Patents
Literature
144 results about "Immunologic assay" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Immunological assays are a biochemical test that measures the presence or concentration of a macromolecule or a small molecule in a solution through the use of an antibody or an antigen.
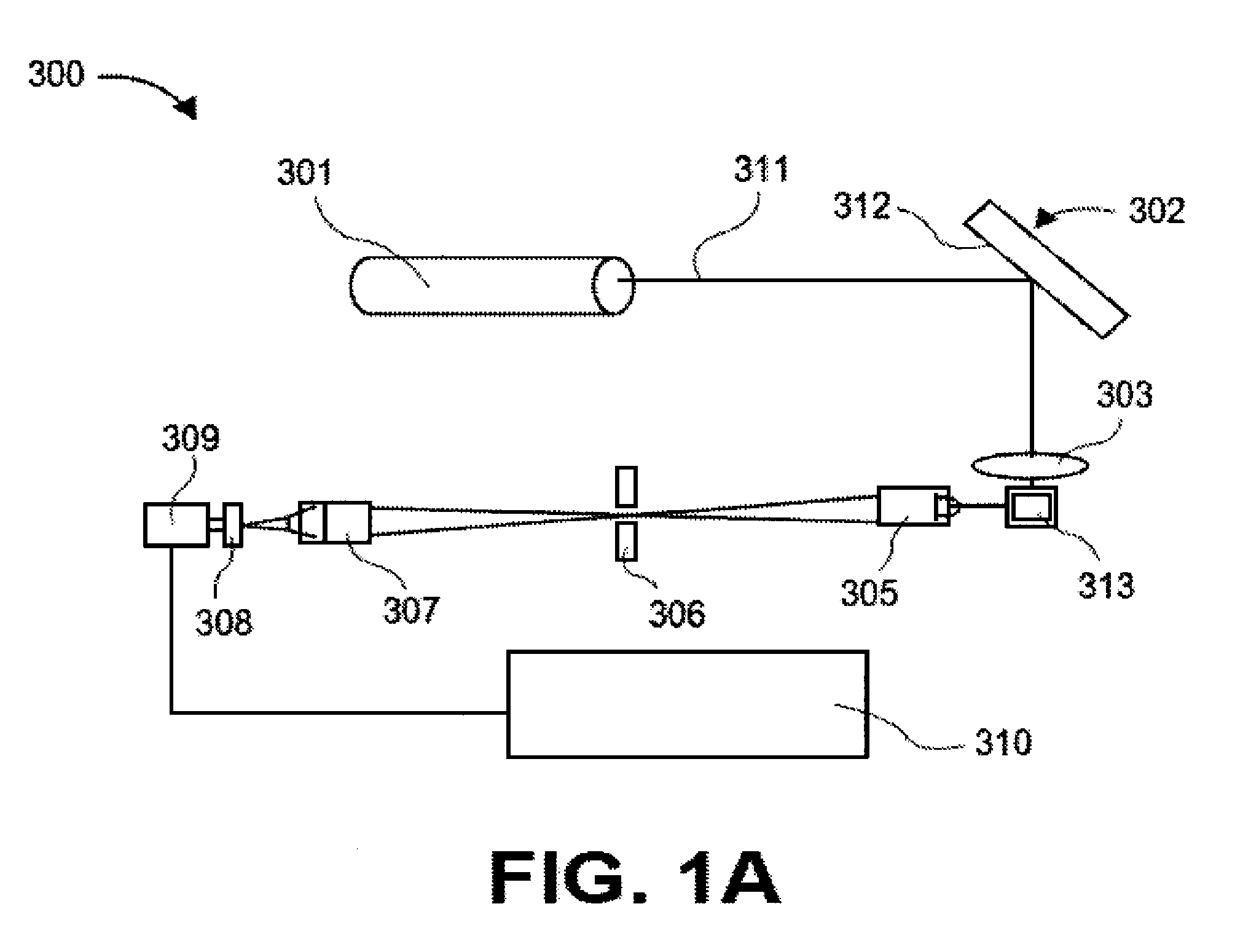
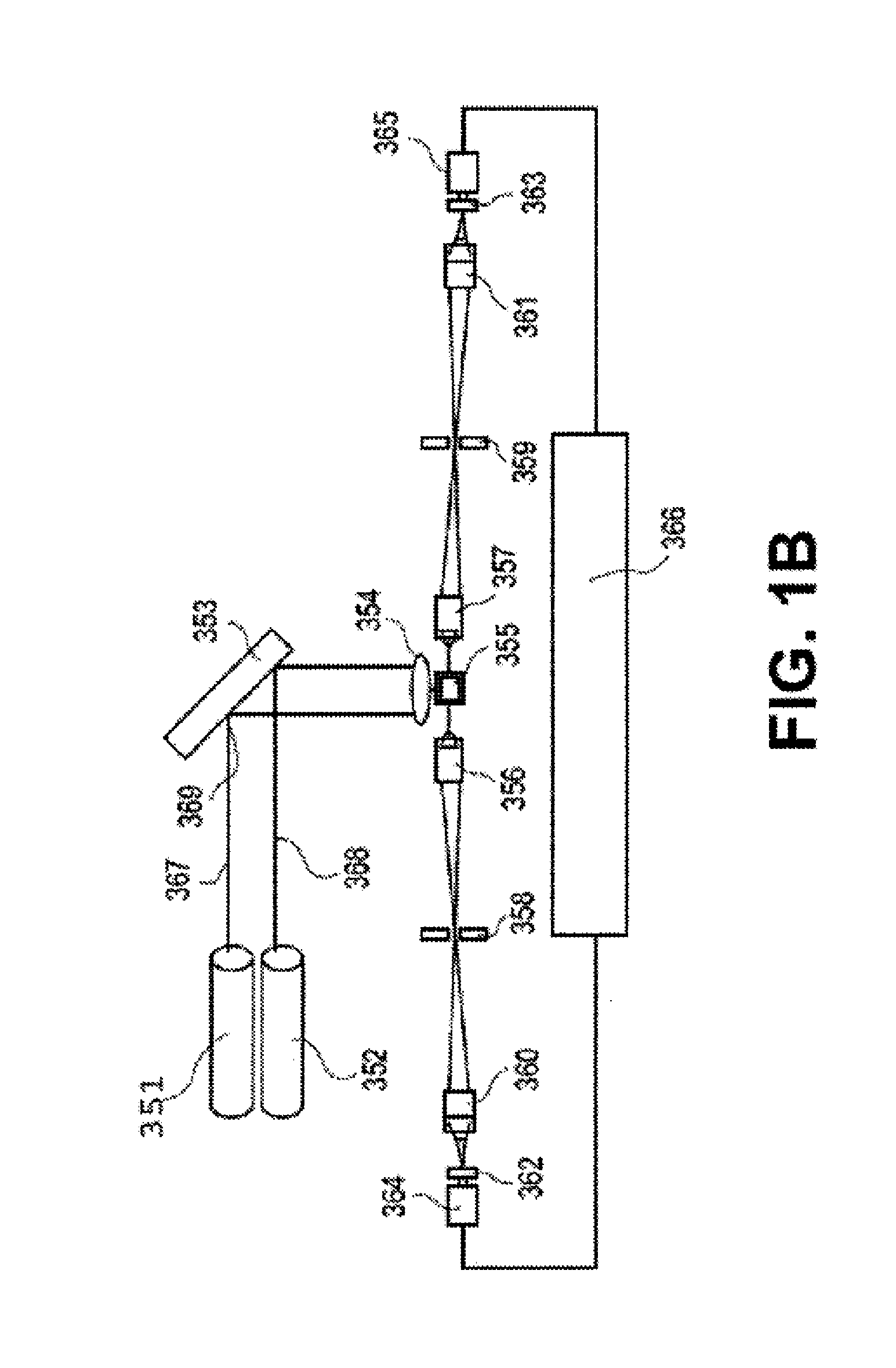
Apparatus and methods for analyte measurement and immuno assay
ActiveUS20030170881A1Avoid disadvantagesBioreactor/fermenter combinationsBiological substance pretreatmentsPoint of careOrganism
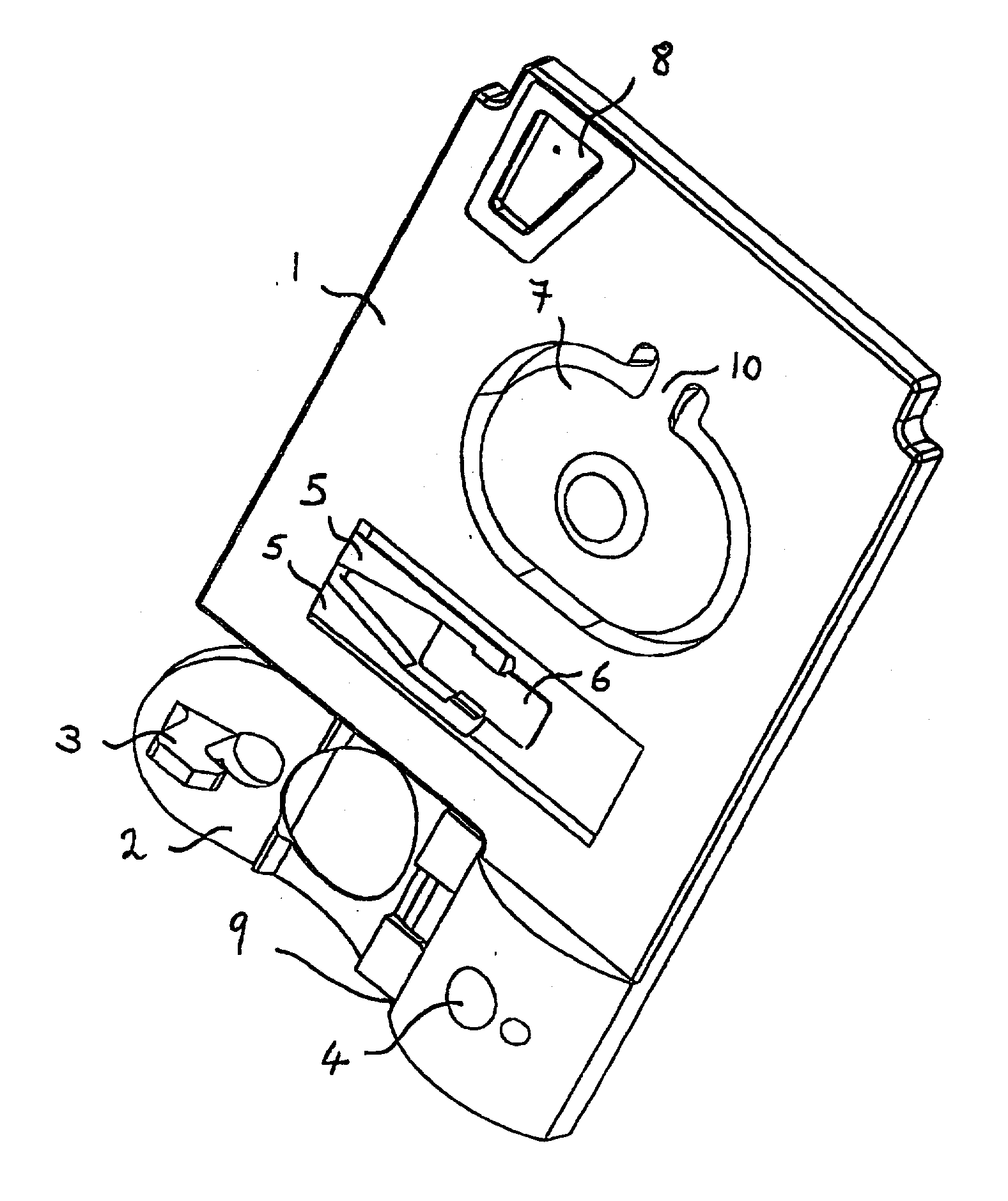
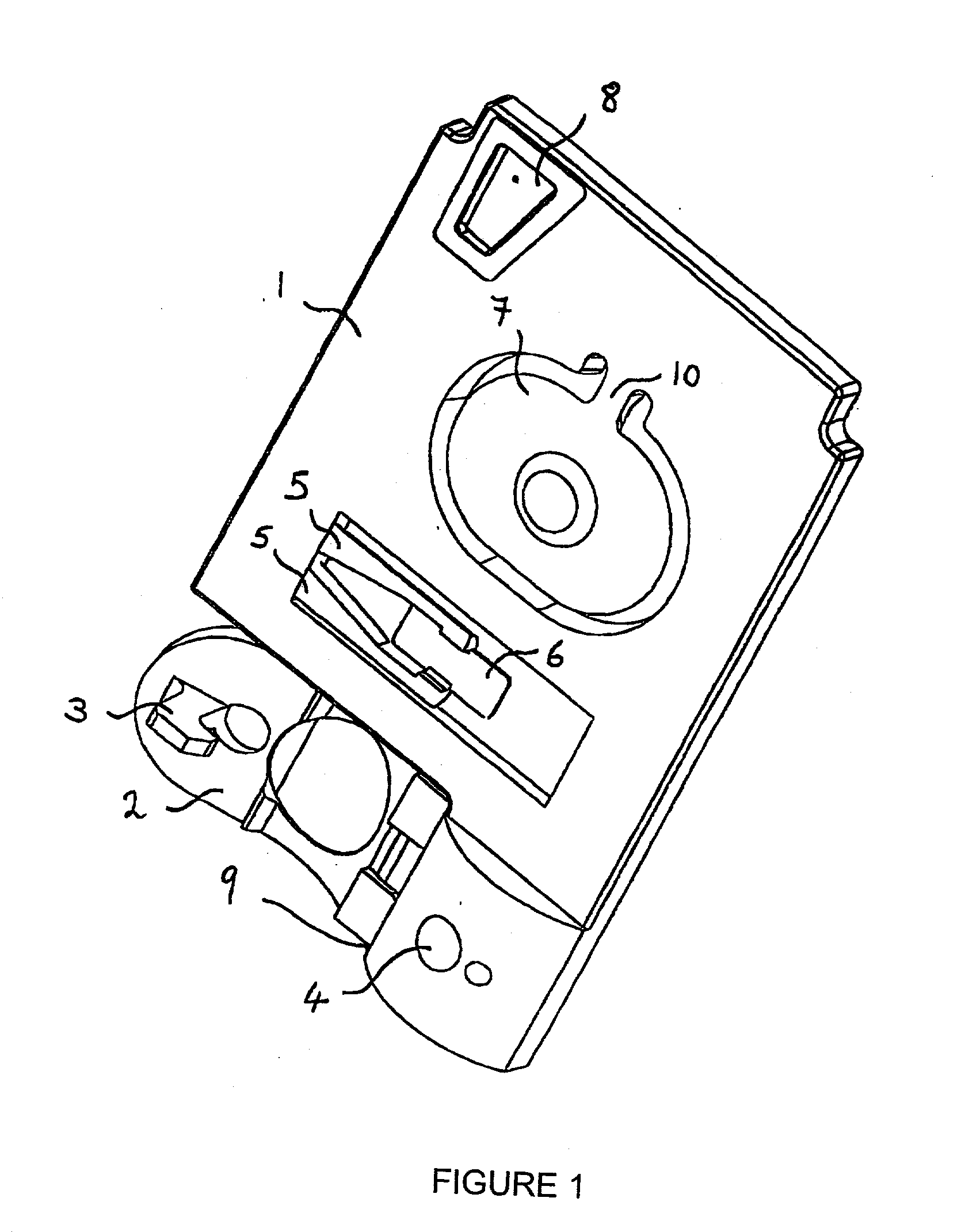
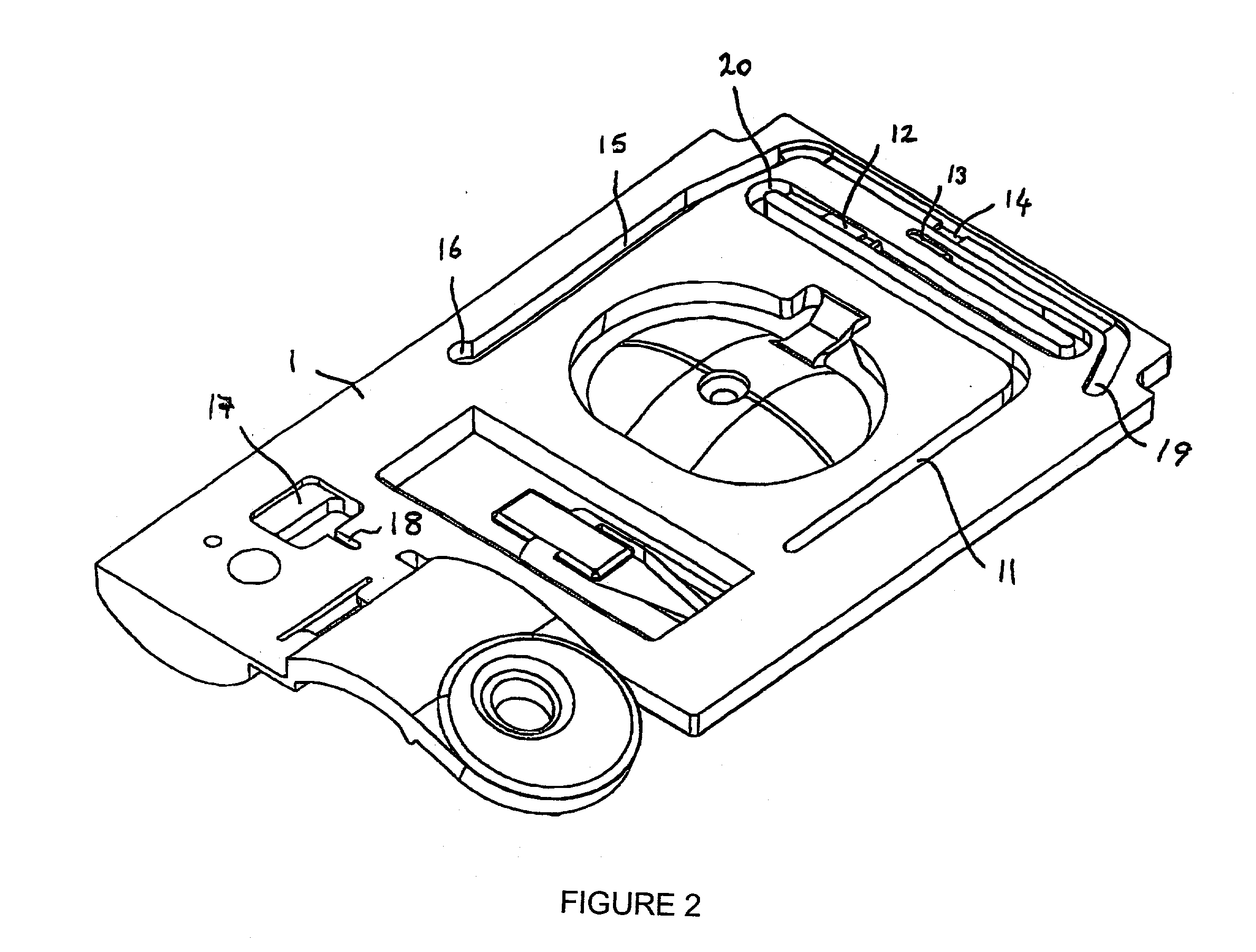
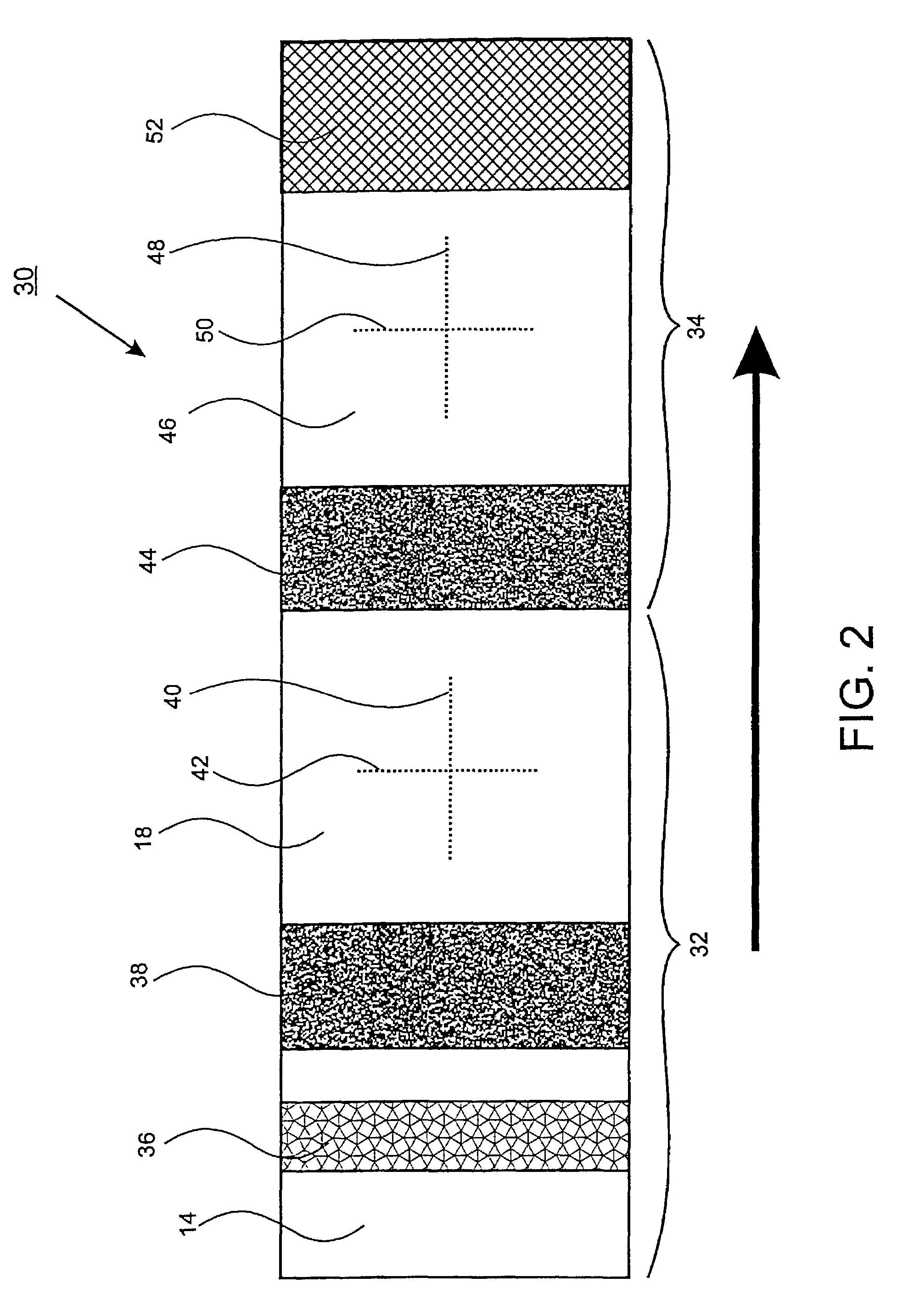
The present invention relates to an apparatus for conducting a variety of assays for the determination of analytes in liquid samples, and relates to the methods for such assays. In particular, the invention relates to a single-use cartridge designed to be adaptable to a variety of real-time assay protocols, preferably assays for the determination of analytes in biological samples using immunosensors or other ligand / ligand receptor-based biosensor embodiments. The cartridge provides novel features for processing a metered portion of a sample, for precise and flexible control of the movement of a sample or second fluid within the cartridge, for the amending of solutions with additional compounds during an assay, and for the construction of immunosensors capable of adaptation to diverse analyte measurements. The disclosed device and methods of use enjoy substantial benefits over the prior art, including simplicity of use by an operator, rapid in situ determinations of one or more analytes, and single-use methodology that minimizes the risk of contamination of both operator and patient. The disclosed invention is adaptable to the point-of-care clinical diagnostic field, including use in accident sites, emergency rooms, surgery, nursing homes, intensive care units, and non-medical environments.
Owner:ABBOTT POINT CARE
Immunoassay that provides for both collection of saliva and assay of saliva for one or more analytes with visual readout
InactiveUS6248598B1Eliminate riskBioreactor/fermenter combinationsBiological substance pretreatmentsAnalyteSaliva sample
A device that provides for both the collection of saliva and detection of at least one analyte therein, e.g., a drug, is provided. This device provides for rapid analysis of saliva samples, while also providing a convenient assay method that does not require the addition of extraneous reagents, or other materials. Thereby, this device can be used by non-laboratory personnel without risk of user introduced errors.
Owner:BOGEMA STUART C
Point of care diagnostic systems
InactiveUS6867051B1Accurate concentrationAccurately presenceComputer-assisted medical data acquisitionMedical imagesPoint of careDiagnostic test
Systems and methods for medical diagnosis or risk assessment for a patient are provided. These systems and methods are designed to be employed at the point of care, such as in emergency rooms and operating rooms, or in any situation in which a rapid and accurate result is desired. The systems and methods process patient data, particularly data from point of care diagnostic tests or assays, including immunoassays, electrocardiograms, X-rays and other such tests, and provide an indication of a medical condition or risk or absence thereof. The systems include an instrument for reading or evaluating the test data and software for converting the data into diagnostic or risk assessment information.
Owner:CYTYC CORP
Coated, resuspendable magnetically responsive, transition metal oxide particles and method for the preparation thereof
InactiveUS6120856AAvoid interferencePromote recoveryMechanical vibrations separationMagnetic materialsCell separationFerroics
The invention relates to an improved method for the manufacture of magnetically responsive particles, also called ferrofluids. The improved method involves a heat treatment step, which may occur at various times during the preparation of the materials, including during subdivision of the magnetic starting material, during the addion of a coating material, after formation of a magnetically responsive particle, or some combination thereof. The materials formed by such a process have numerous advantages over materials formed by other processes, including enhanced salt stability, increased coating uptake, and increased binding capacity. These ferrofluids have applications in a variety of preparative and diagnostic techniques, including immunoassay, cell separations, toxicity testing, food testing, environmental analysis, and MRI.
Owner:JANSSEN DIAGNOSTICS LLC
Point of care diagnostic systems
InactiveUS6936476B1Accurate concentrationAccurately presenceScattering properties measurementsComputer-assisted medical data acquisitionPoint of careDiagnostic test
Systems and methods for medical diagnosis or risk assessment for a patient are provided. These systems and methods are designed to be employed at the point of care, such as in emergency rooms and operating rooms, or in any situation in which a rapid and accurate result is desired. The systems and methods process patient data, particularly data from point of care diagnostic tests or assays, including immunoassays, electrocardiograms, X-rays and other such tests, and provide an indication of a medical condition or risk or absence thereof. The systems include an instrument for reading or evaluating the test data and software for converting the data into diagnostic or risk assessment information.
Owner:CYTYC CORP
Highly Sensitive Biomarker Panels
InactiveUS20110003707A1Detection moreImprove the level ofLibrary screeningDisease diagnosisVascular inflammationBiomarker panel
Cardiovascular disease, e.g., congestive heart failure, is often first diagnosed after the onset of clinical symptoms, eliminating potential for early intervention. The invention provides a multi-marker immunoassay, including cardiac pathology and vascular inflammation biomarkers, yielding a more sensitive assay for early detection of CHF in plasma. A panel consisting of cardiac pathology (cTnI, BNP) and vascular inflammation (IL-6, TNFα, IL-17a) biomarkers provided a sensitivity of 94% for association with CHF.
Owner:SINGULEX
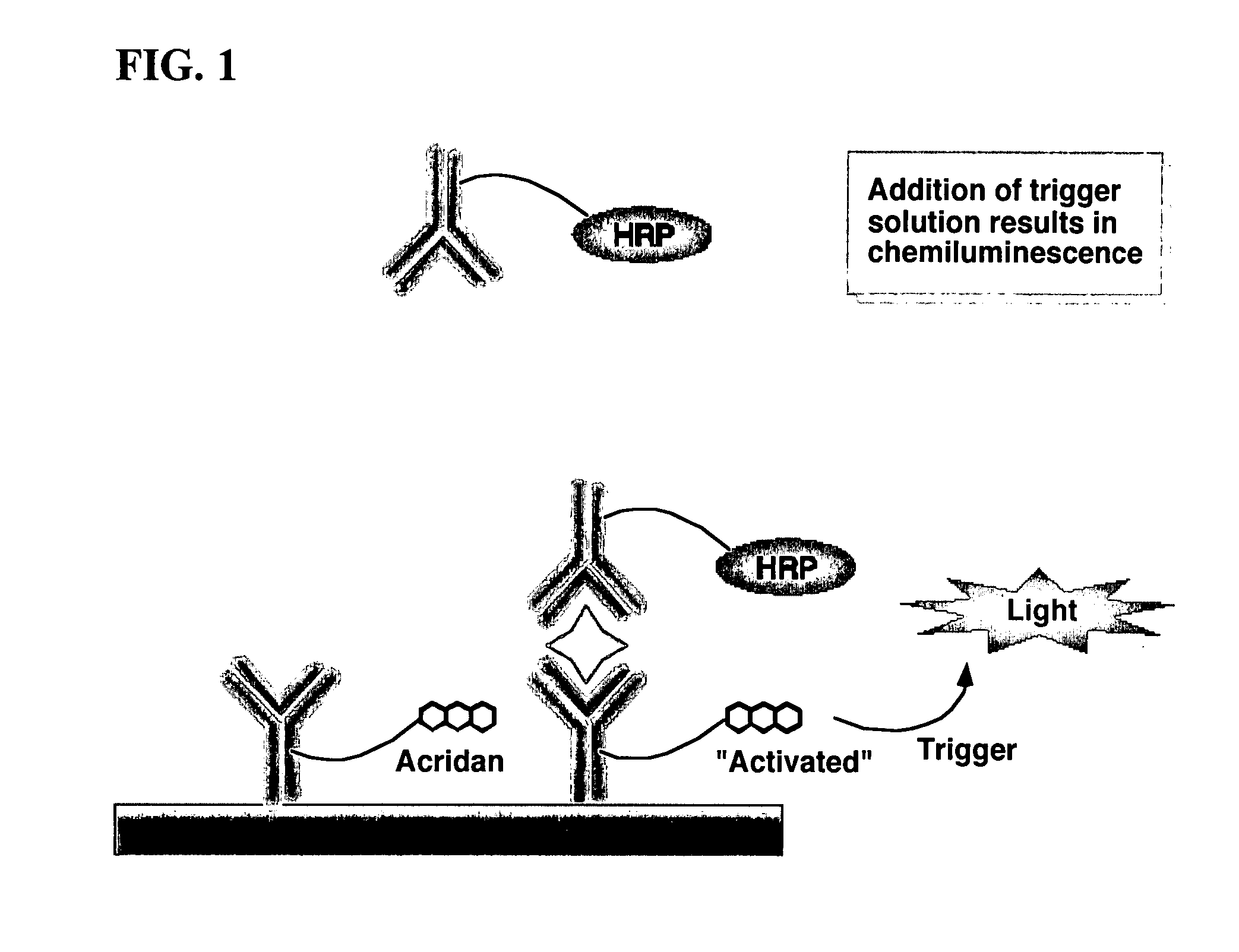
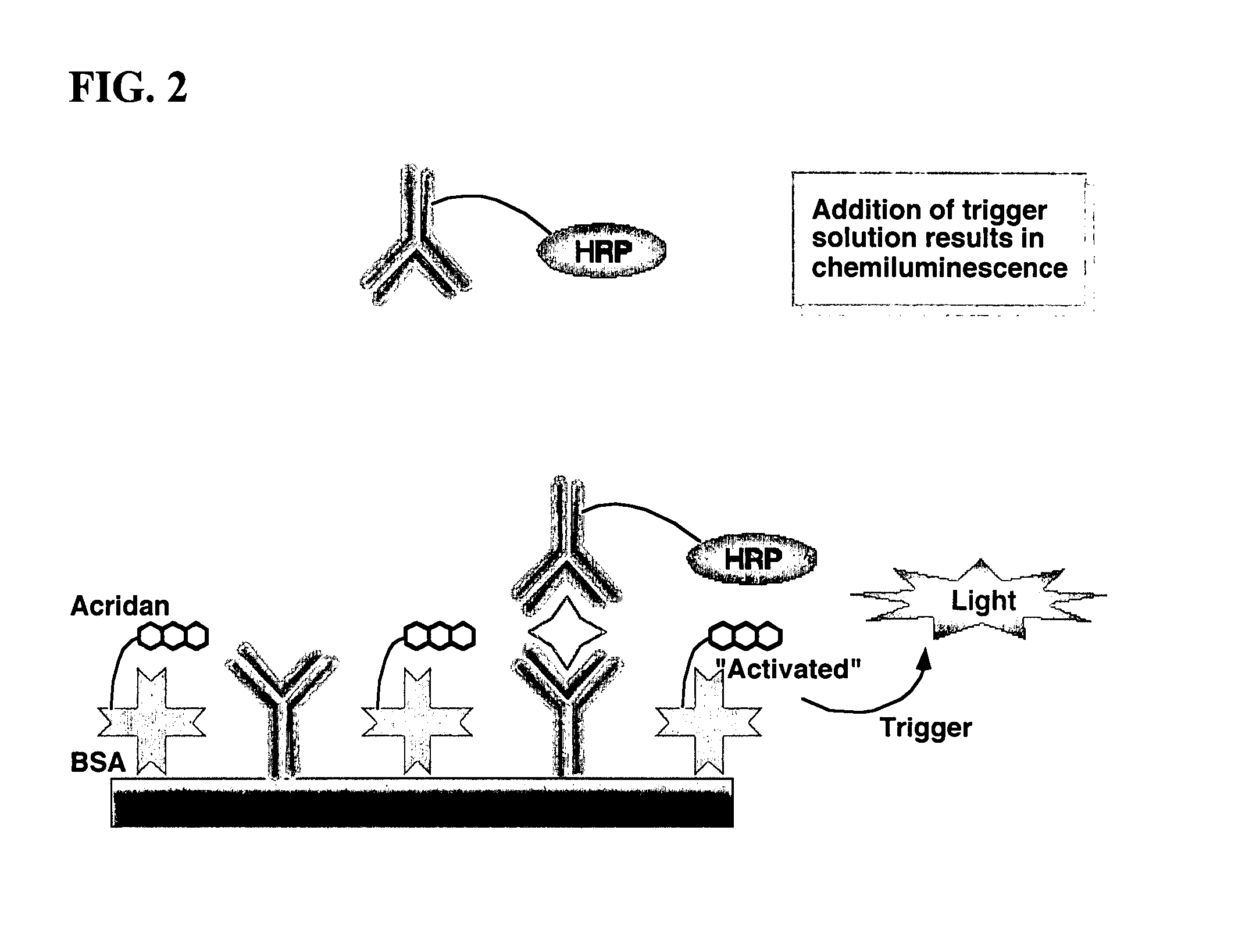
Nonseparation assay methods
ActiveUS20070264665A1Simplifying assayAnalysis using chemical indicatorsMicrobiological testing/measurementAnalyteAssay
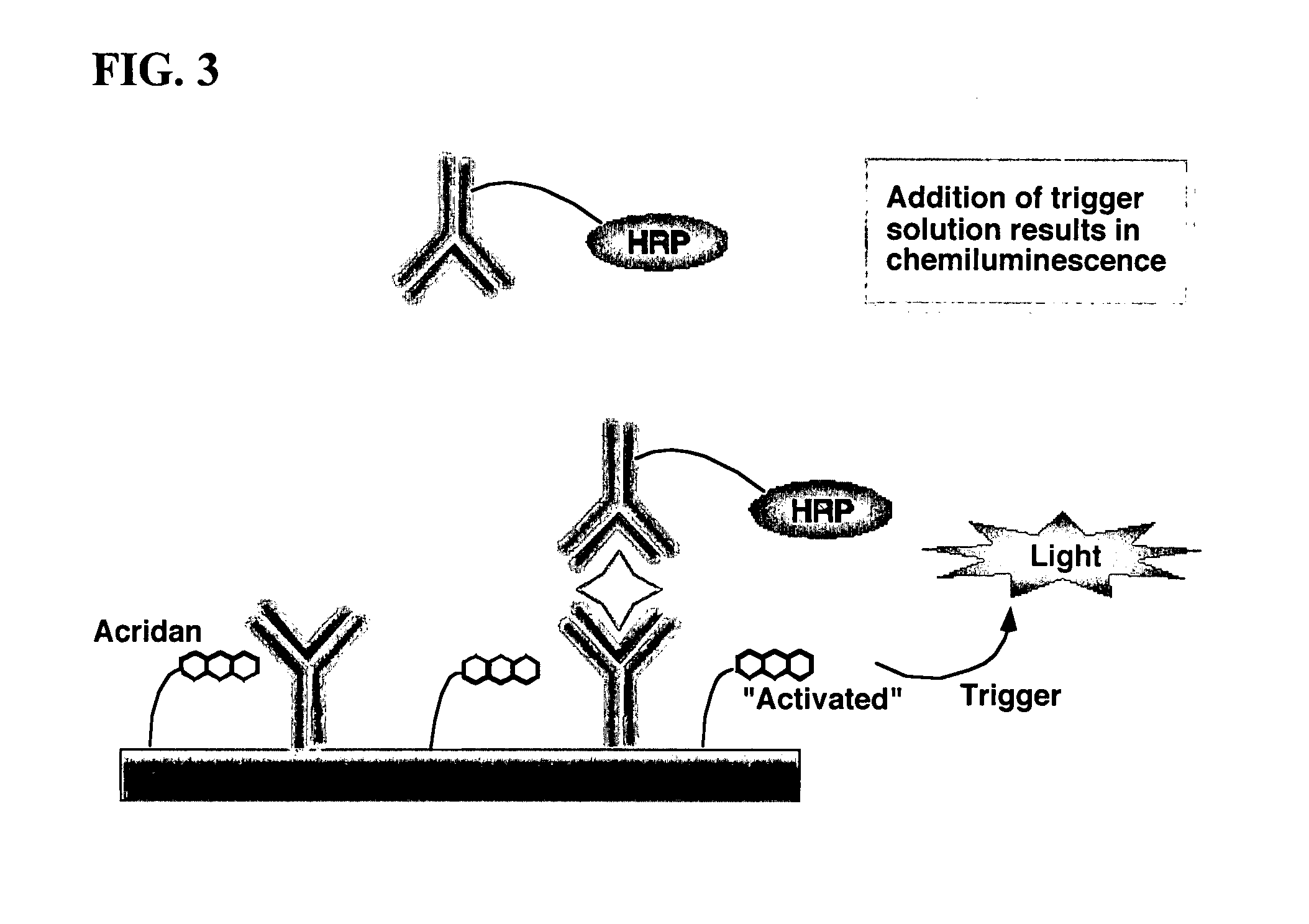
Assay methods are disclosed involving specific binding reactions which are simplified compared to known methods. A compound capable of producing chemiluminescence is immobilized on a solid support as is a member of a specific binding pair for capturing an analyte from a sample. An activator compound that activates the chemiluminescent compound and is conjugated to a specific binding pair member is added in excess along with the sample to the solid support. Addition of a trigger solution causes a chemiluminescent reaction at the sites where the activator conjugate has been specifically bound. The assay methods are termed non-separation assays because they do not require removal or separation of excess detection label (activator conjugate) prior to the detection step. The methods are applicable to various types of assays including immunoassays, receptor-ligand assays and nucleic acid hybridization assays.
Owner:BECKMAN COULTER INC
Immunoassays for human brain natriuretic peptide
Immunoassays and antibodies useful in conducting them for human and canine brain natriuretic peptide are described.
Owner:SCIOS
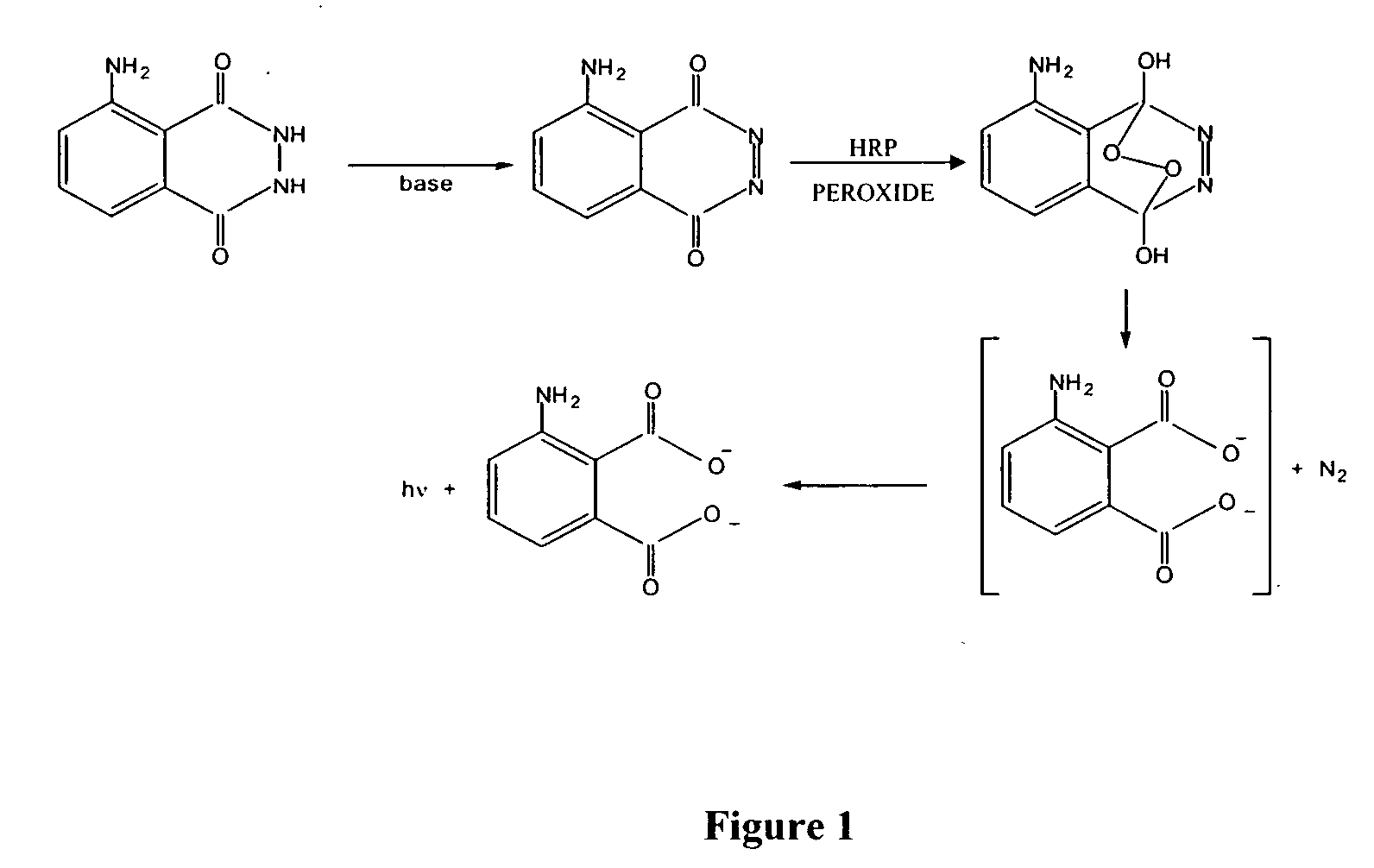
Method for analyzing acridine ester labeled antigen or acridine ester labeled antibody
InactiveCN101609090AReduce consumptionProduct quality and safetyChemiluminescene/bioluminescenceBiological testingChemical structureAntigen
The invention relates to a chemiluminescent immunoassay method, in particular to a method for analyzing an acridine ester labeled antigen or an acridine ester labeled antibody and an immunoassay kit prepared by same. An acridine ester label is provided with a special luminescent group in a chemical structure, and the special group directly participates in luminescent reaction in the analyzing process of luminescent immunization; the substance does not have background luminescence usually, can be used for detecting a sample with low concentration or trace concentration in the reaction and is a luminescent agent with high luminescent efficiency; and molecules of acridine ester I and acridine ester II and acridine amide III can be combined with an antibody (antigen) to generate the labeled antibody with high chemiluminescent activity and immunological reaction specificity.
Owner:BEIJING ELCOTEQ BIO TECH
Anti-mesothelin antibodies useful for immunological assays
InactiveUS20090047211A1Growth inhibitionAntibody mimetics/scaffoldsImmunoglobulins against cell receptors/antigens/surface-determinantsAnti-Mesothelin AntibodyAssay
The present invention provides antibodies that have a surprisingly good combination of affinity for mesothelin and ability to be used in immunological assays for detecting the presence of mesothelin in biological samples. The invention further provides methods of using the antibodies, and kits comprising them. The antibodies can also be used to target toxins and other agents to cells expressing mesothelin, and can be used in methods and medicaments for inhibiting the growth of such cells.
Owner:UNITED STATES OF AMERICA
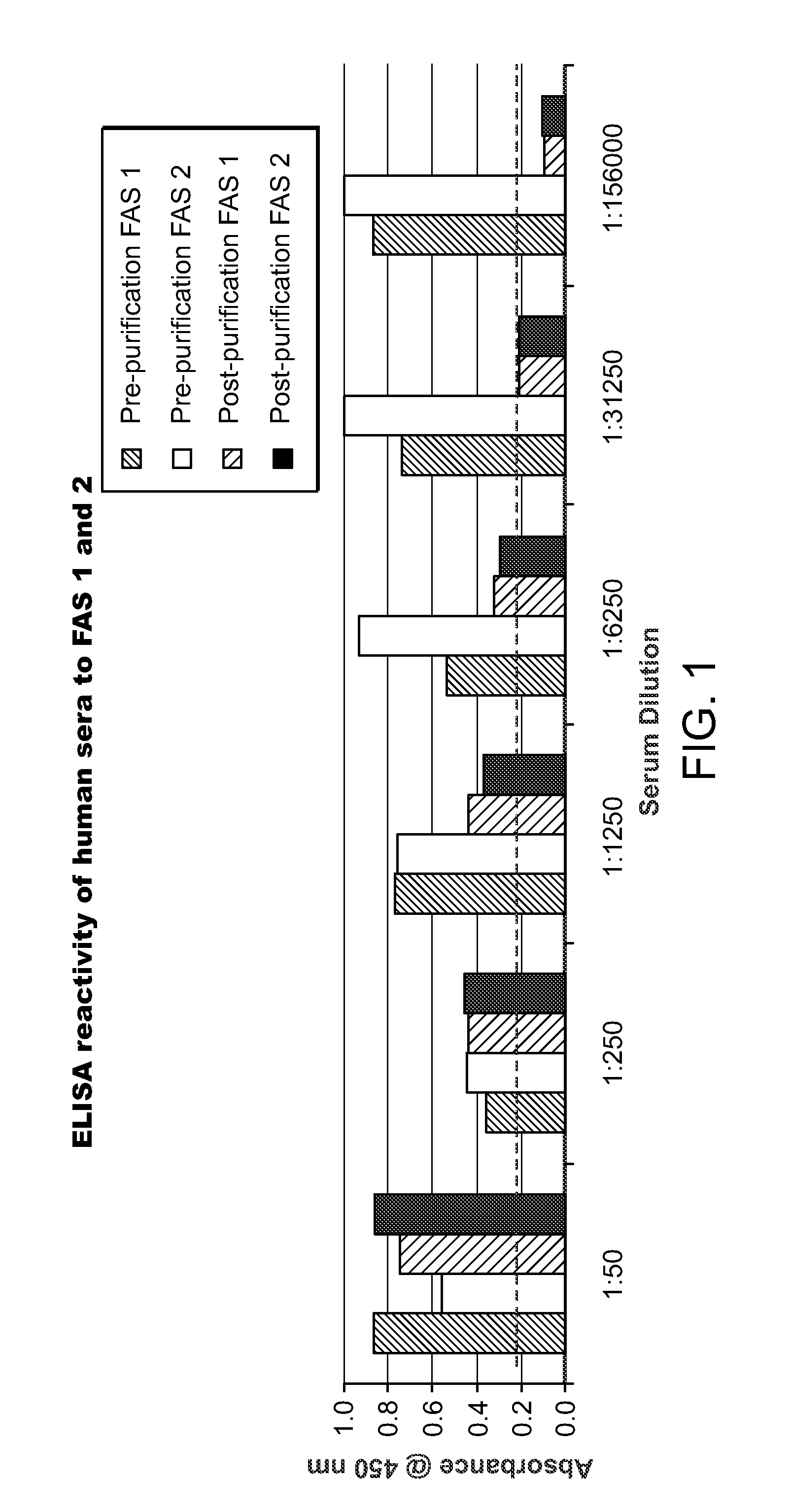
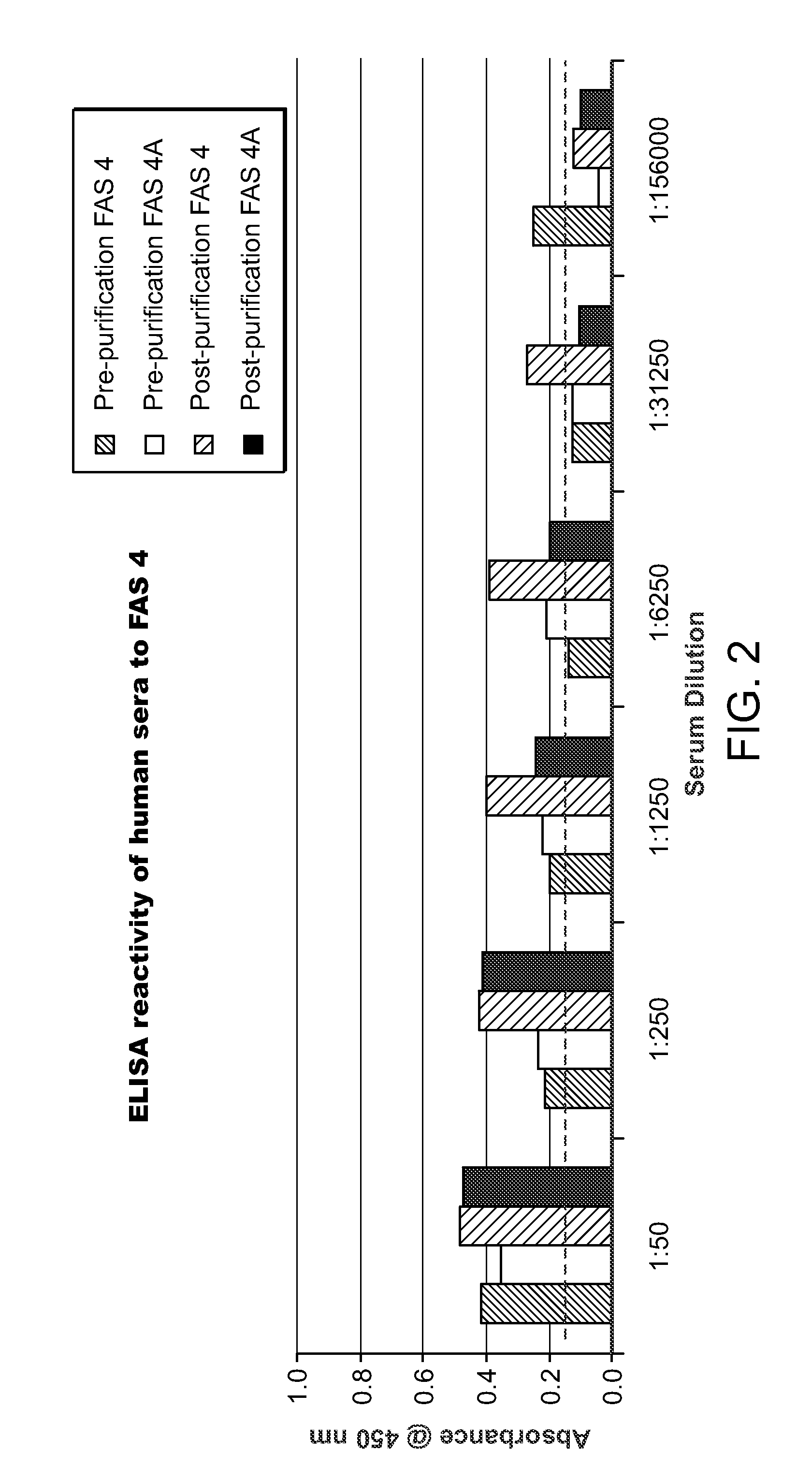
Method and assay for determining FAS expression
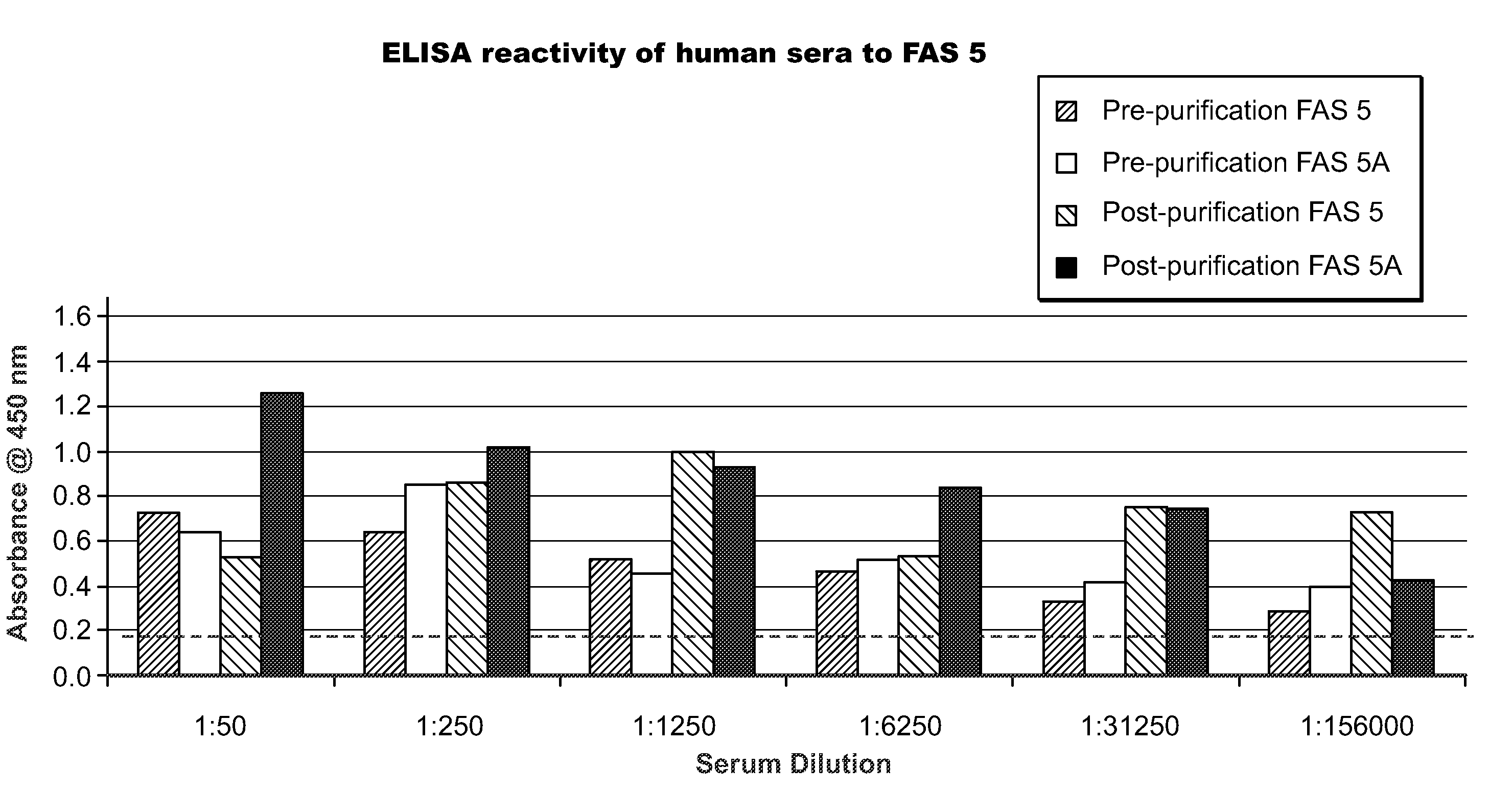
Methods and immunoassays for the determination of fatty acid synthase (FAS) expression in patients having or suspected of having a proliferative disorder, especially prostate cancer, are disclosed. The sensitive method and assay detect the level of expression of FAS in a biological sample using antibodies that are highly specific for FAS. The method and assay can be used to monitor the progression of cancer, and / or to predict the efficacy of certain treatments or the likelihood of recurrence of the cancer.
Owner:NUCLEA BIOMARKERS
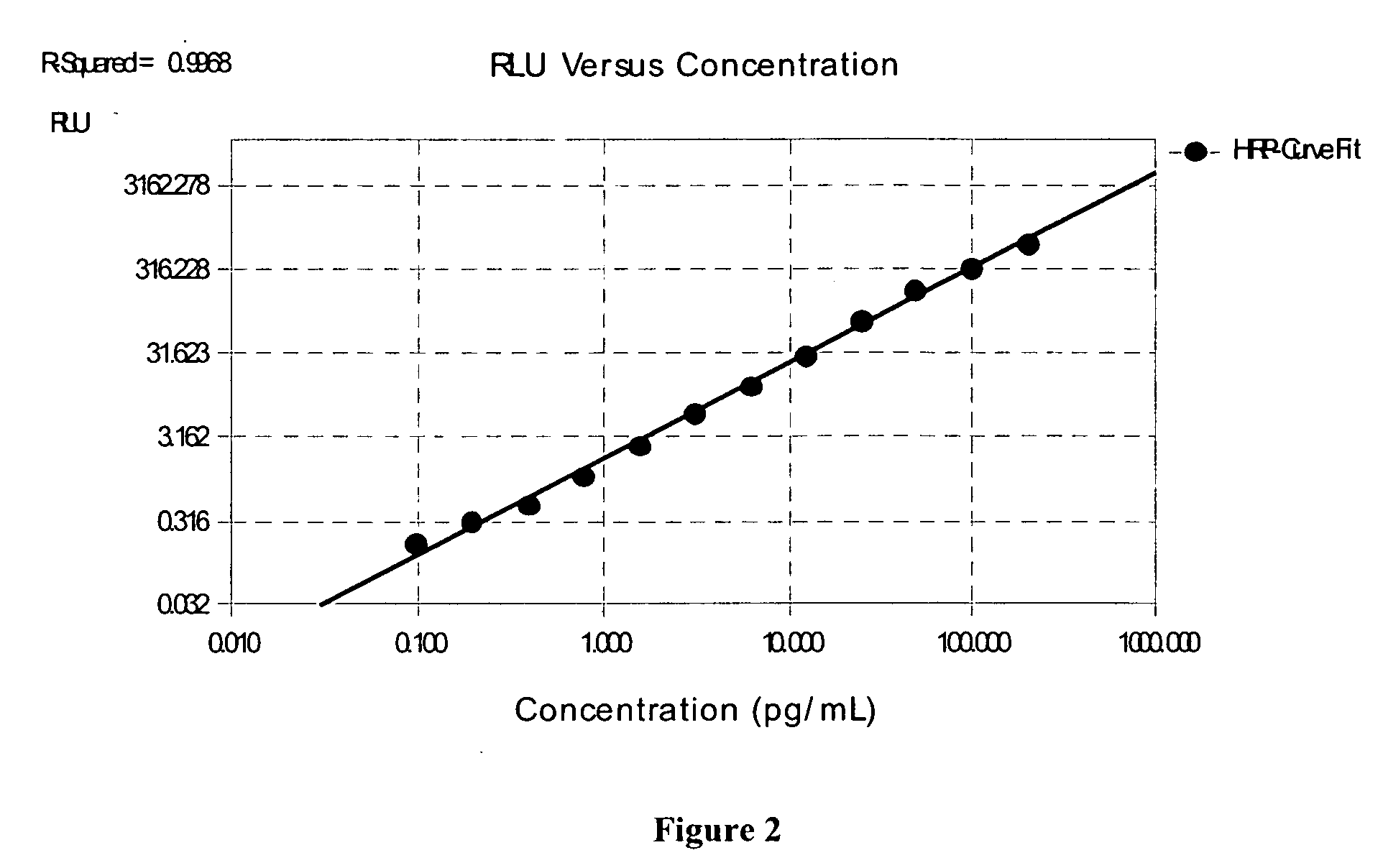
Stabilized two component system for chemiluminescent assay in immunodiagnostics
The present invention provides stabilized chemiluminescent formulations for use in in vitro diagnostics, including competitive as well as sandwich-type immunological assays. The stabilized assay system may be composed of two components, where the first component may contain a chemiluminescent organic compound, an enhancer, a homogenizing agent, and a suitable buffer with formulations having a pH range from about 7.2 to about 12, and optionally a solubilizing agent. The chemiluminescent system of the present invention is useful in immunoenzymatic analytical procedures, such as immunometric, competitive binding and sandwich type assays. In such immunoassays employing the chemiluminescent system of the present invention, the detectable light signal shows a proportional decay with time in the test samples and standards, so that the decay of the light emitted does not effect the concentration of the analyte measured over the entire analyte measurement range of the immunoassay. This allows accurate measurement of analyte concentrations in a test sample over extended periods of time.
Owner:KALRA BHANU +1
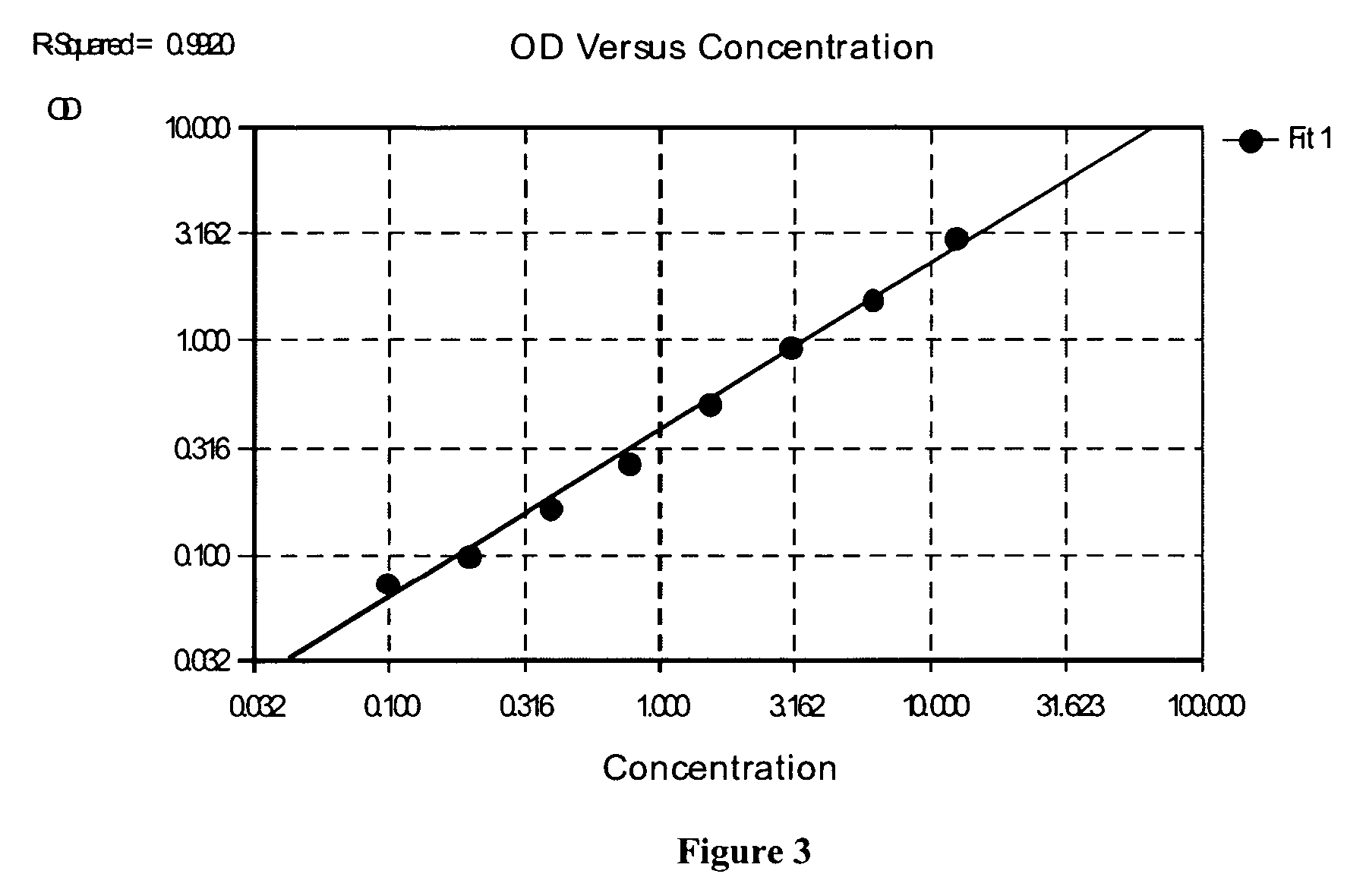
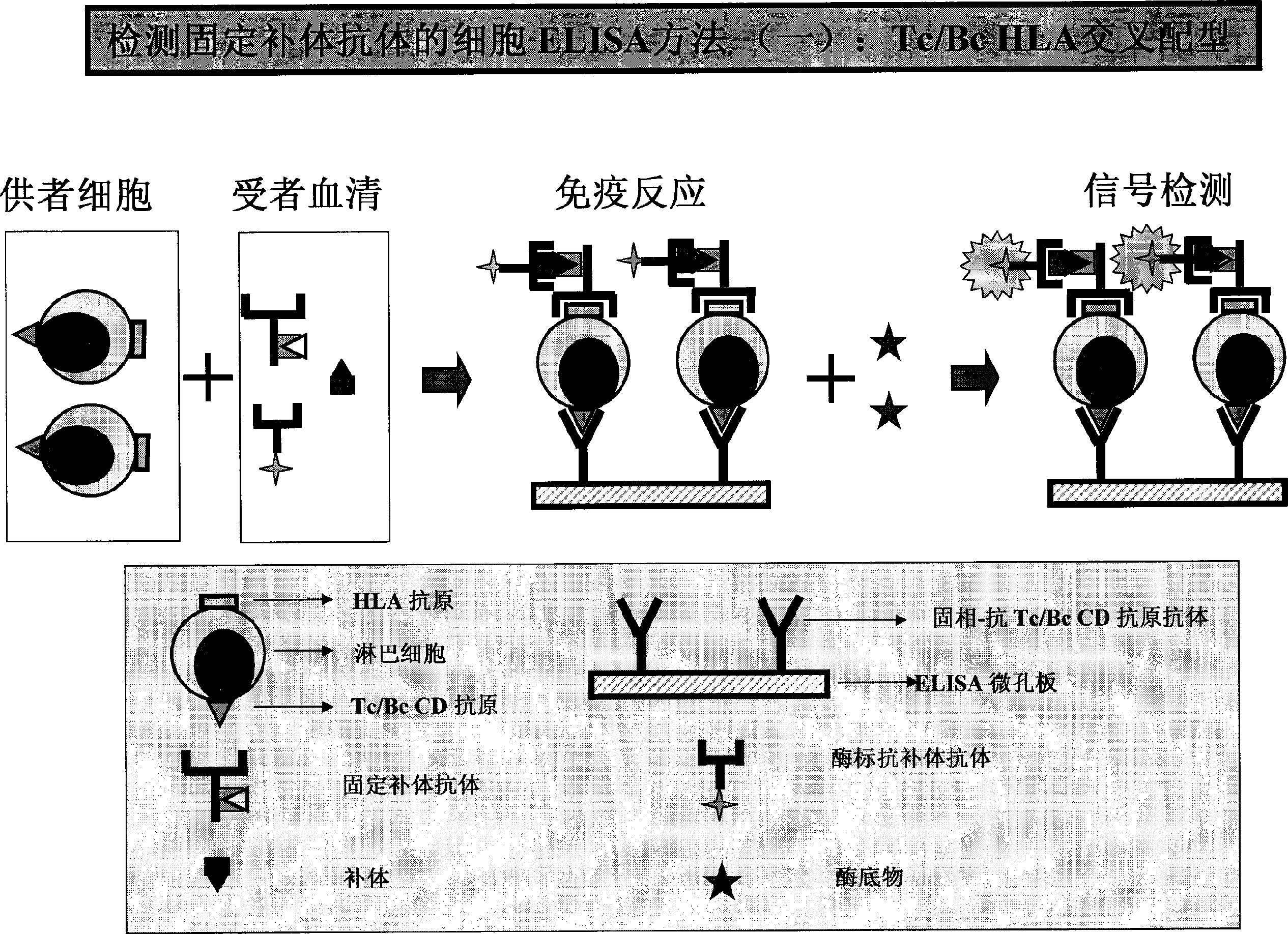
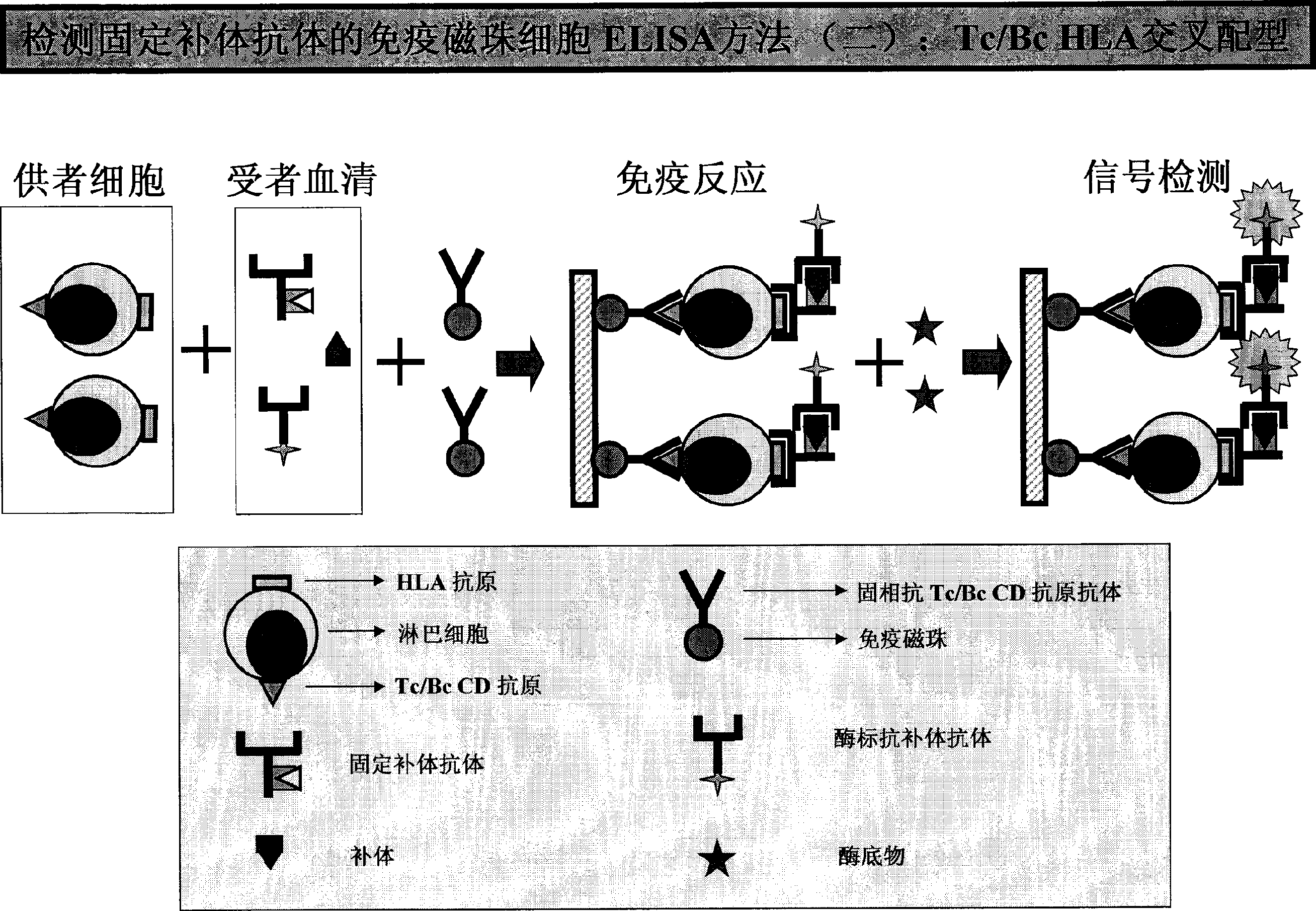
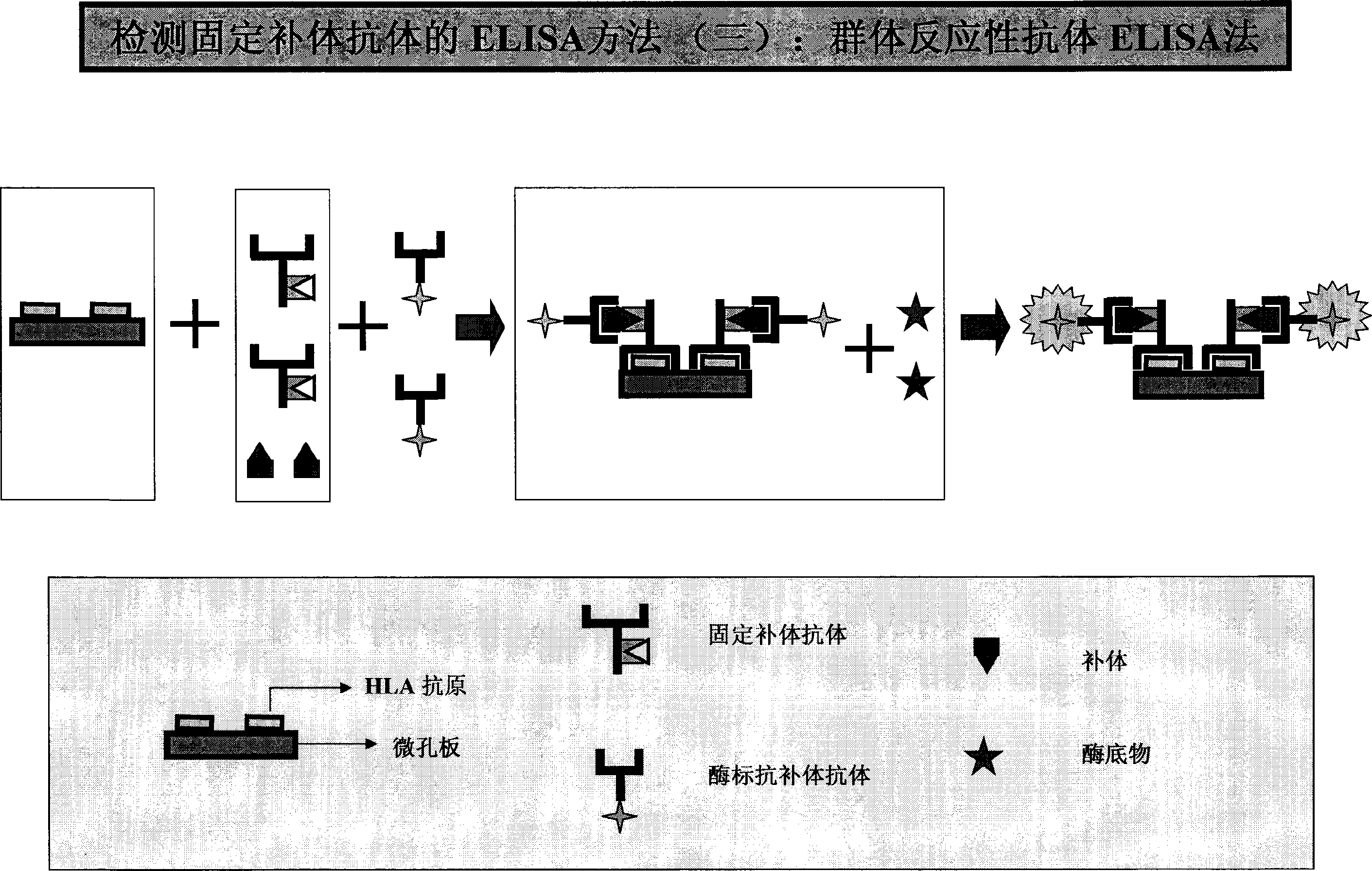
HLA complement-dependent cytotoxcity antibody detection method using ELIA as basis and its kit
InactiveCN1444044AShort reaction timeHigh sensitivityBiological testingComplement-dependent cytotoxicityCytotoxic antibody
According to the complement-dependent cytotoxicity (CDC) reaction principle in the immunology said invention creates an in-vitro enzyme-linked immunoreactino method for assaying HLA complement fixingantibodies (CFAbs) and its kit. The reaction system is formed from solid-phase HLA antigen or target cell with HLA antigen and liquid-phase enzyme-labelled ligand. CFAbs in the tested sample and solid-phase HLA antigen or HLA antigen of target cell are combined, and simultaneously fixed and existed in enzyme-labelled complement or the enzyme-labelled complement anti body is combined with complement fixed in HLA antigen-CFAb composite, then the correspondent enzyme substrate can be added to produce zymolygical color development reaction.
Owner:PEL FREEZ BIOTECH BEIJING
Use of cytokines secreted by dendritic cells
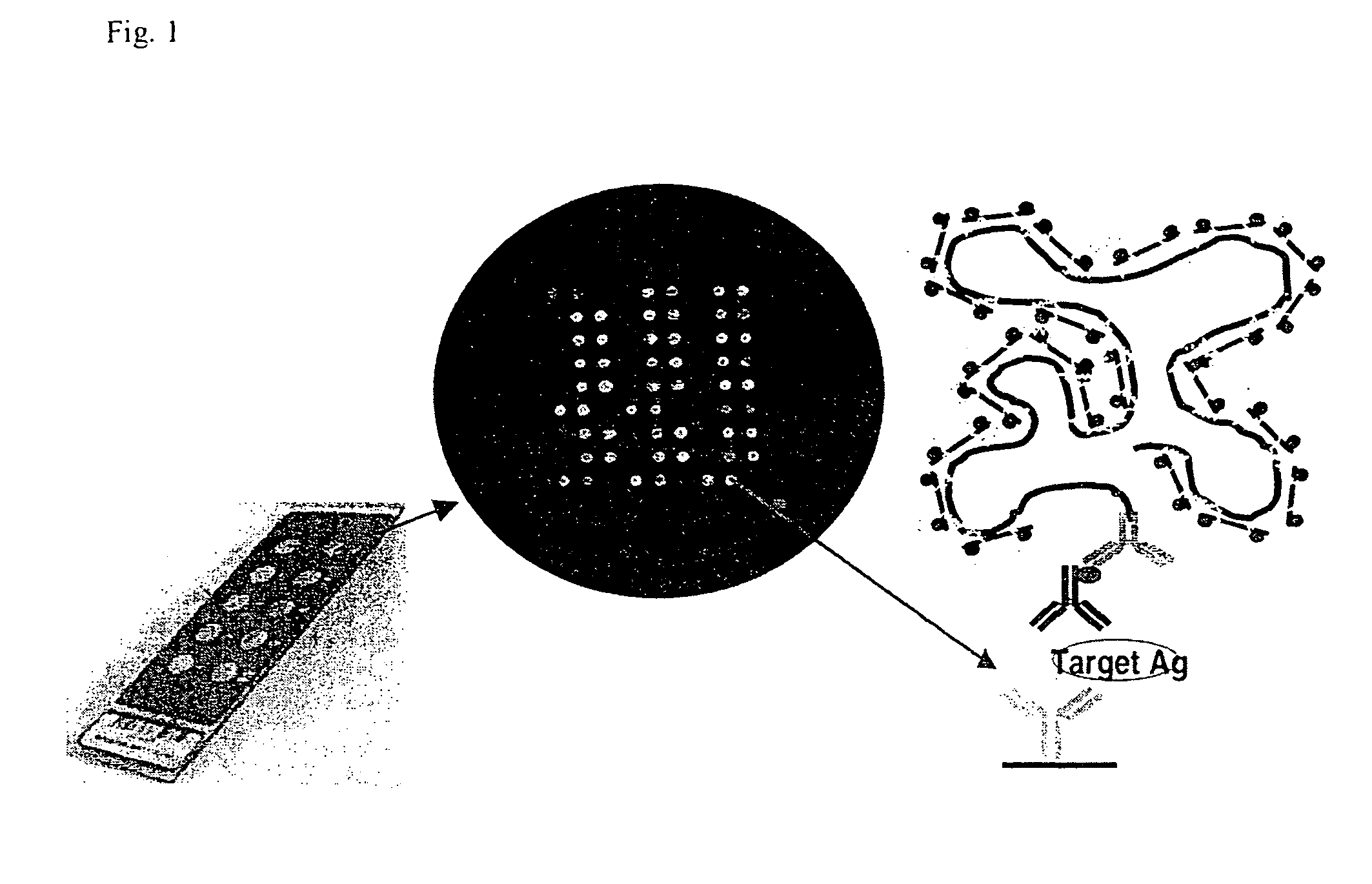
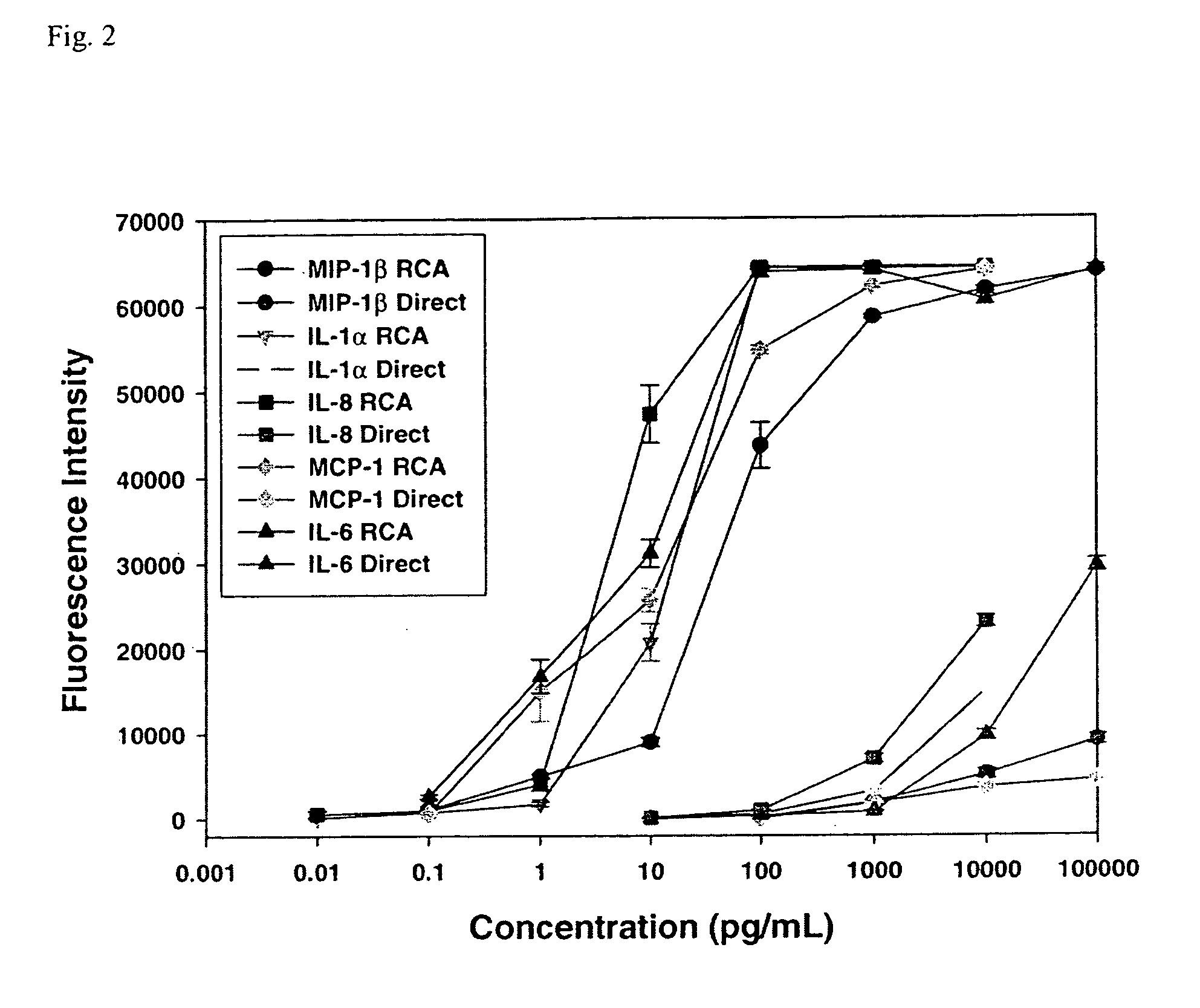
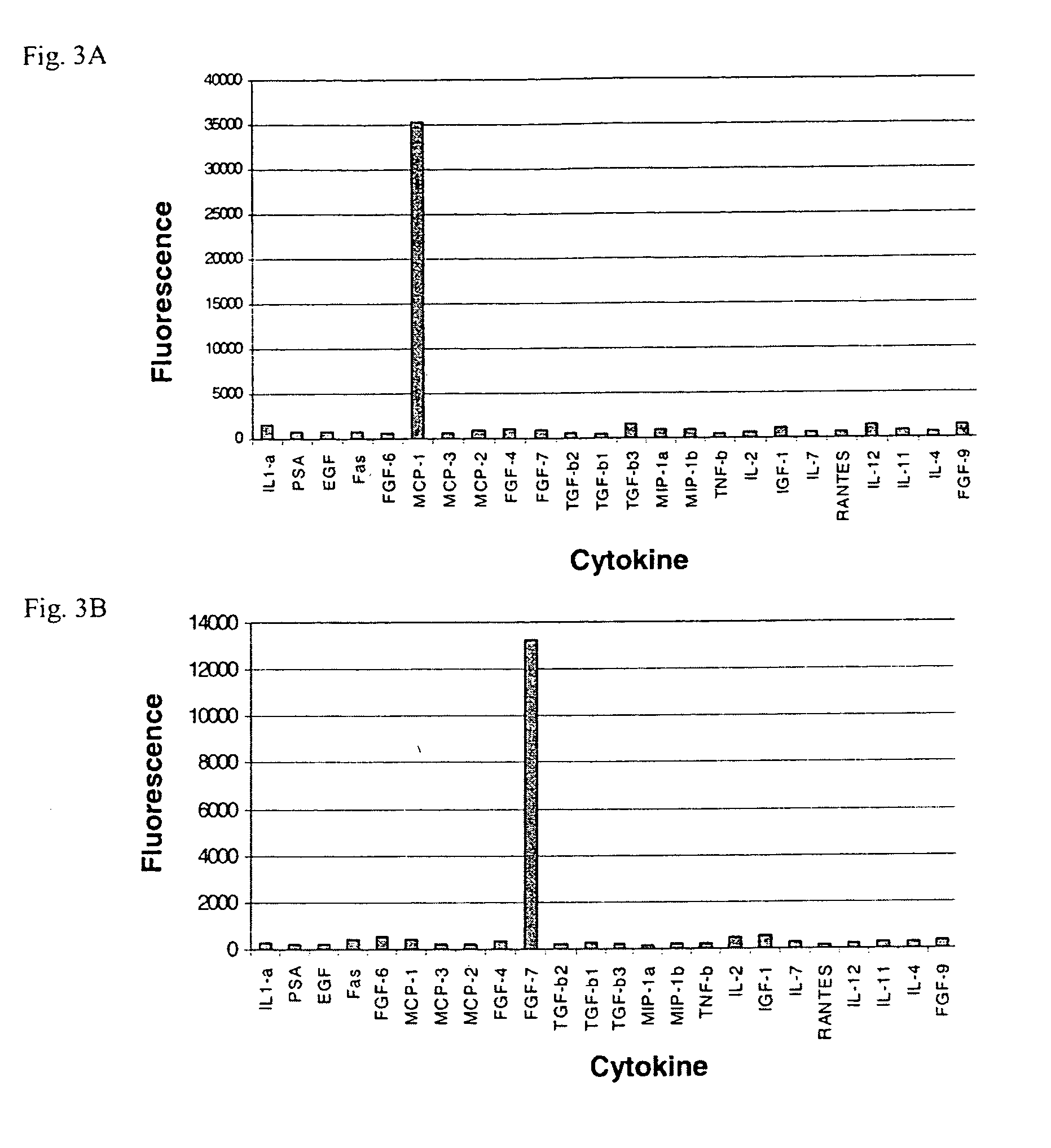
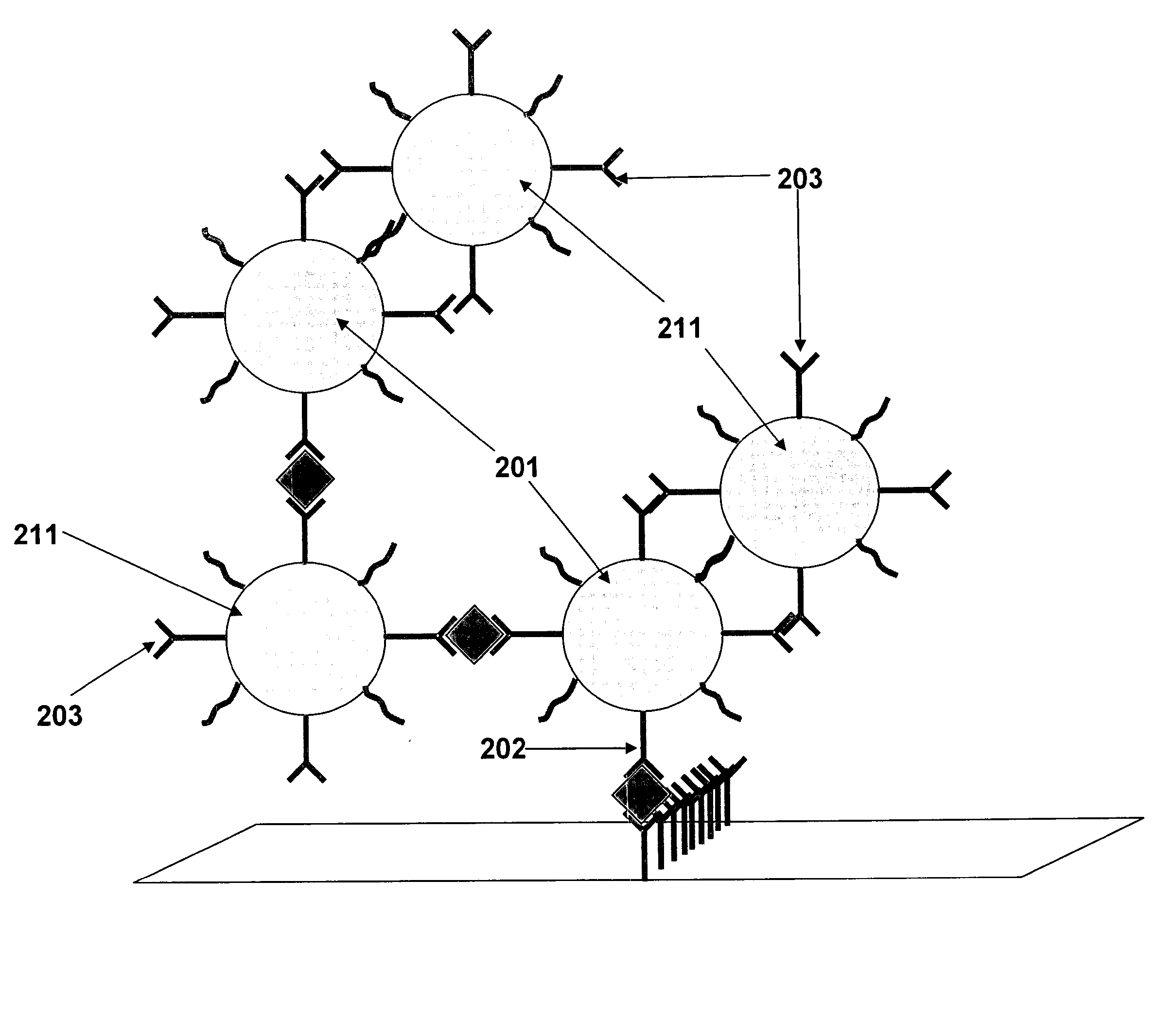
A prerequisite of proteomics is the ability to quantify many selected proteins simultaneously. Fluorescent sandwich immunoassays on microarrays hold appeal for such studies, since equipment and antibodies are readily available, and assays are simple, scalable and reproducible. To attain adequate sensitivity and specificity, however, a general method of immunoassay amplification is required. Coupling of isothermal rolling circle amplification (RCA) to universal antibodies can be used for this purpose: RCA on a synthetic DNA circle is initiated by a complementary oligonucleotide attached to an anti-biotin antibody; single-stranded RCA product remains attached to the antibody, and is detected by hybridization of complementary, fluorescent oligonucleotides. 51 cytokines were measured simultaneously on microarrays with signal amplification by RCA with high specificity, femtomolar sensitivity and 4 log quantitative range. This cytokine microarray was used to measure secretion from human Dendritic cells (DCs) induced by lipopolysaccharide (LPS) or tumor necrosis factor-alpha (TNF-(alpha). Rapid secretion of inflammatory cytokines such as MIP-1beta, IL-8, and IP-10 was induced by LPS. Eotaxin-2 and I-309 were found to be induced by LPS, and MDC, TARC, sIL-6R, and sTNF-RI were found to be induced by TNF-alpha. Since microarrays can accommodate ~1000 sandwich immunoassays of this type, a relatively small number of RCA microarrays appears to offer a tractable approach for proteomic surveys.
Owner:MOLECULAR STAGING
Fluorescent latex granular immune chromatography by time resolution
InactiveCN1818653AFast flowResolving directly coated antibodiesFluorescence/phosphorescenceAntigenFluorescence
A microparticle immune chromatography of time-resolution fluorescence emulsion includes preparing biotinylation antibody and preparing immune time-resolution fluorescence microparticles, using Fusion 5 film to prepare avidin solid phase film detection line and quality control line by utilizing avidin to envelop said film, finally using double-antibody sandwich method to detect antigen quickly.
Owner:SHANGHAI JIAOTONG UNIV SCHOOL OF MEDICINE
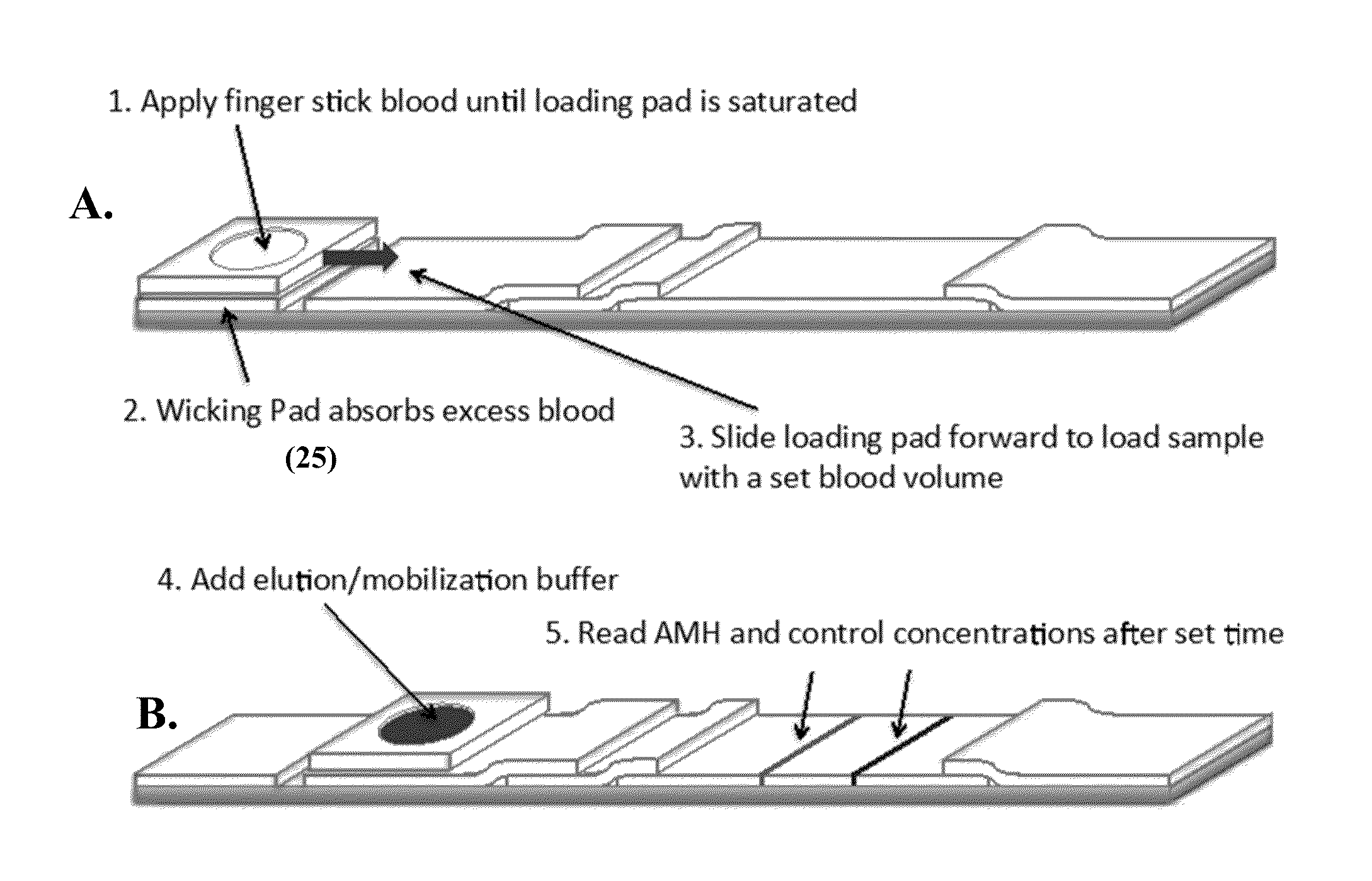
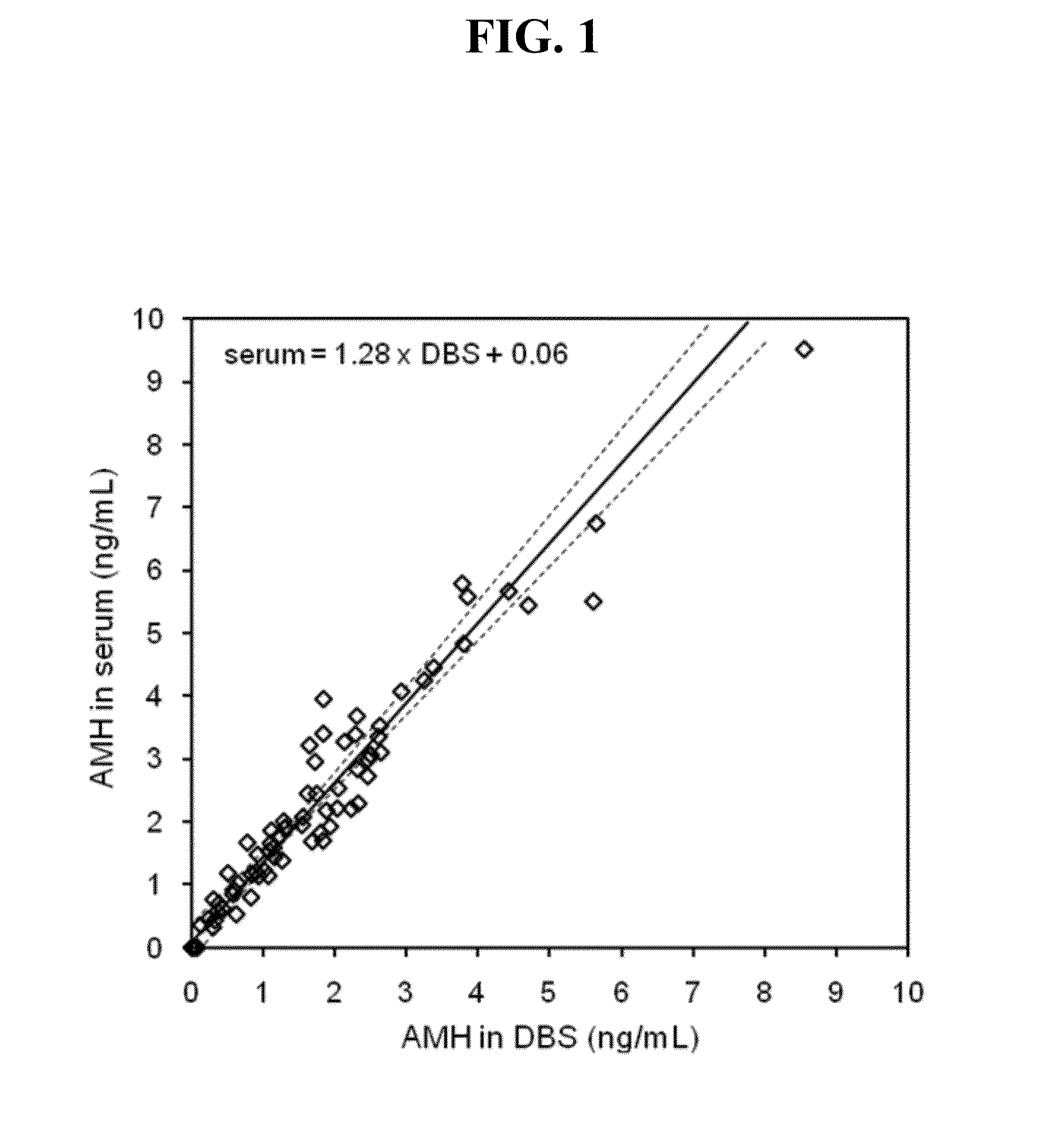
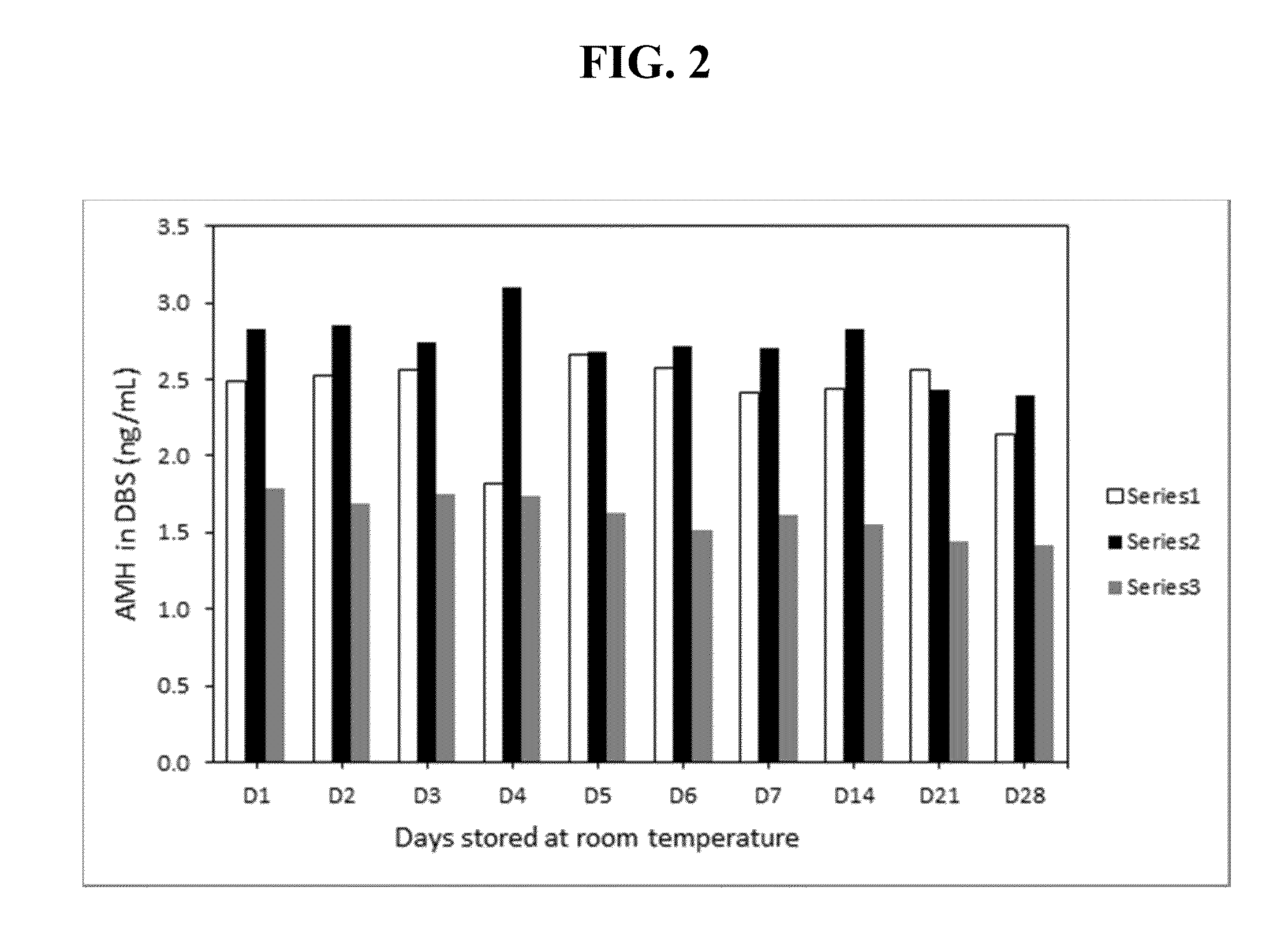
Anti-mullerian hormone detection in whole blood
InactiveUS20130224771A1Bioreactor/fermenter combinationsBiological substance pretreatmentsAntibody conjugatePhysiology
The present invention provides methods, kits, compositions, and devices for detecting Anti-Mullerian hormone (AMH) in whole blood samples. In certain embodiments, the methods, kits, compositions, and devices employ immunoassays that generate a colorimetric or fluorescent signal (e.g., using antibodies conjugated to gold nanoparticles or fluorescent particles) where the signal generated is proportional to the approximate concentration of AMH in a whole blood sample. In particular embodiments, the present invention provides quantitative or semi-quantitative lateral flow immunoassay devices and kits for detecting AMH at home (e.g., in order for women to estimate their ovarian age or diagnose polycystic ovarian syndrome).
Owner:NORTHWESTERN UNIV
Tumour marker proteins and uses thereof
ActiveUS20060094069A1Easy to disassembleIncrease concentrationTumor rejection antigen precursorsTumor specific antigensAutoantibody productionExcretion process
The invention relates to tumour marker proteins and their preparation from fluids from one or more cancer patients, wherein said fluids are those which collect in a body cavity or space which is naturally occurring or which is the result of cancer or medical intervention for cancer. The invention also relates to preparation of tumour marker proteins from excretions taken from patients with cancer. The tumour marker proteins are useful as immunoassay reagents in the detection of cancer-associated anti-tumour marker autoantibodies.
Owner:ONCIMMUNE
Methods and device for detecting prostate specific antigen (PSA)
An immunoassay device and assay to detect an antigen, such as PSA, in a biological sample. The device comprises a solid support having multiple reaction zones containing capture antibodies directed to the antigen. Exposure of a test sample to a mixture of incubation antibodies with known and different concentrations prior to exposure to the capture antibodies in the reaction zones facilitates determination of a range of concentrations of the antigen.
Owner:WANG TANG J
Immune diagnostic assay to diagnose and monitor tuberculosis infection
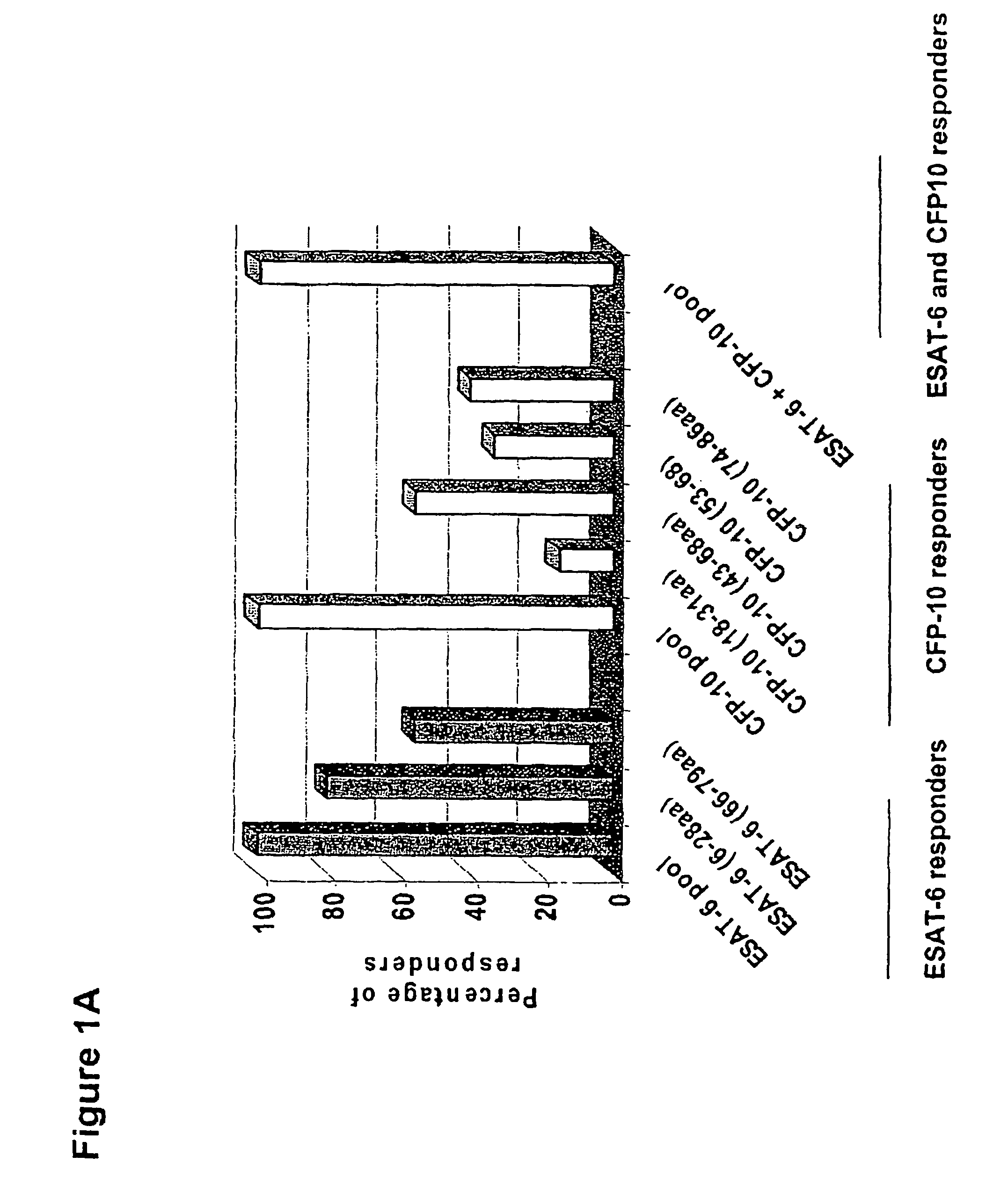
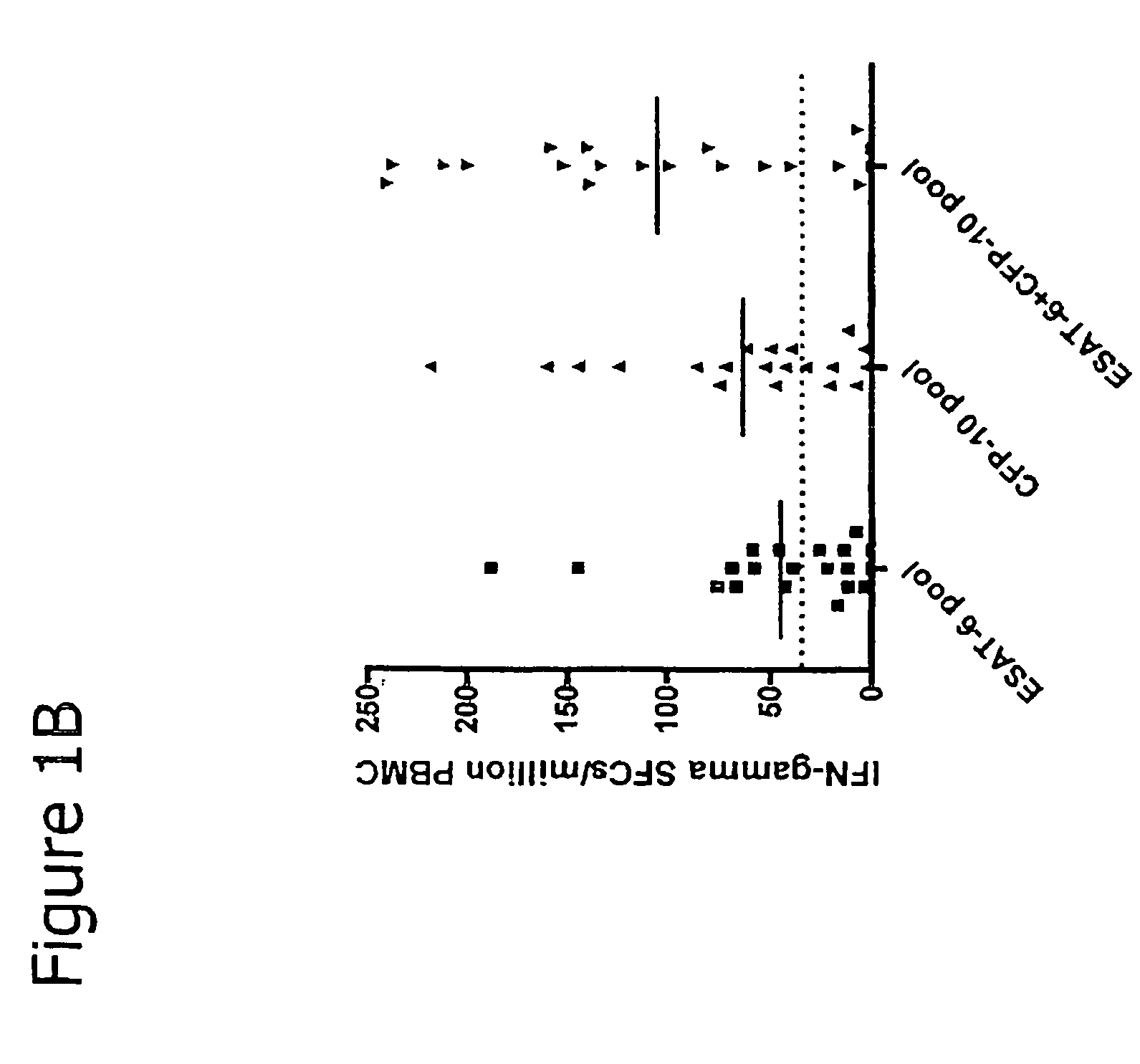
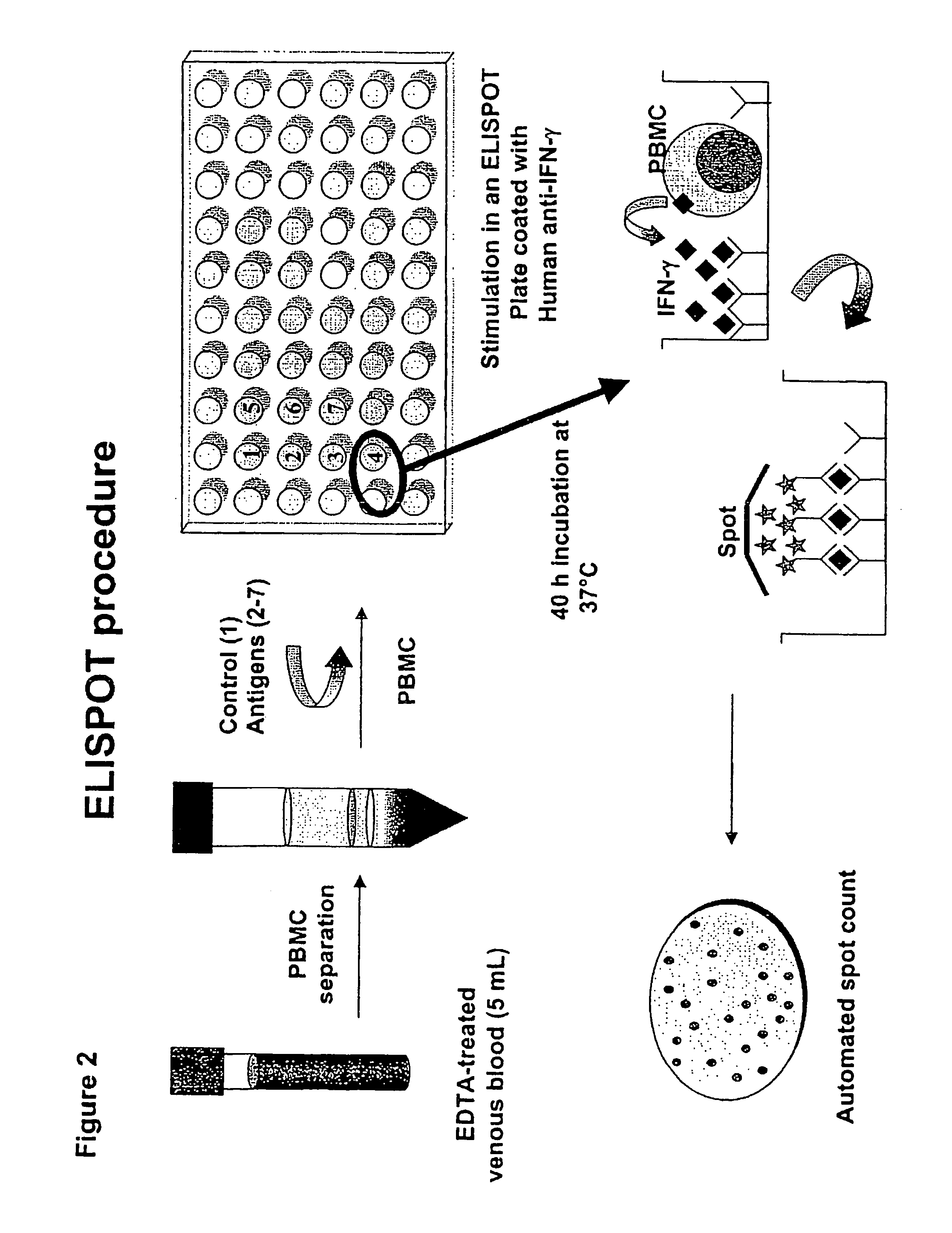
InactiveUS7785607B2High detection sensitivityHigh sensitivityBacterial antigen ingredientsPeptide/protein ingredientsDiseaseT lymphocyte
The present invention relates to a method of diagnosing and monitoring various distinct presentations of tuberculosis: active tuberculosis disease, latent tuberculosis infection and recent tuberculosis infection. The rapid immune assay is based on the evaluation of the frequency of Interferon (IFN) gamma-producing antigen-specific T lymphocytes responding to selected peptide sequences from Mycobacterium tuberculosis, selected for their immunogenicity. The invention concerns also immunogenic and vaccine compositions based on these specific peptide sequences.
Owner:INST NAT PER LE MALATTIE INFETTIVE LAZZARO SPALLANZANI IRCCS
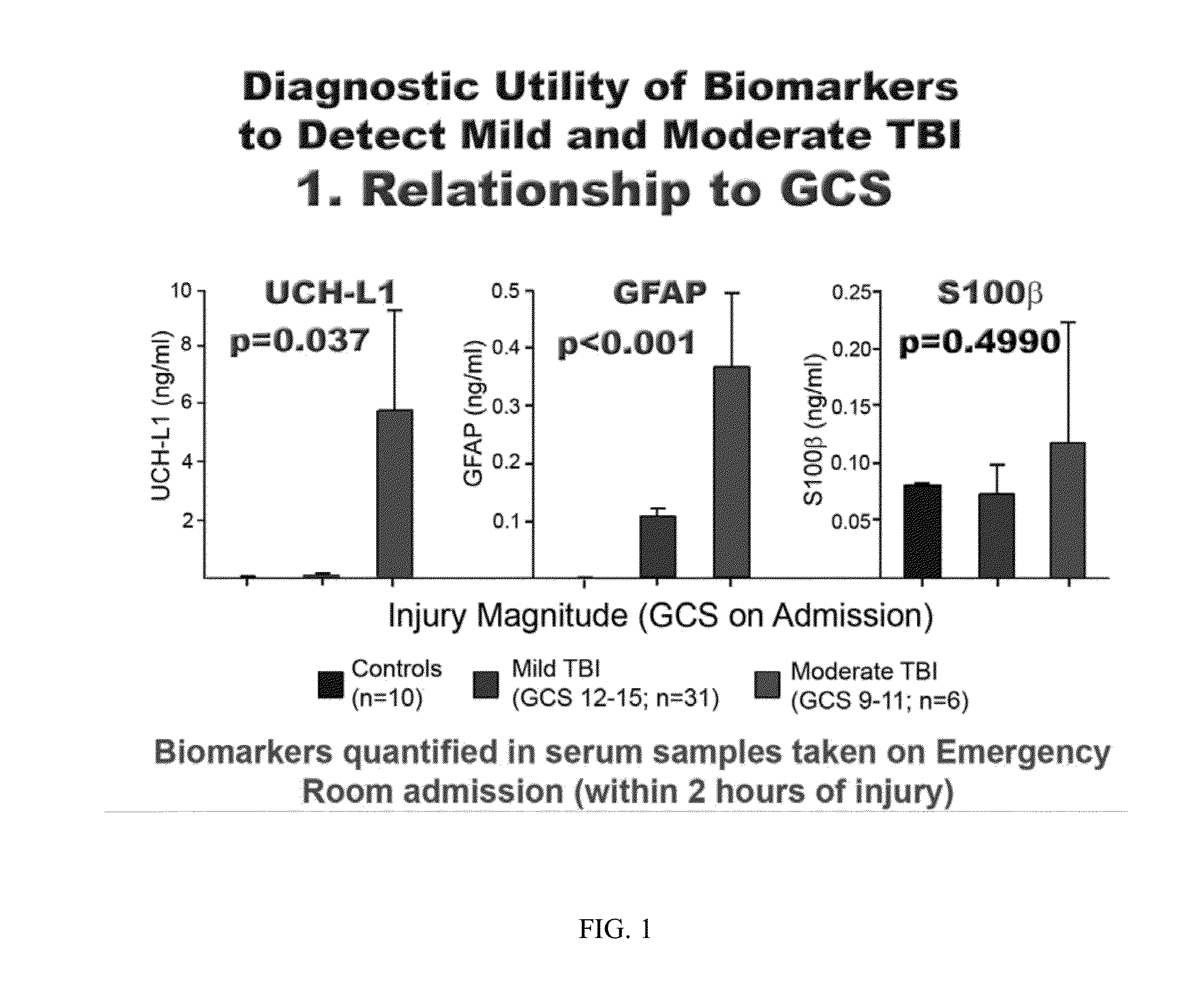
Neural specific s100b for biomarker assays and devices for detection of a neurological condition
InactiveUS20150141528A1Auxiliary diagnosisBiocideBioreactor/fermenter combinationsDisplay deviceNeuronal disease
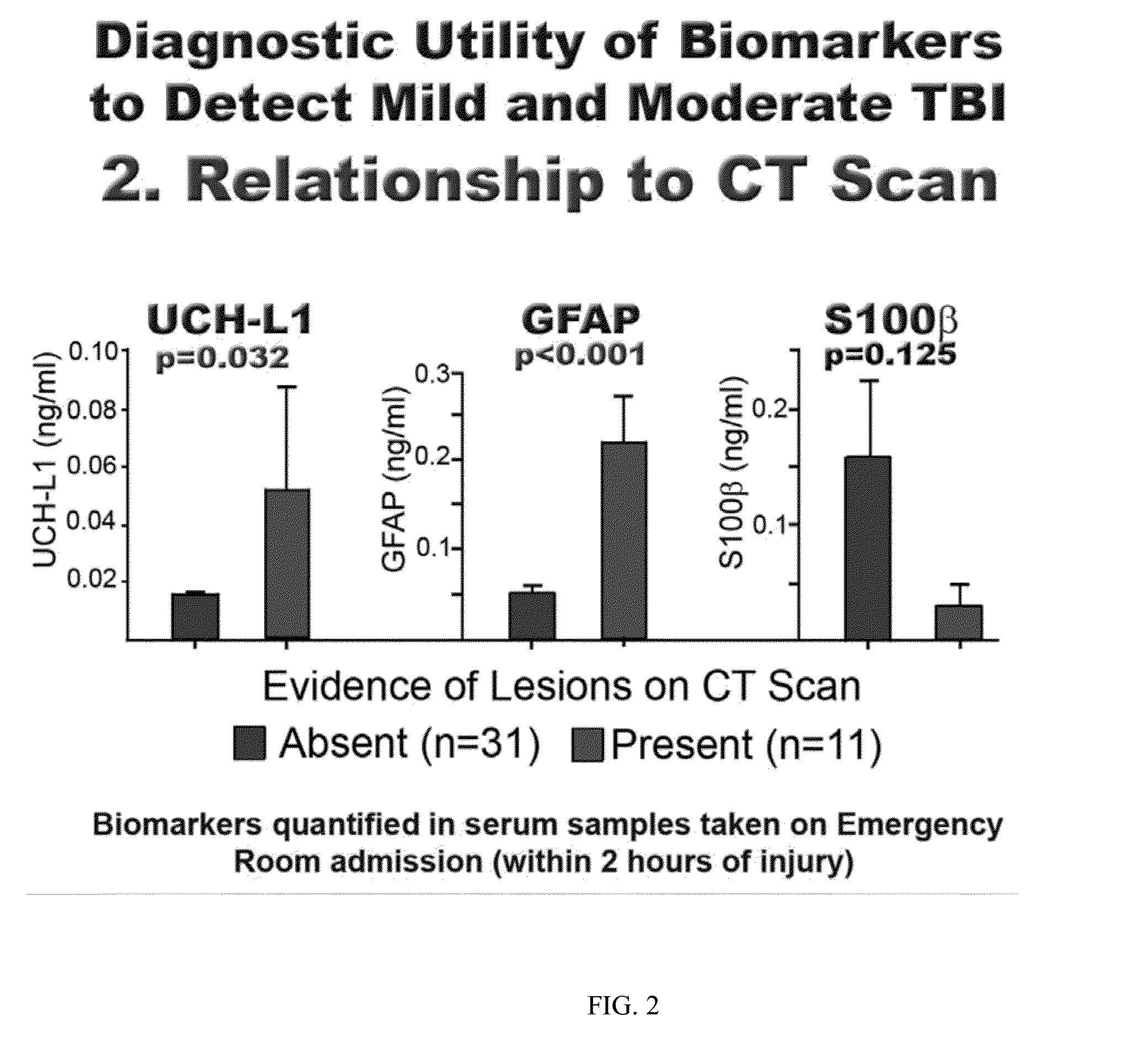
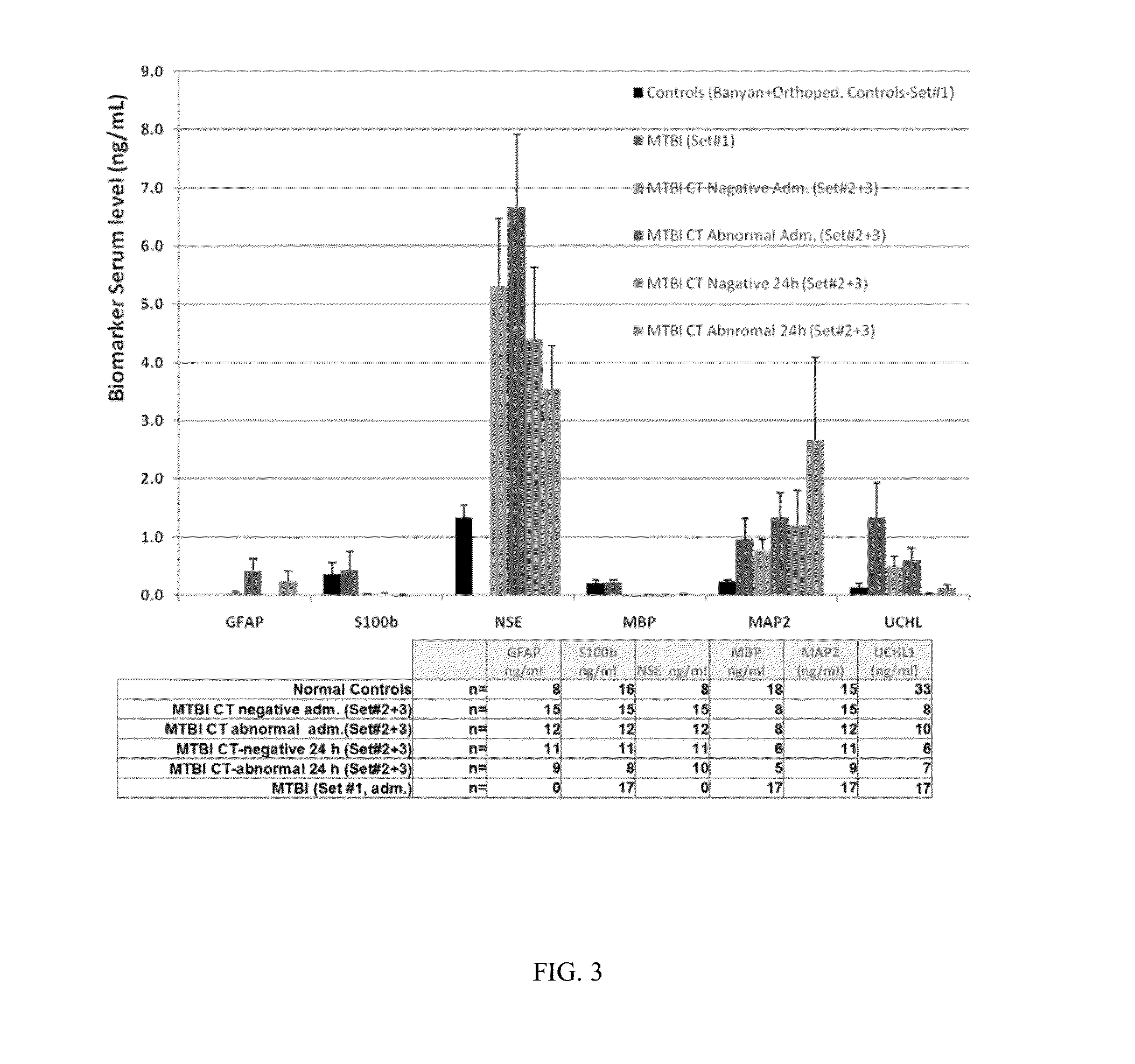
An in vitro diagnostic (IVD) device is used to detect the presence of and / or severity of neural injuries or neuronal disorders in a subject. The IVD device relies on an immunoassay which identifies biomarkers that are diagnostic of neural injury and / or neuronal disorders in a biological sample, such as whole blood, plasma, serum, and / or cerebrospinal fluid (CSF). An IVD device may measure one or more of several neural specific markers in a biological sample and output the results to a machine readable format, either to a display device or to a storage device internal or external to the IVD.
Owner:BANYAN BIOMARKERS INC
Turbidimetric immunoassay for assessing human cystatin c
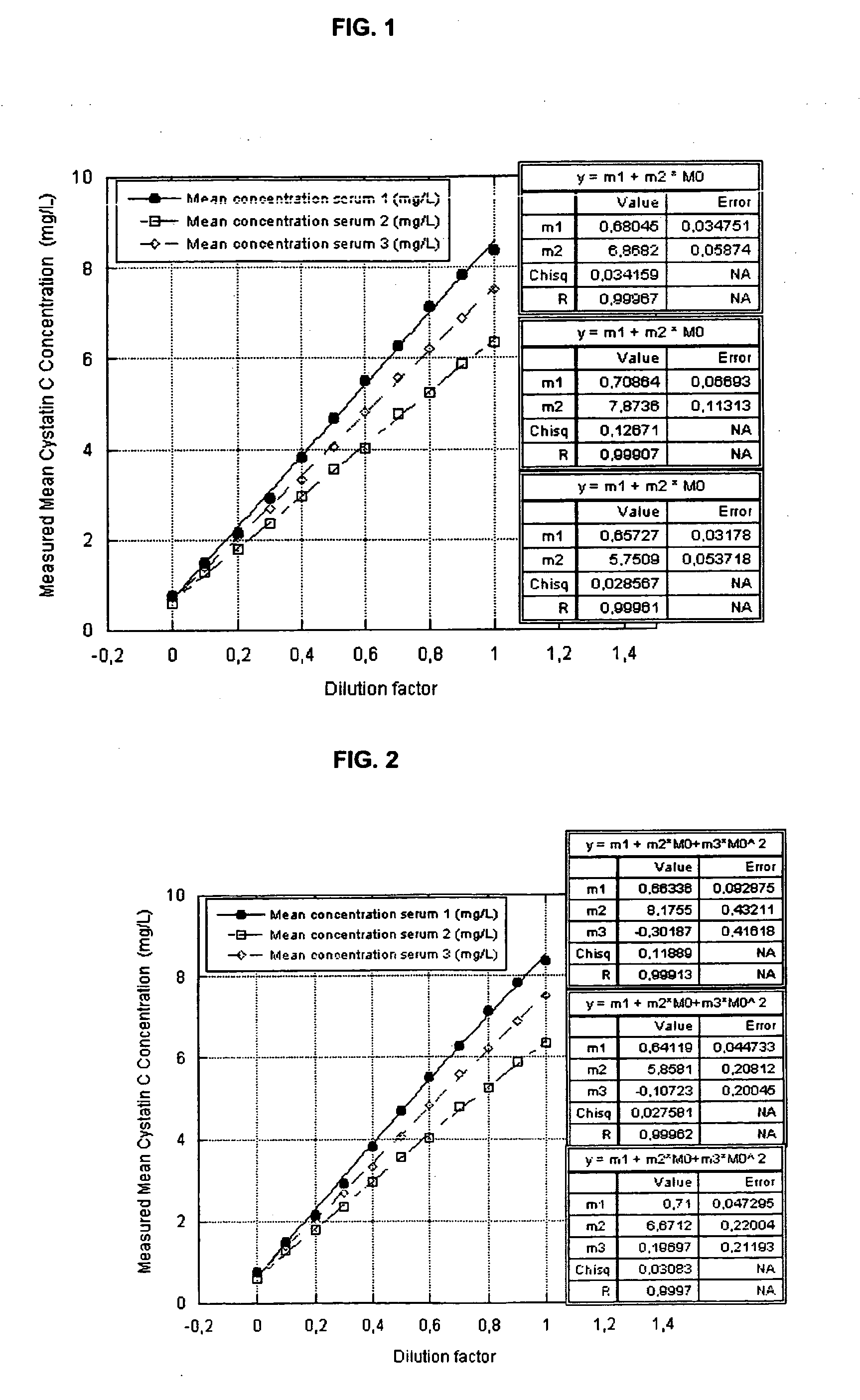
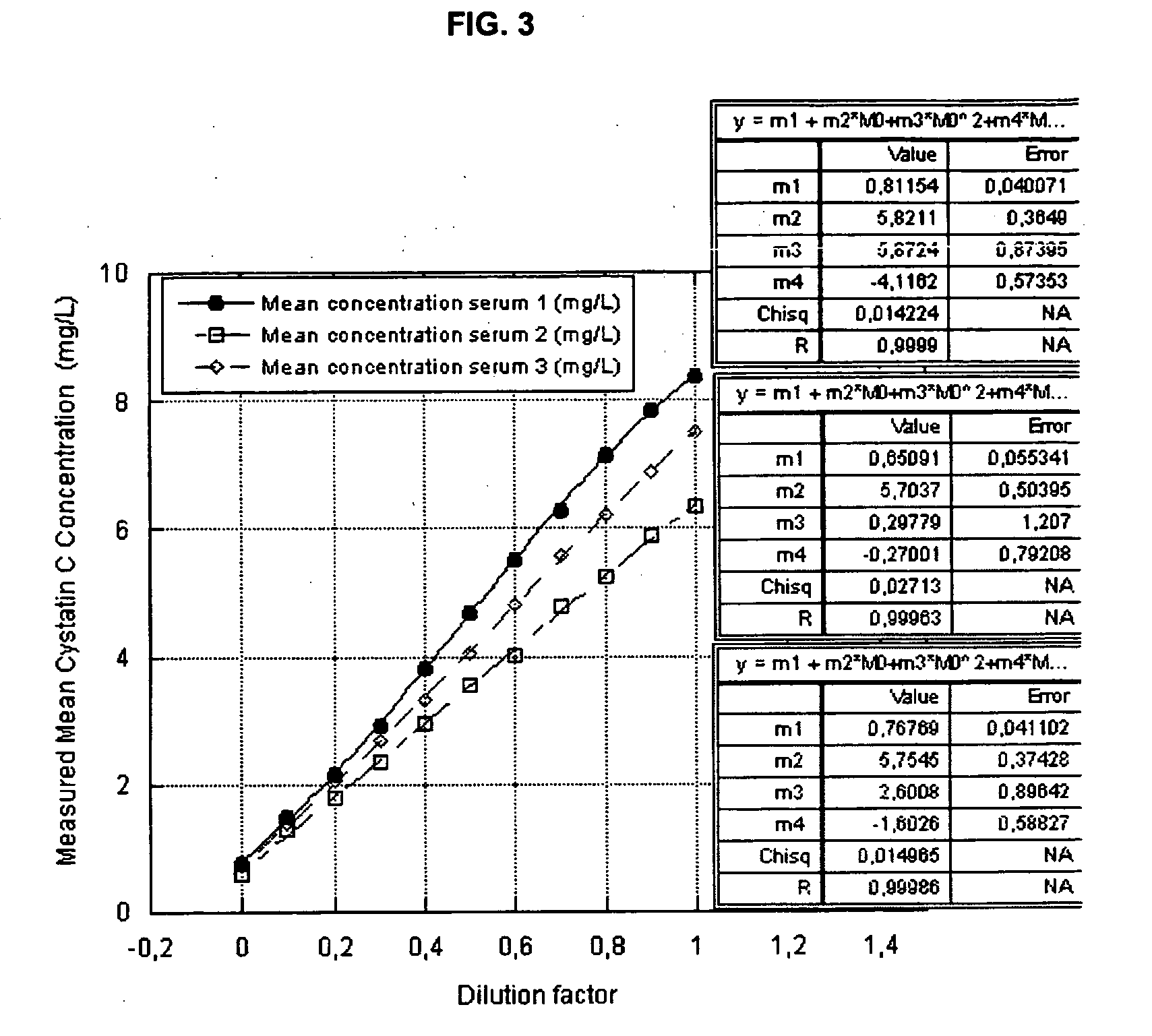
ActiveUS20100047922A1Strong and faster signalReduce distractionsDisease diagnosisImmunoglobulinsTurbidityBody fluid
There is a demand for improved turbidimetric immunoassays for human Cystatin C in biological samples, especially in human clinical samples of body fluids. The present invention provides a turbidimetric immunoassay method and reagent set enabling measurement of human Cystatin C by turbidimetric methods, resulting in a surprisingly stronger and faster turbidimetric signal than in the present state of the art. The increased and faster signal is accomplished by the use of new reagents and compositions, and enables shorter assay times and kinetic reading with a stronger signal, improving overall assay speed and quality. Improved robustness to lipid interference and improved linearity is achieved.
Owner:GENTIAN AS
Ileitis diagnostic assay
Improved immunoassays for the protection of antibodies against Lawsonia intracellularis are provided which permit rapid, easy detection of low concentrations of anti-Lawsonia antibodies in animal-derived specimens. The preferred assay is an ELISA assay employing an antigenic extract of L. intracellularis lipopolysaccharide.
Owner:BOEHRINGER INGELHEIM ANIMAL HEALTH USA INC
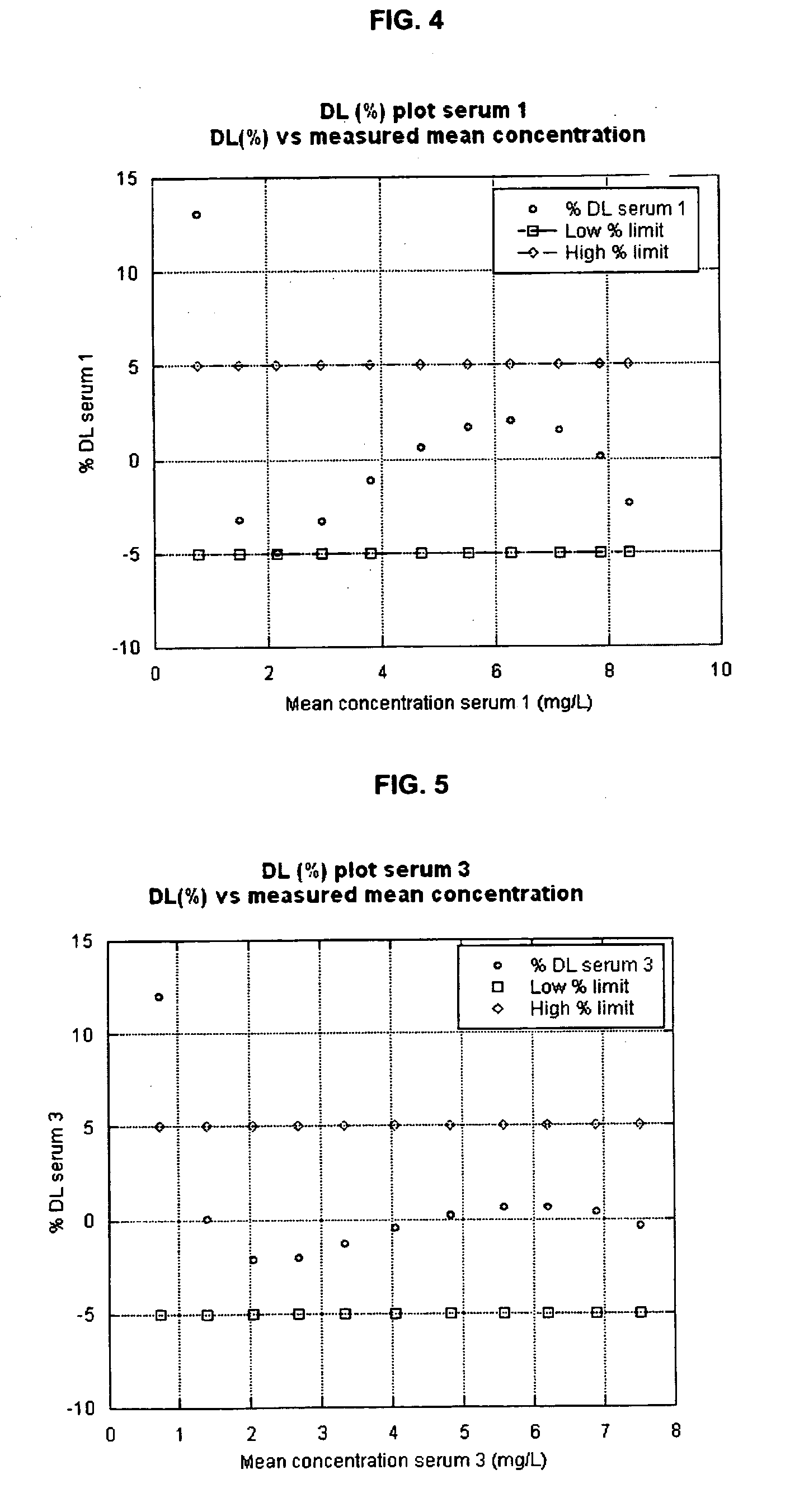
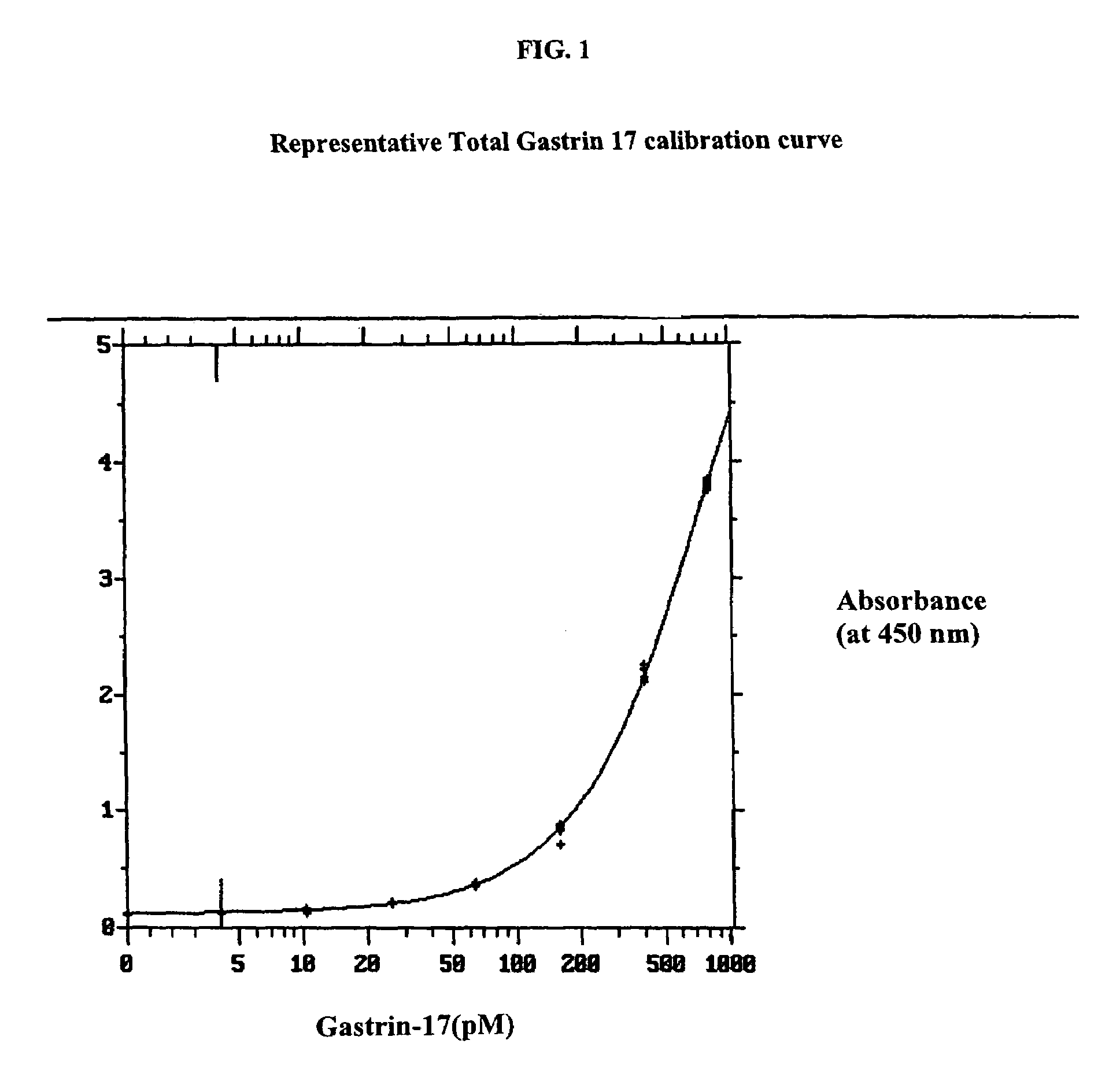
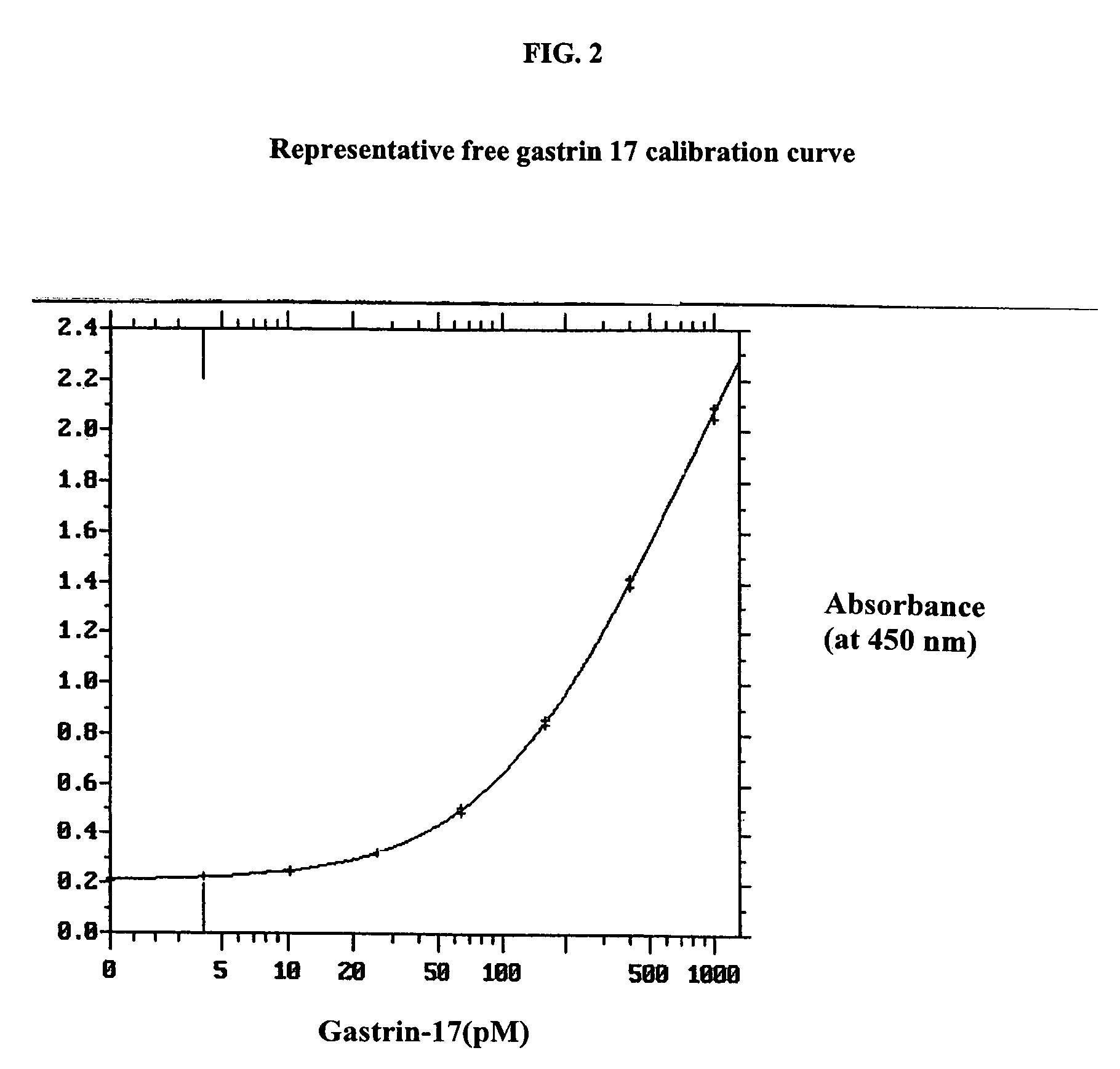
Gastrin hormone immunoassays
ActiveUS7235376B2Biological material analysisImmunoglobulins against hormonesDiseaseHormones regulation
The invention provides assay methods for the detection and quantitation of gastrin hormones, including total and free gastrin hormone in a sample. ELISA-type heterogeneous phase assays suitable for use with biological fluid samples such as blood, plasma or other bodily fluids of a mammal, particularly a human subject are provided. The method provides a precise assay for the amounts of free and total G17 and G34 in biological fluid samples, as well as the amounts of free and total Gly-extended G17, and Gly-extended G34. Also provided are methods of determining suitable treatment for patient suffering from a gastrin hormone-mediated disease or condition employing gastrin hormone immunoassays.
Owner:CANCER ADVANCES INC
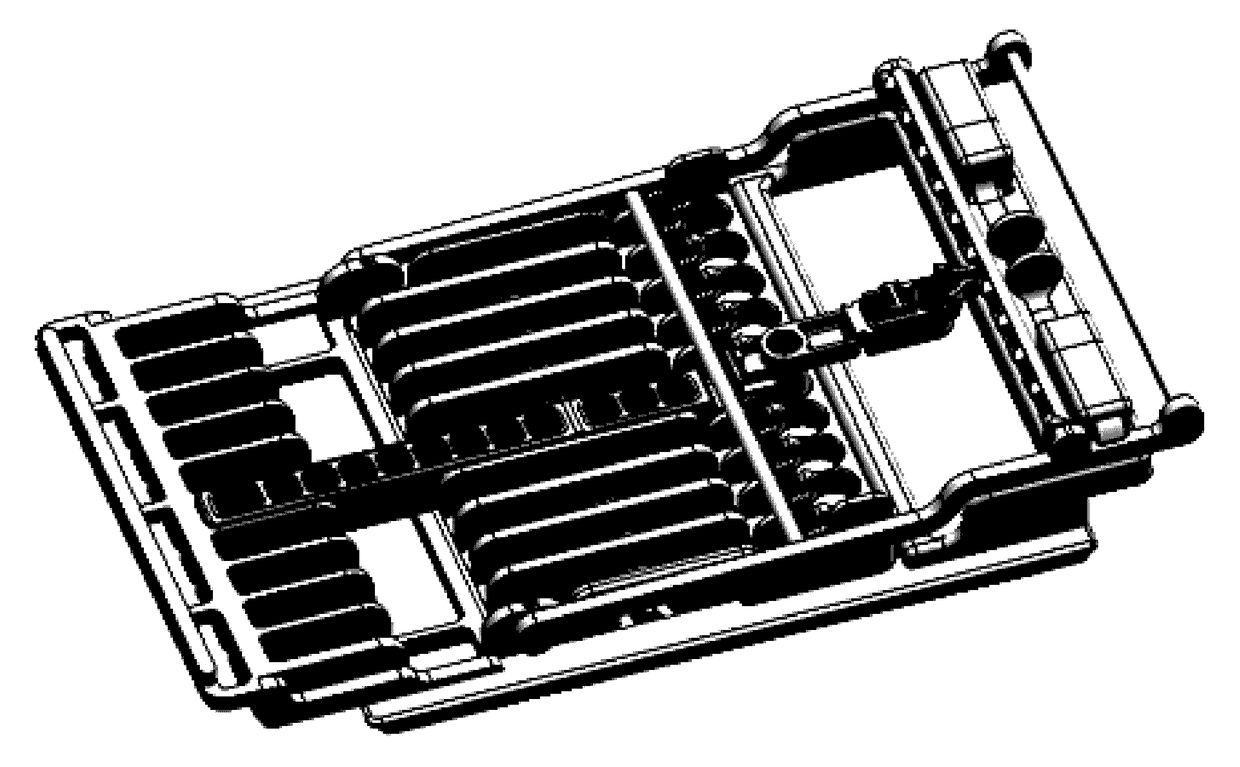
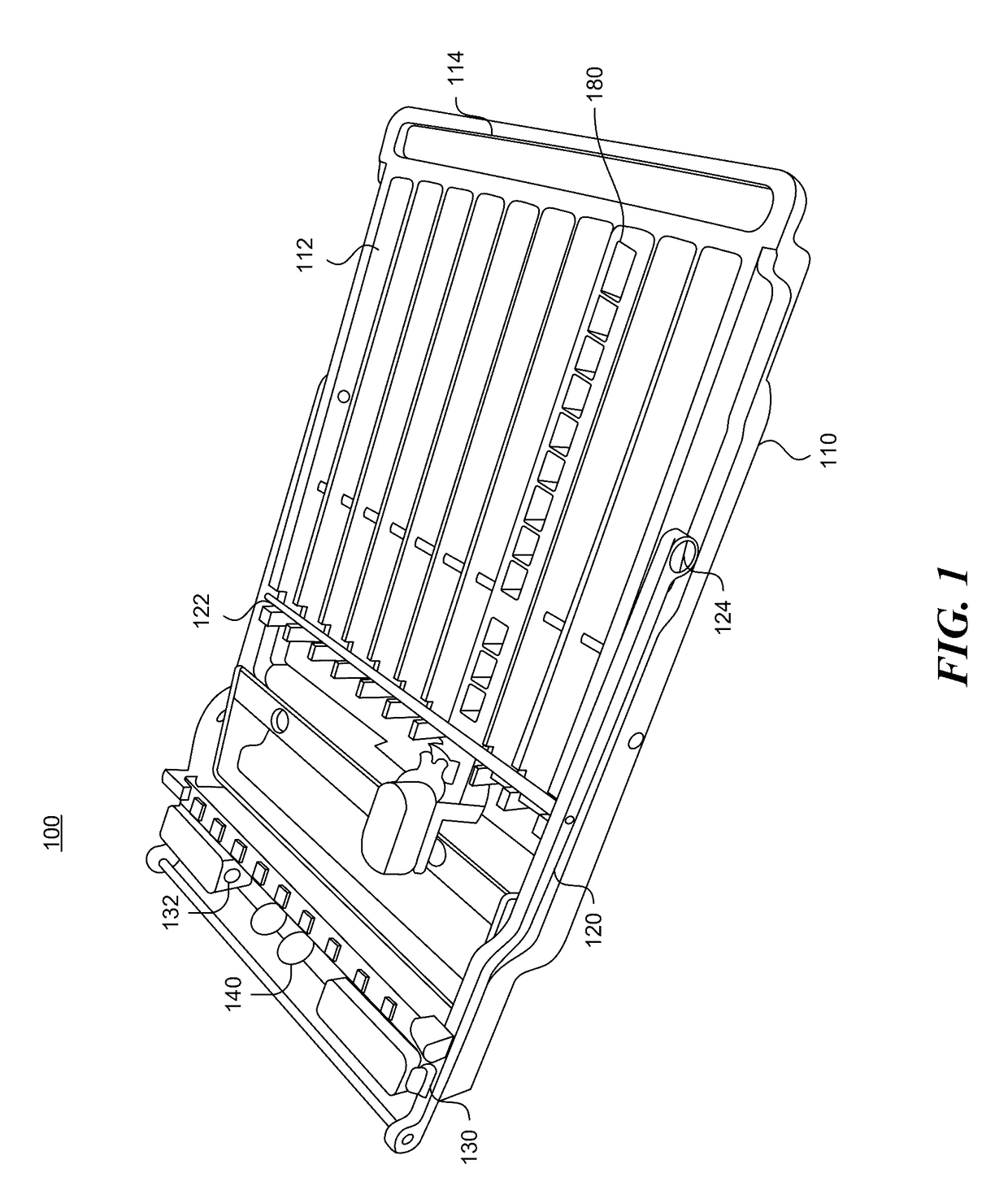
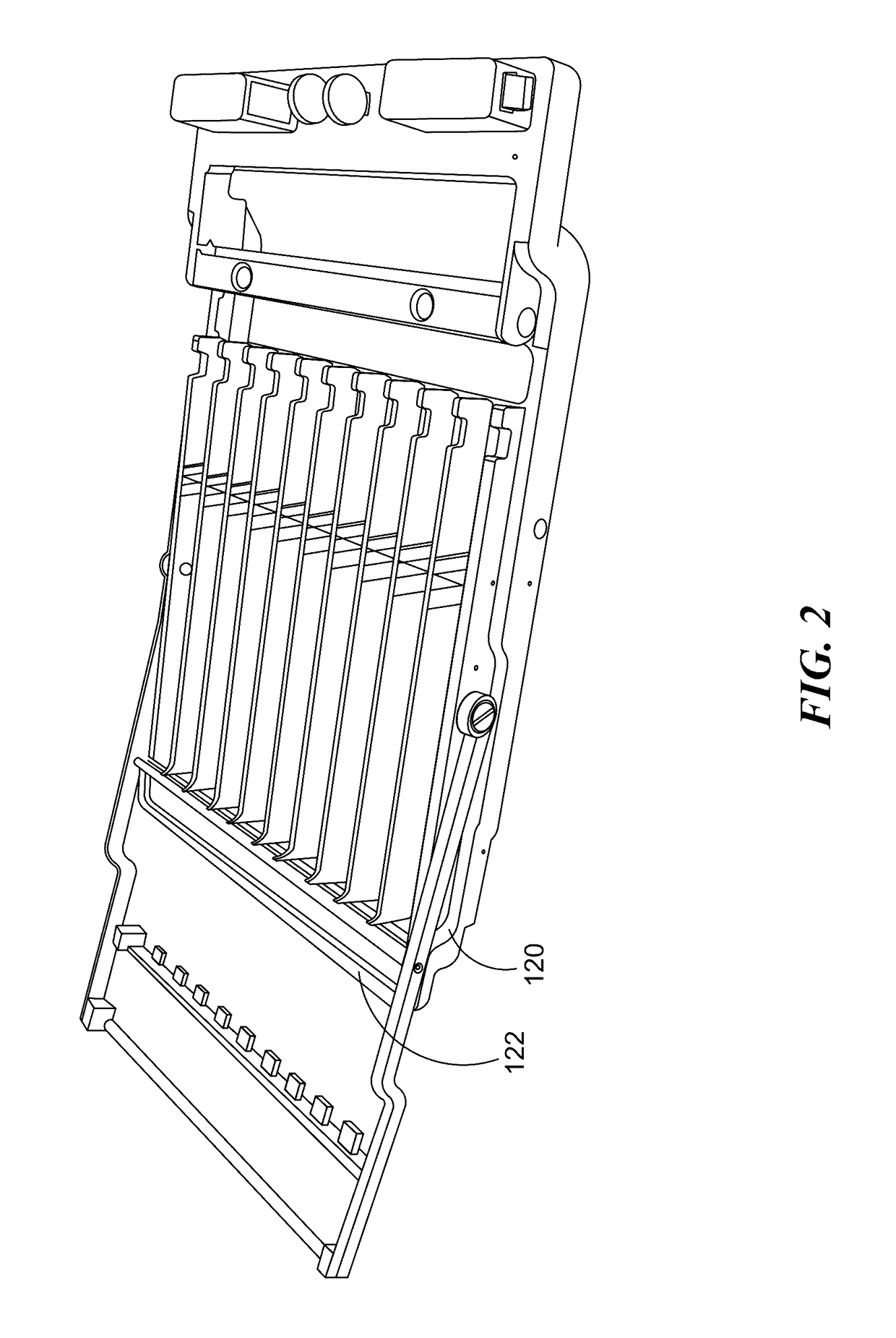
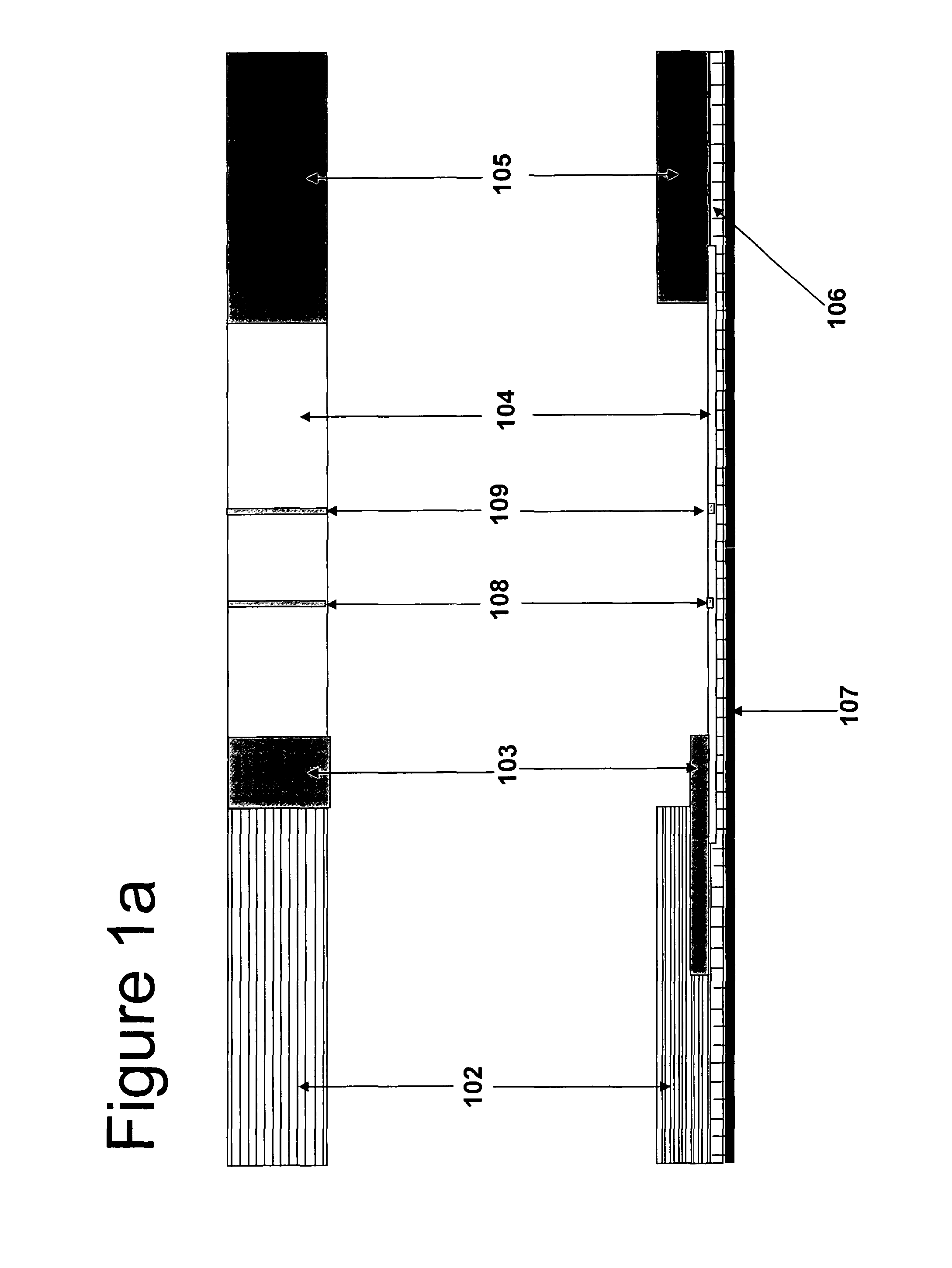
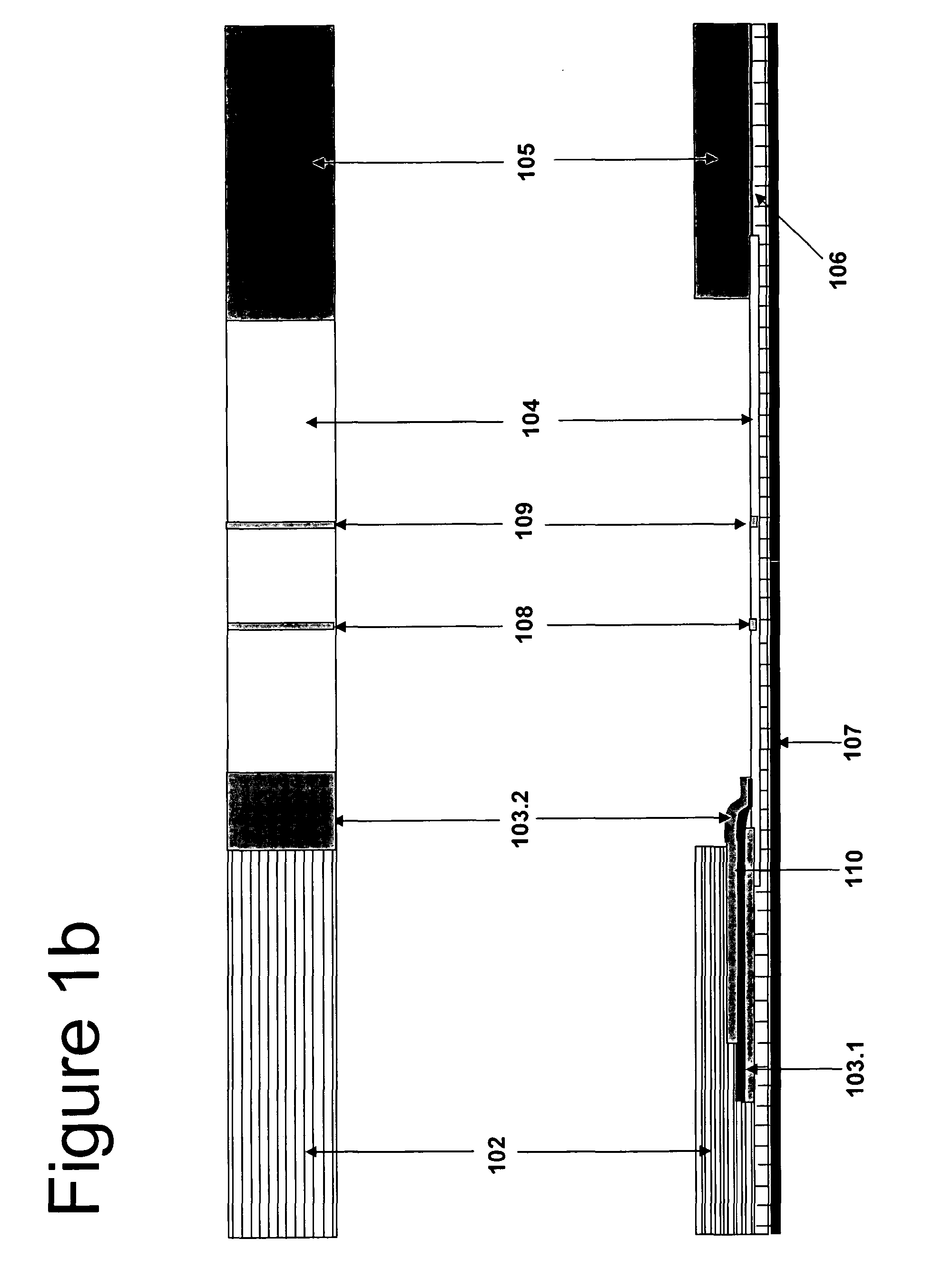
Cartridge assembly tray for immunoassay tests
This invention relates to a cartridge assembly tray for conducting automated biochemical tests, such as immunoassay tests. The tray comprises a base member, a hinged frame and a locking mechanism. The base member includes a plurality of slots within the base member. Each of the plurality of slots is to receive a test cartridge. The hinged frame is coupled to the base member. The hinged frame is capable to rotate to an opened position or a closed position. The hinged frame includes a horizontal push bar configured to apply a downward force to test cartridges received in the plurality of slots when the hinged frame is in the closed position. The locking mechanism is to lock the hinged frame in the closed position when the hinged frame rotates to the closed position.
Owner:ACCESS MEDICAL SYSTEMS LTD
Bovine pregnancy test
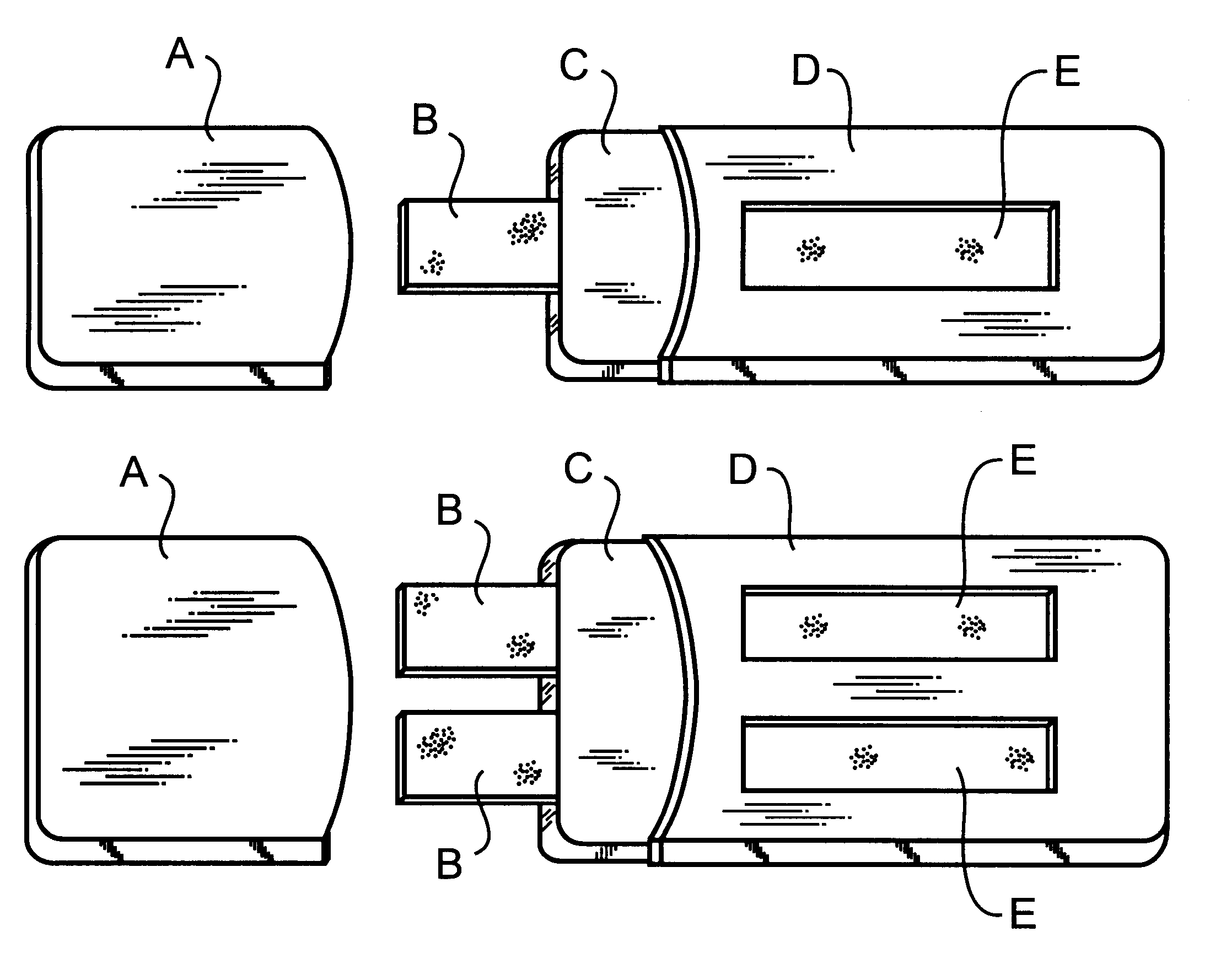
InactiveUS7393696B2Ensure high efficiency and accuracyAnimal reproductionBioreactor/fermenter combinationsAntigenPregnancy tests
This invention provides bovine pregnancy test methods and devices. The test is also suitable for other ruminant and / or ungulate animals. Antigens from Group A (early pregnancy antigens), and / or Group B (mid-pregnancy antigens), and Group C (early, mid- and late pregnancy antigens) are detected in a fluid from the animal, and pregnancy is reliably determined. The pregnancy assays of this invention are preferably carried out using immunoassay devices which provide immediate results in the field.
Owner:ASPENBIO PHARMA
Rapid immunochromatographic detection by amplification of the colloidal gold signal
InactiveUS20100068727A1Improve rapid immunochromatographic detection of targetHigh sensitivityComponent separationPreparing sample for investigationColloidSpecific antibody
The present invention relates to a method for rapid immunochromatographic detection of a target in a sample by double sandwich immunoassay detection, wherein the target is an antibody and / or an antigen, using different colloidal gold conjugates conjugated with a first and a second specific antibody or antigen and with at least one oligonucleotide and its at least one complementary oligonucleotide and / or at least one non-specific antibody and its related non-specific antigen, to a rapid immunochromatographic detection device, to uses of the method for detecting diseases or specific conditions, and to a method for the manufacture of the device as well as to a kit which comprises the device.
Owner:ARAGEN BIOTECH
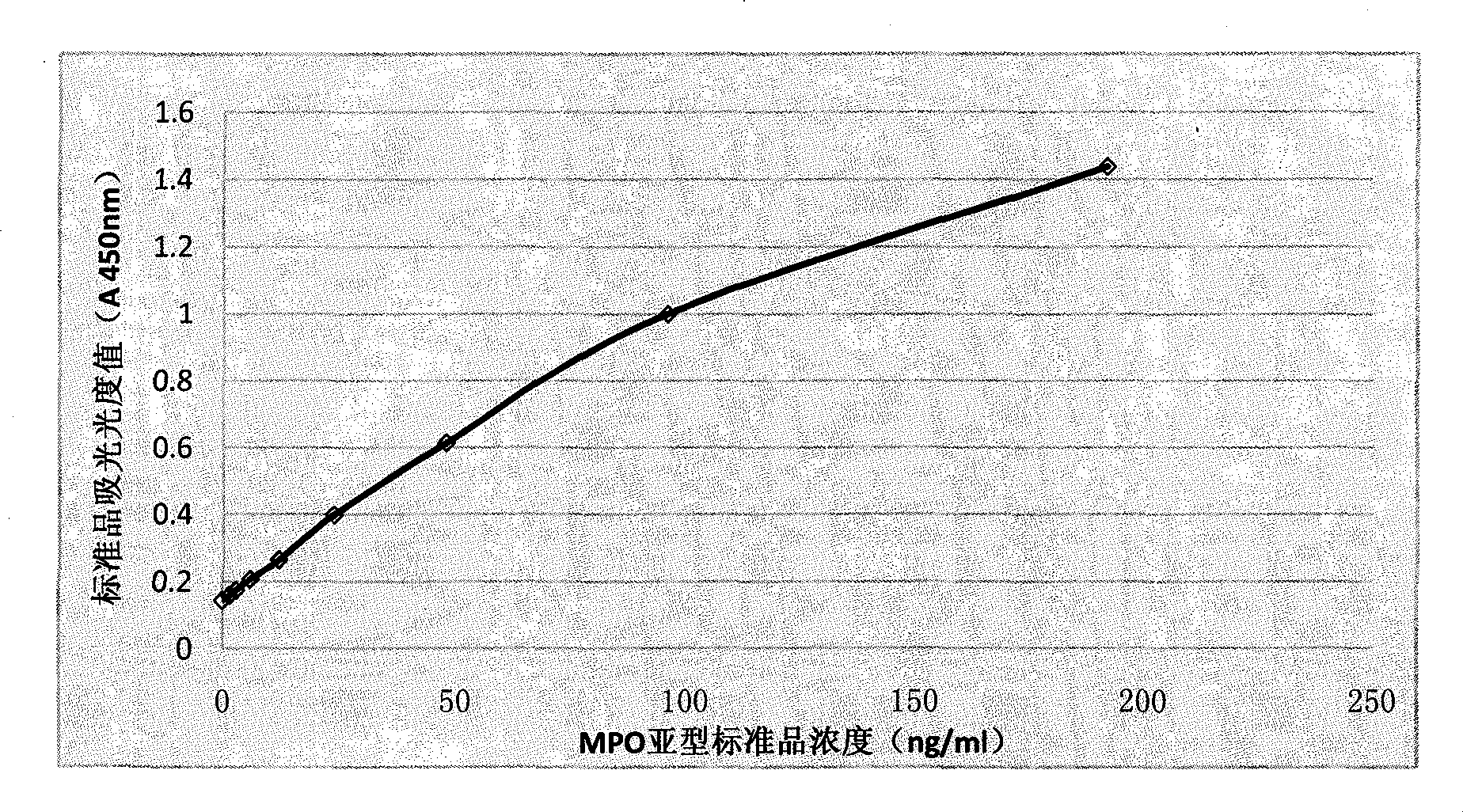
Method and kit for detecting myeloperoxidase (MPO)
The invention provides a method for detecting either homotype or subtype MPO or a composition of MPO, in particular to an immune detection method with high specificity and flexibility through one but not all of either homotype or subtype MPO or the composition of MPO. The method is used for carrying out auxiliary diagnoses on whether the patients are suffered from an anaphylactic disease, an inflammation disease, a cardiovascular disease or an autoimmune disease or not, or analyzing the risk that the prognostic patients are developed to the anaphylactic disease, the inflammation disease, the cardiovascular disease or the autoimmune disease. The invention further provides a kit for implementing the method.
Owner:杭州华得森生物技术有限公司
Tumour marker proteins and uses thereof
InactiveUS8592169B2Easy to disassembleIncrease concentrationTumor rejection antigen precursorsTumor specific antigensExcretion processAutoantibody production
The invention relates to tumor marker proteins and their preparation from fluids from one or more cancer patients, wherein said fluids are those which collect in a body cavity or space which is naturally occurring or which is the result of cancer or medical intervention for cancer. The invention also relates to preparation of tumor marker proteins from excretions taken from patients with cancer. The tumor marker proteins are useful as immunoassay reagents in the detection of cancer-associated anti-tumor marker autoantibodies.
Owner:ONCIMMUNE
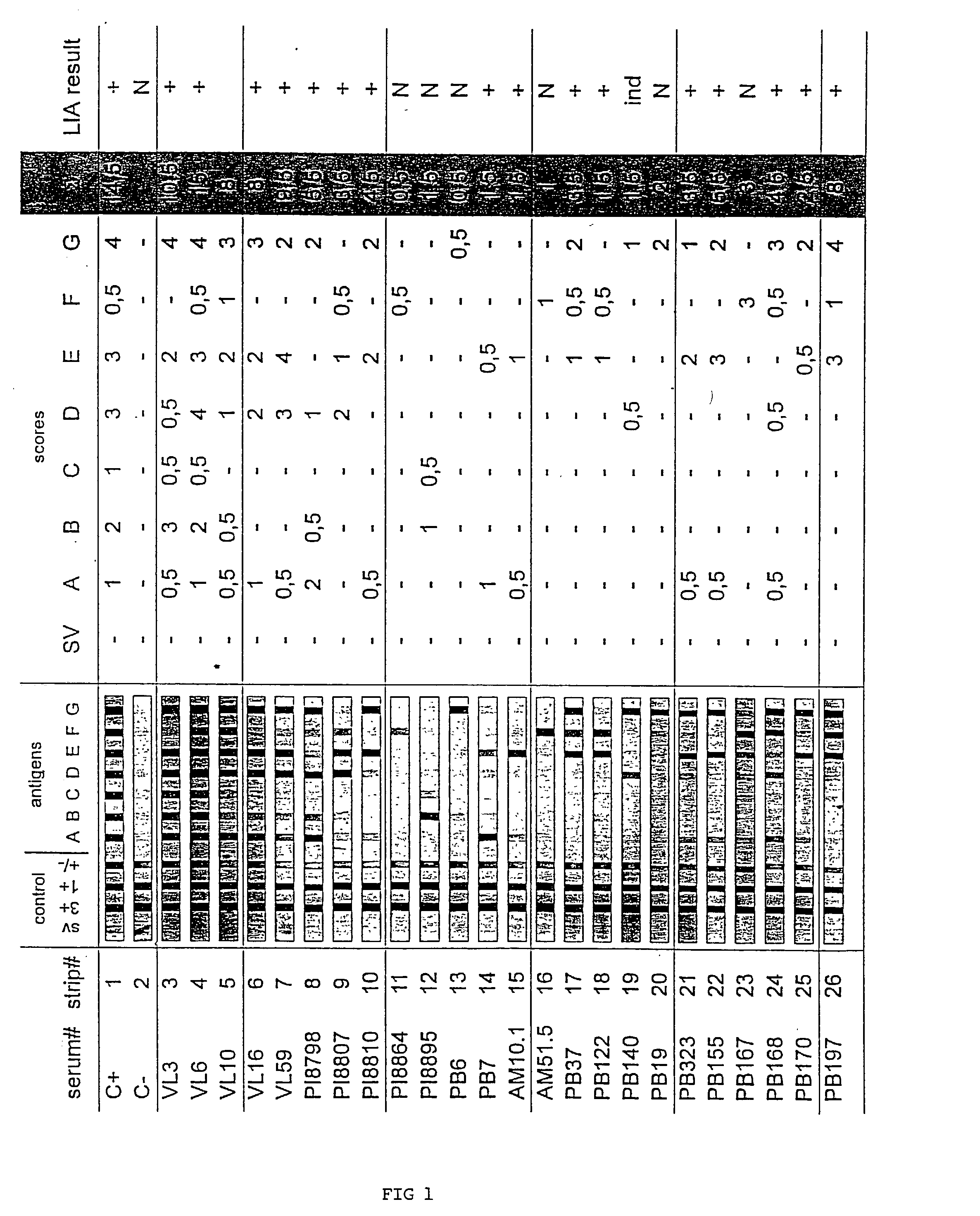
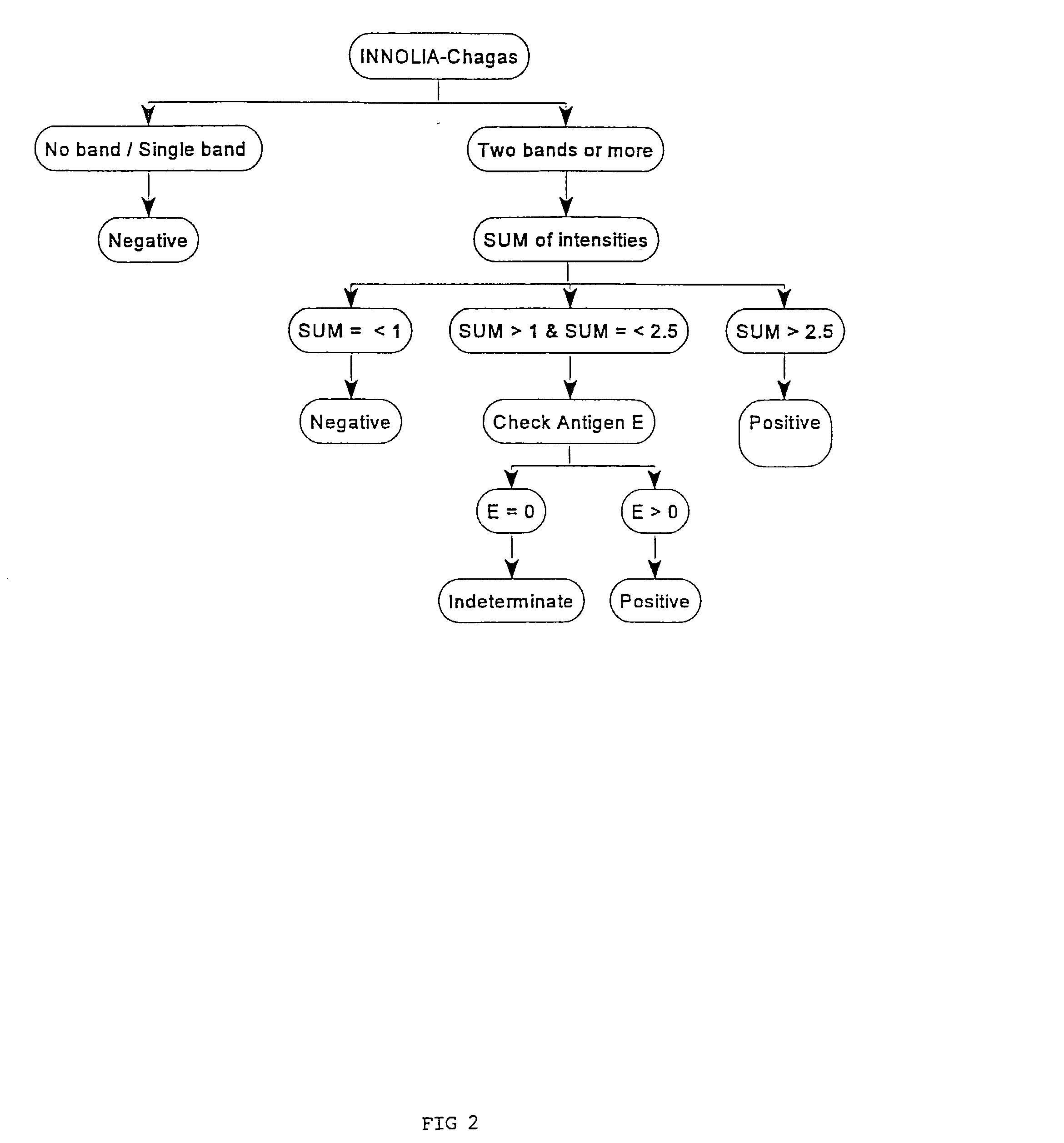
Antigens and immunoassays for diagnosing Chagas' disease
Transfusion of contaminated blood has become the major route of transmission for Chagas' disease. Current screening tests are insensitive and yield conflicting results, while confirmatory assays do not exist. The present invention relates to antigens and their use for serological diagnosis of Chagas' disease. More specifically, the present invention concerns assays which are able to reliably and accurately detect the presence of antibodies to various specific antigens of Trypanosoma cruzi in a highly sensitive and specific manner.
Owner:INNOGENETICS NV
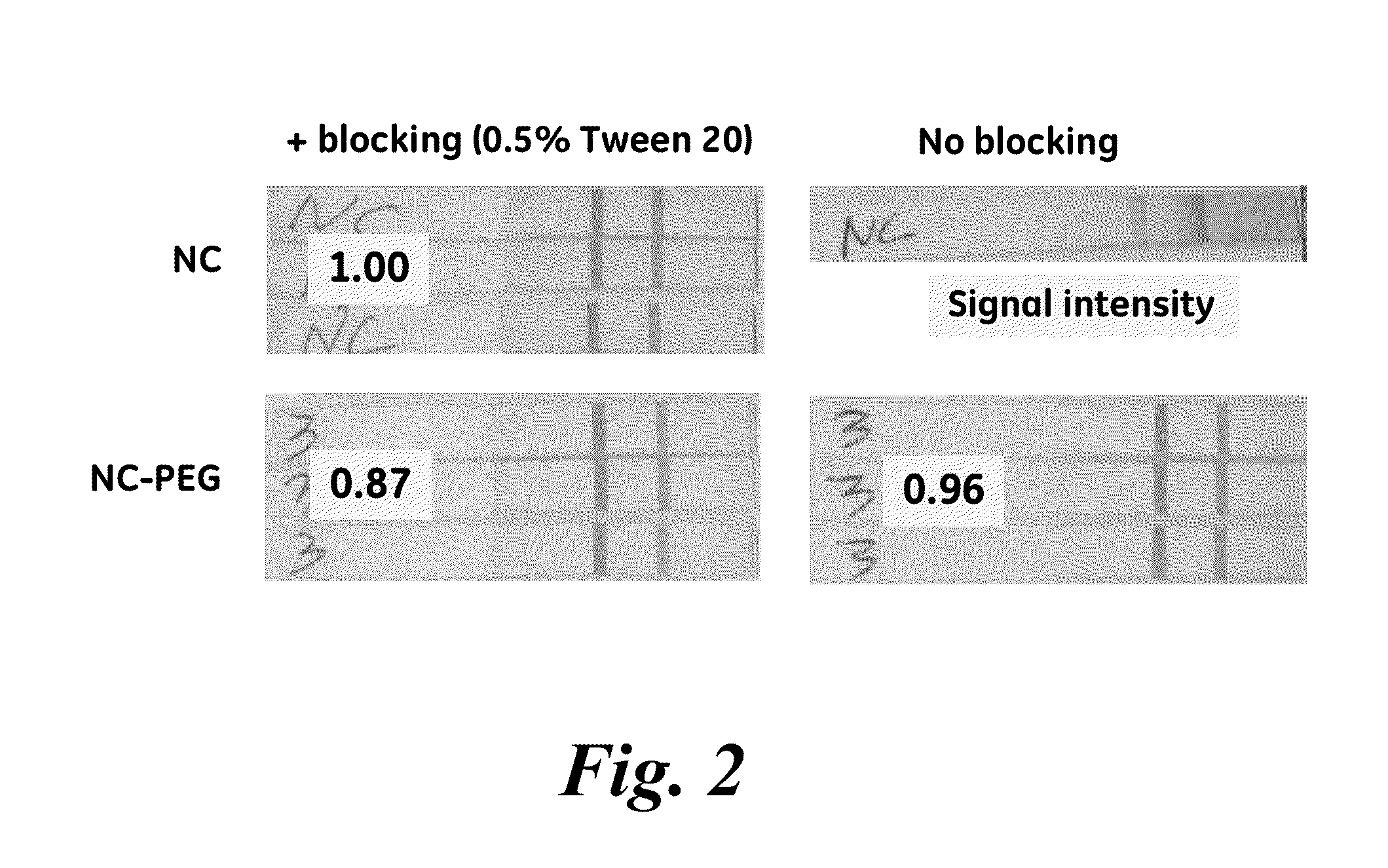
Porous membranes having a hydrophilic coating and methods for their preparation and use
InactiveUS20130171619A1Microbiological testing/measurementChemiluminescene/bioluminescenceHydrophilic coatingSignal-to-noise ratio (imaging)
A modified porous membrane comprising a polymeric hydrophilic coating grafted to a porous membrane is described. The polymeric hydrophilic coatings grafted to the porous membranes comprise, for example, a PEG moiety such as a PEGMA, a PEGDA, or a TMPET, wherein the polymeric hydrophilic coating on the porous membrane decreases non-specific binding of unwanted material to the porous membrane and increases the signal to noise ratio in immunoassays, in vitro diagnostic tests, and point of care tests. Methods of making these modified porous membranes are also disclosed.
Owner:GENERAL ELECTRIC CO
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com