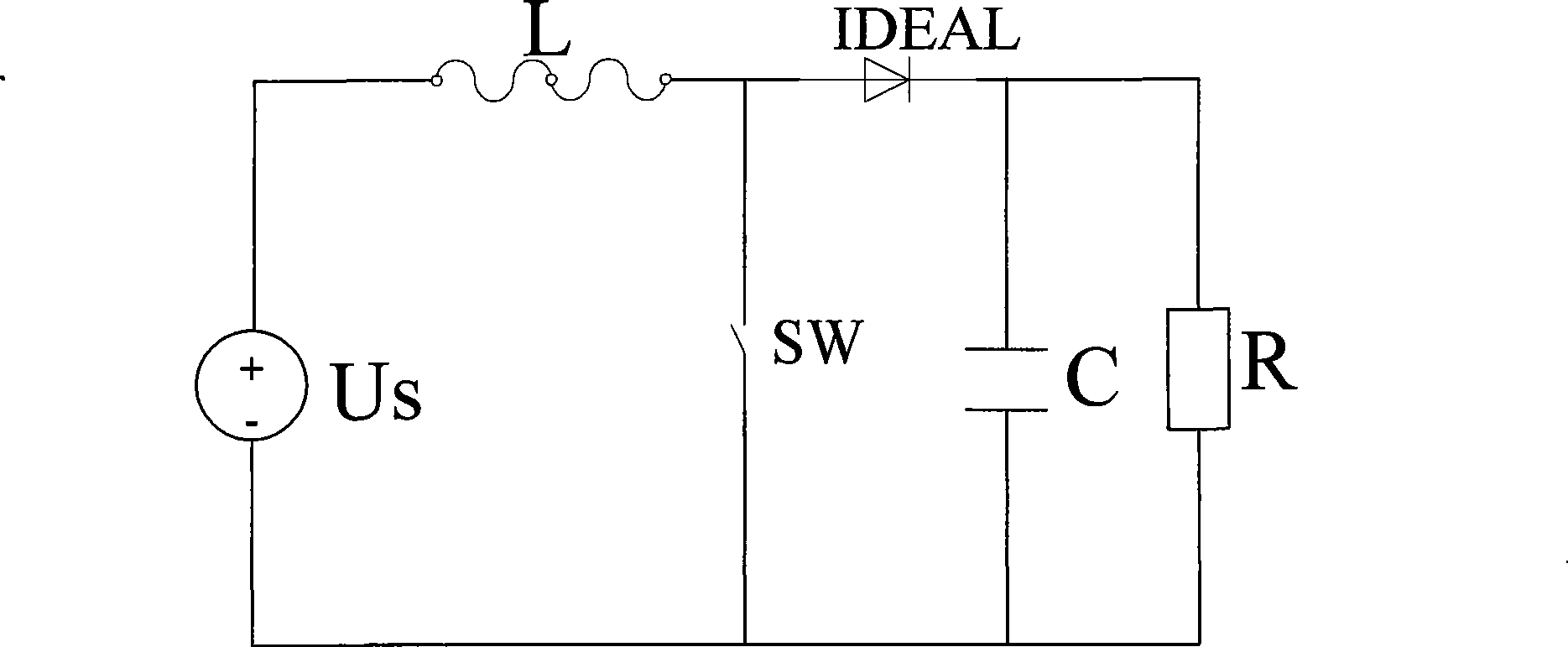
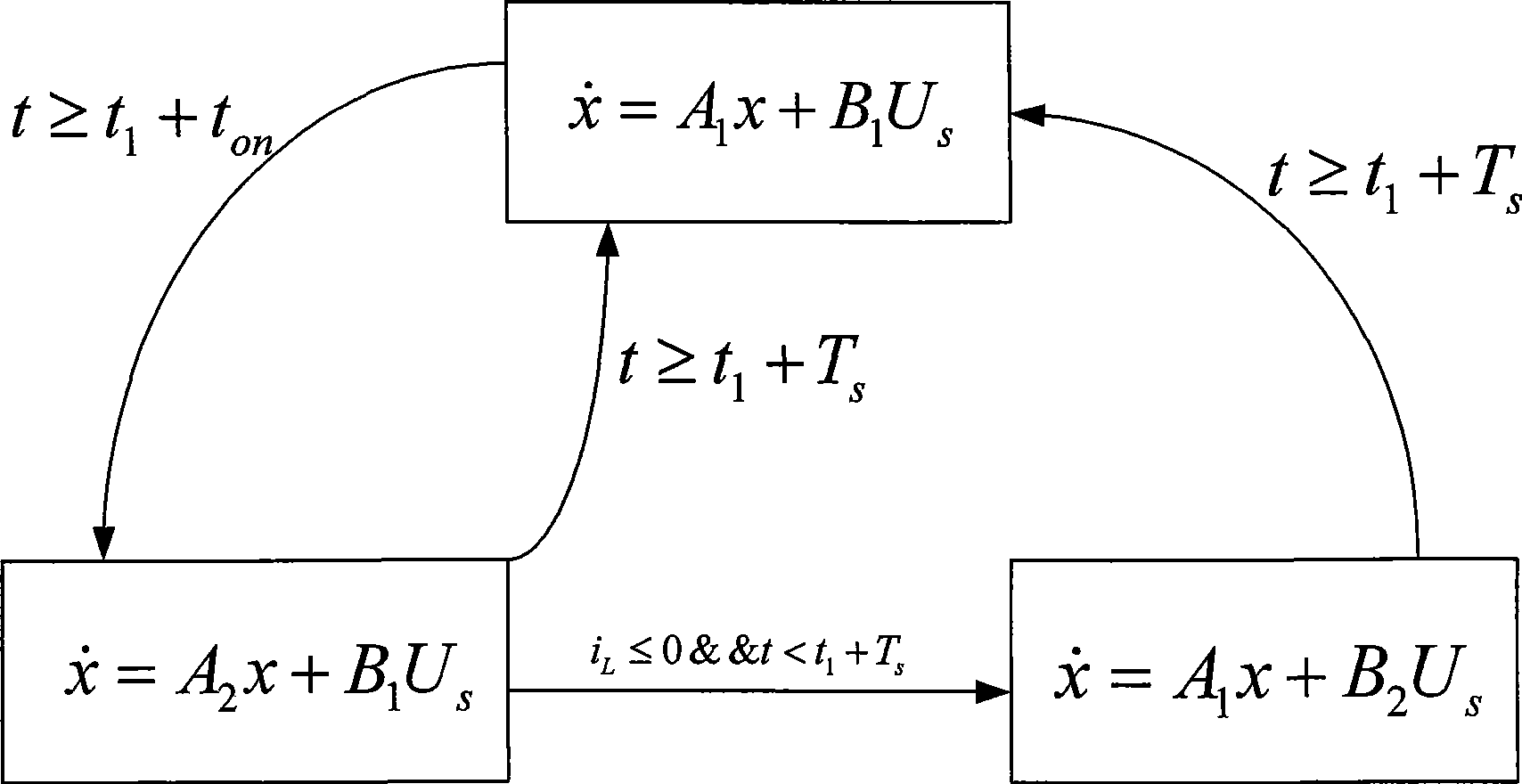
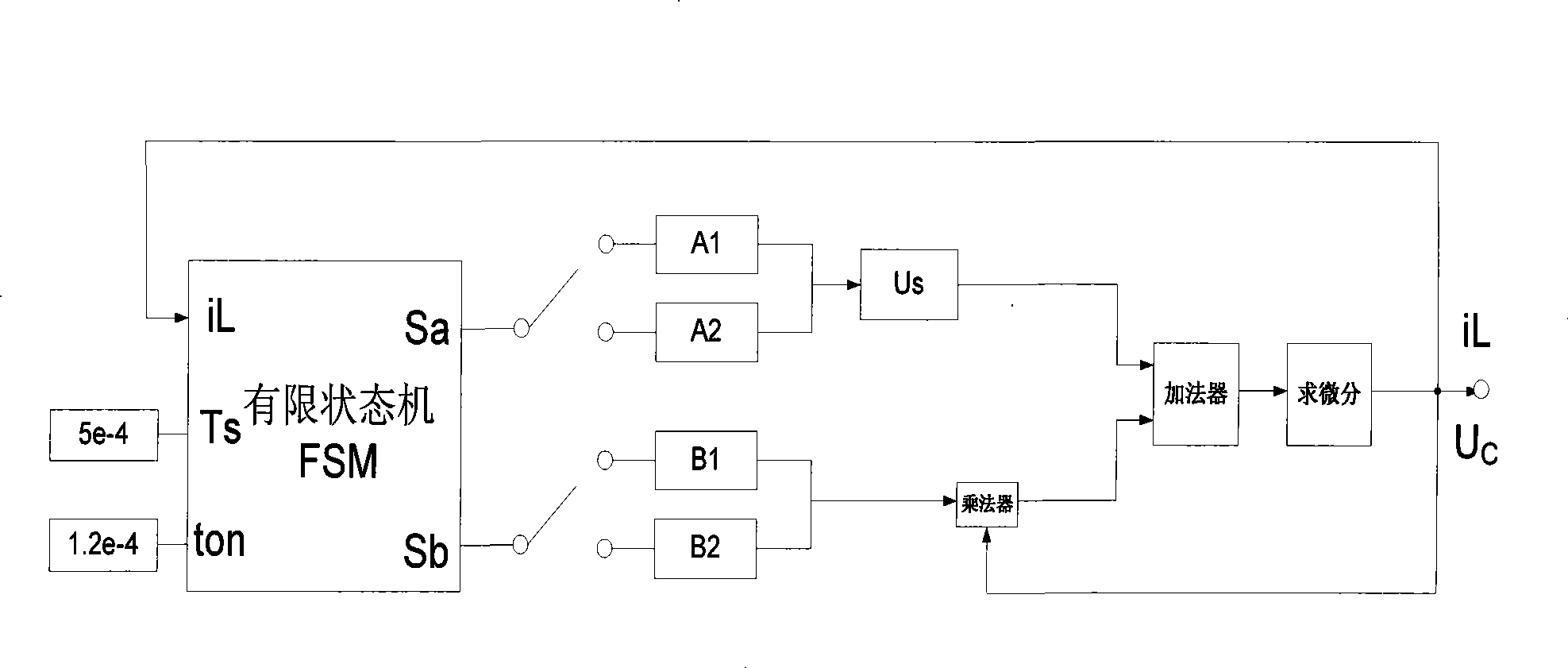
The invention provides a finite state machine-based Boost circuit modeling method and application thereof. The modeling method is characterized by comprising the following steps: (1) determining the number of operational states; (2) determining conversion conditions among the operational states; (3) establishing a mathematical equation; (4) determining a matrix according to parameters; and (5) building a model. The modeling method has the advantages that the Boost circuit model built by using the finite state machine is very precise and is easy to analyze the control characteristics of a system, especially a plurality of complicated characteristics caused by the switching of a switch in a converter system. The Boost circuit has the advantages of small volume, simple structure, high conversion efficiency and so on.
Owner:TIANJIN UNIVERSITY OF TECHNOLOGY
System and method for providing an interactive visual learning environment for creation, presentation, sharing, organizing and analysis of knowledge on subject matter
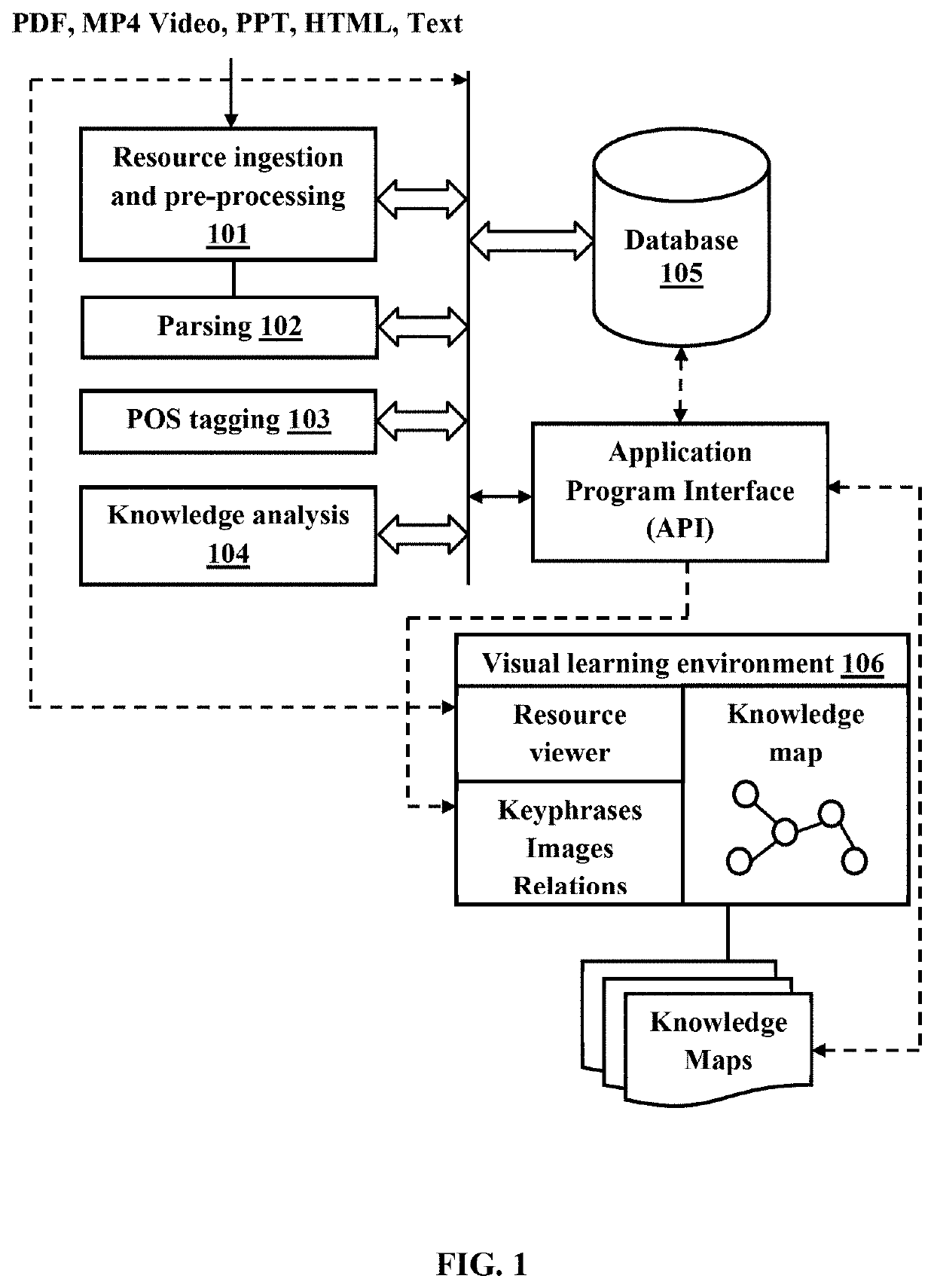
ActiveUS20210065569A1Easy to learnEnhance memorySemantic analysisElectrical appliancesPersonalizationSubject matter
The embodiments herein disclose a system and a method for providing an online web-based interactive audio-visual platform for note creation, presentation, sharing, organizing, and analysis. The system provides a conceptual and interactive interface to content; analyses a student's notes and instantly determines the accuracy of the conceptual connections made and a student's understanding of a topic. The system enables the student to add and use audio, visual, drawing, text notes, and mathematical equations in addition to those suggested by the note taking solution; to collate notes from various sources in a meaningful manner by grouping concepts using colors, images, and text; and to personalize other maps developed within the same environment while maintaining links back to the original source from which the notes are derived. The system highlights keywords in conjunction with spoken text to complement the advantages of using visual maps to improve learning outcomes.
Owner:IDEAPHORA INDIA PTE LTD
Methods and systems for the identification of components of mammalian biochemical networks as targets for therapeutic agents
InactiveUS20030215786A1Low costAvoid delayData processing applicationsMicrobiological testing/measurementCell biochemistryAnalysis method
Systems and methods for modeling the interactions of the several genes, proteins and other components of a cell, employing mathematical techniques to represent the interrelationships between the cell components and the manipulation of the dynamics of the cell to determine which components of a cell may be targets for interaction with therapeutic agents. A first such method is based on a cell simulation approach in which a cellular biochemical network intrinsic to a phenotype of the cell is simulated by specifying its components and their interrelationships. The various interrelationships are represented with one or more mathematical equations which are solved to simulate a first state of the cell. The simulated network is then perturbed by deleting one or more components, changing the concentration of one or more components, or modifying one or more mathematical equations representing the interrelationships between one or more of the components. The equations representing the perturbed network are solved to simulate a second state of the cell which is compared to the first state to identify the effect of the perturbation on the state of the network, thereby identifying one or more components as targets. A second method for identifying components of a cell as targets for interaction with therapeutic agents is based upon an analytical approach, in which a stable phenotype of a cell is specified and correlated to the state of the cell and the role of that cellular state to its operation. A cellular biochemical network believed to be intrinsic to that phenotype is then specified by identifying its components and their interrelationships and representing those interrelationships in one or more mathematical equations. The network is then perturbed and the equations representing the perturbed network are solved to determine whether the perturbation is likely to cause the transition of the cell from one phenotype to another, thereby identifying one or more components as targets.
Owner:CORNELL RES FOUNDATION INC +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com