Patents
Literature
62results about How to "Accurately tailored" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
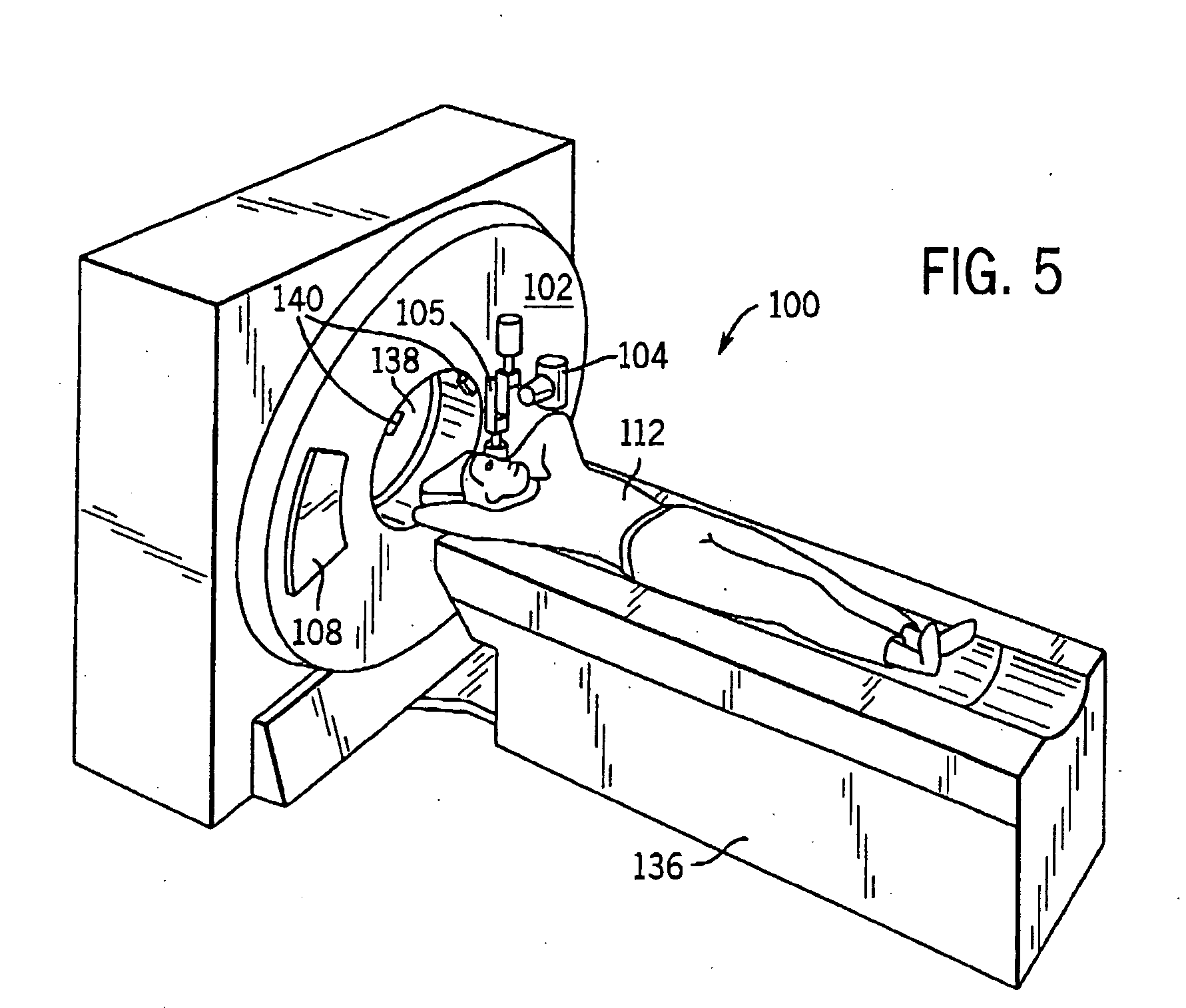
System and method of determining a center of mass of an imaging subject for x-ray flux management control
InactiveUS7068751B2Optimize radiation exposureAccurately tailoredMaterial analysis using wave/particle radiationRadiation/particle handlingUltrasound attenuationX-ray
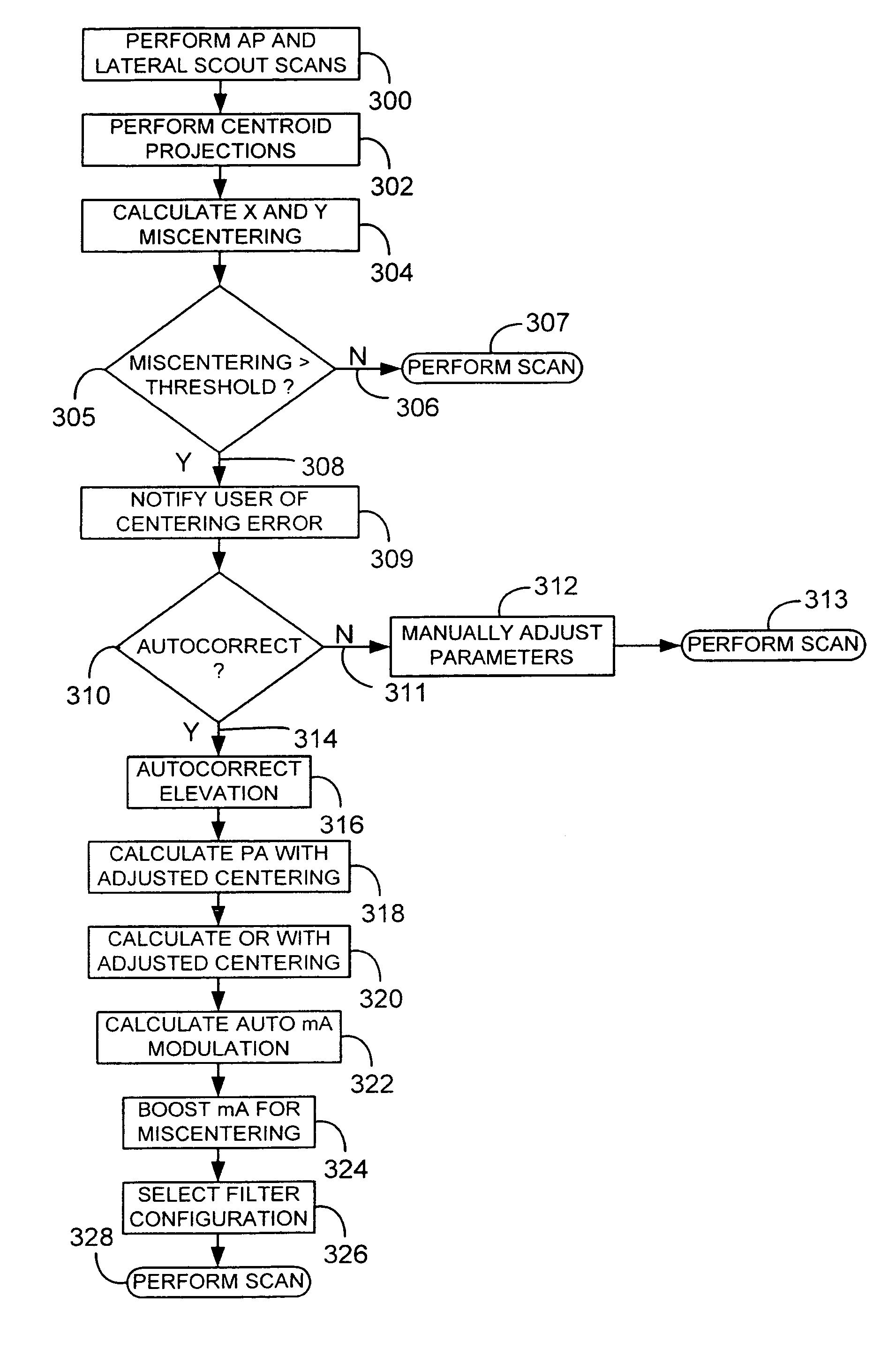
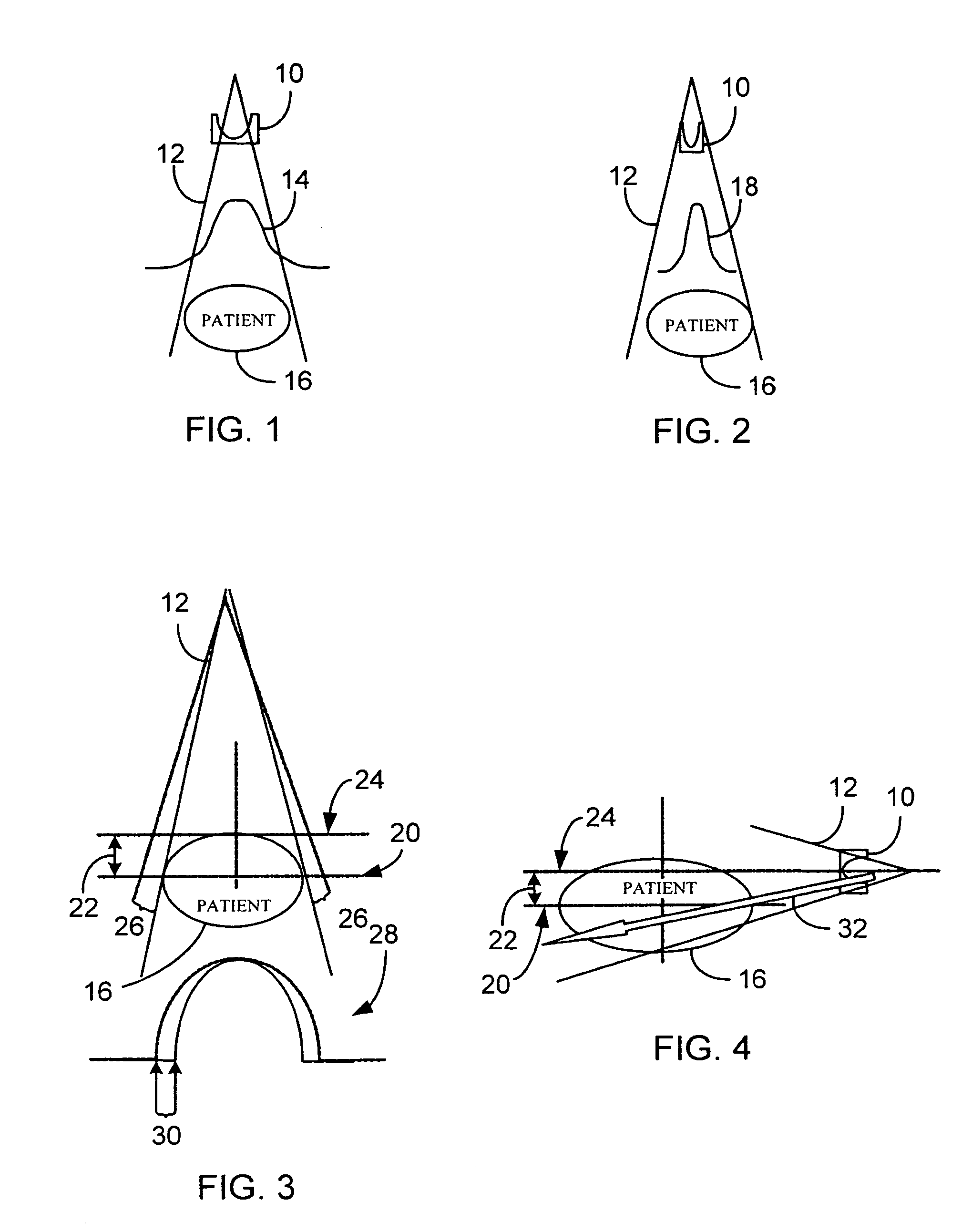
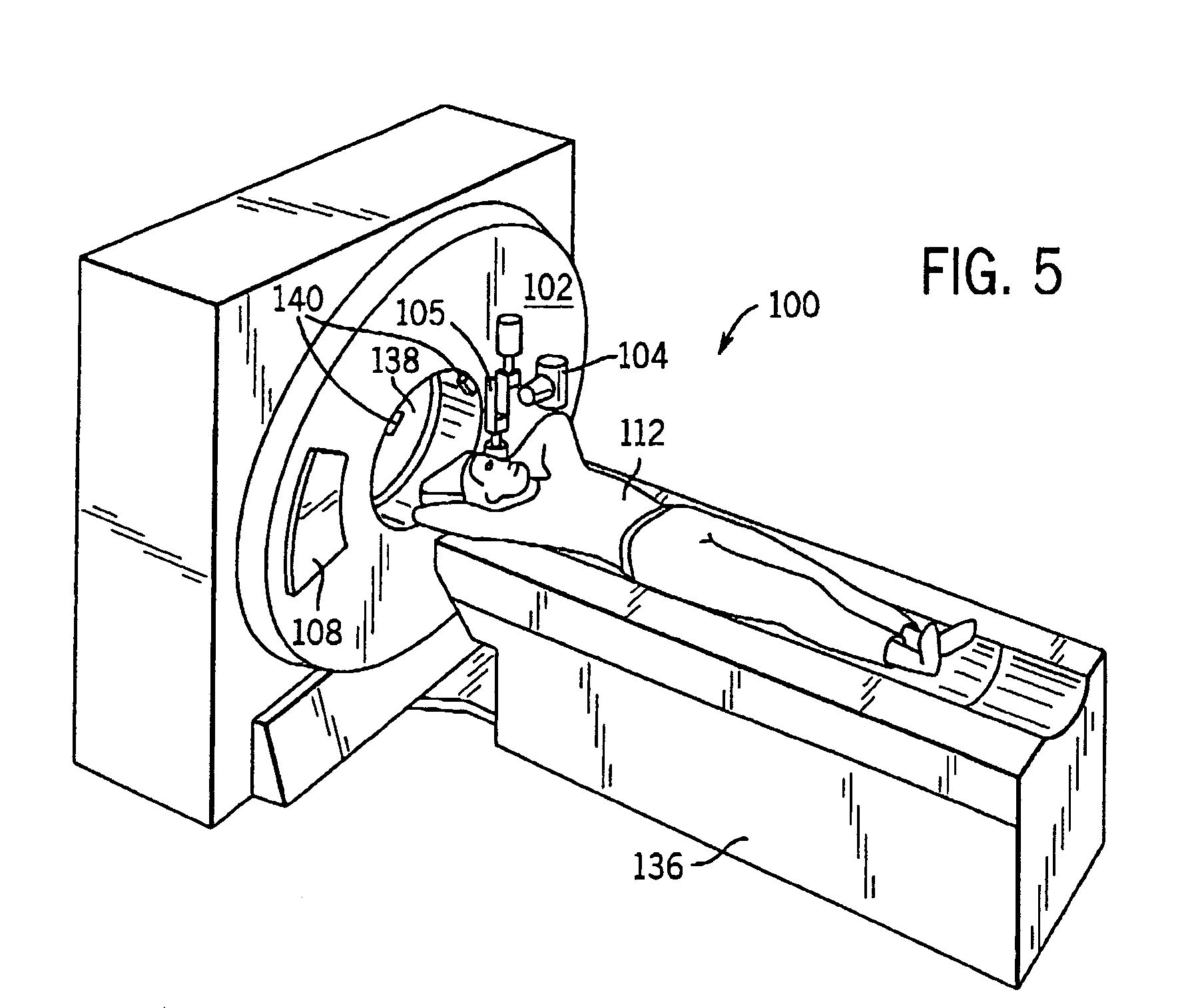
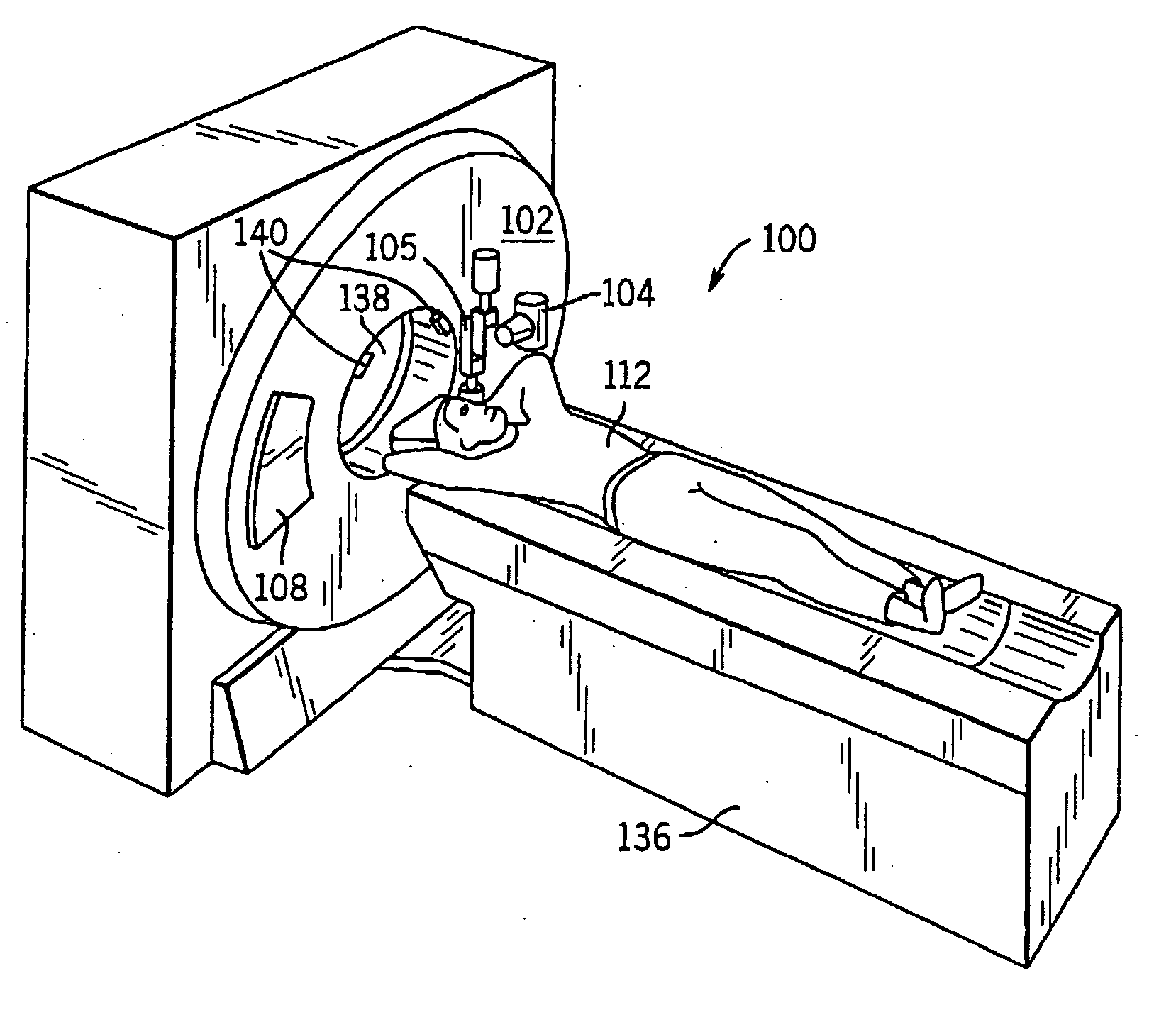
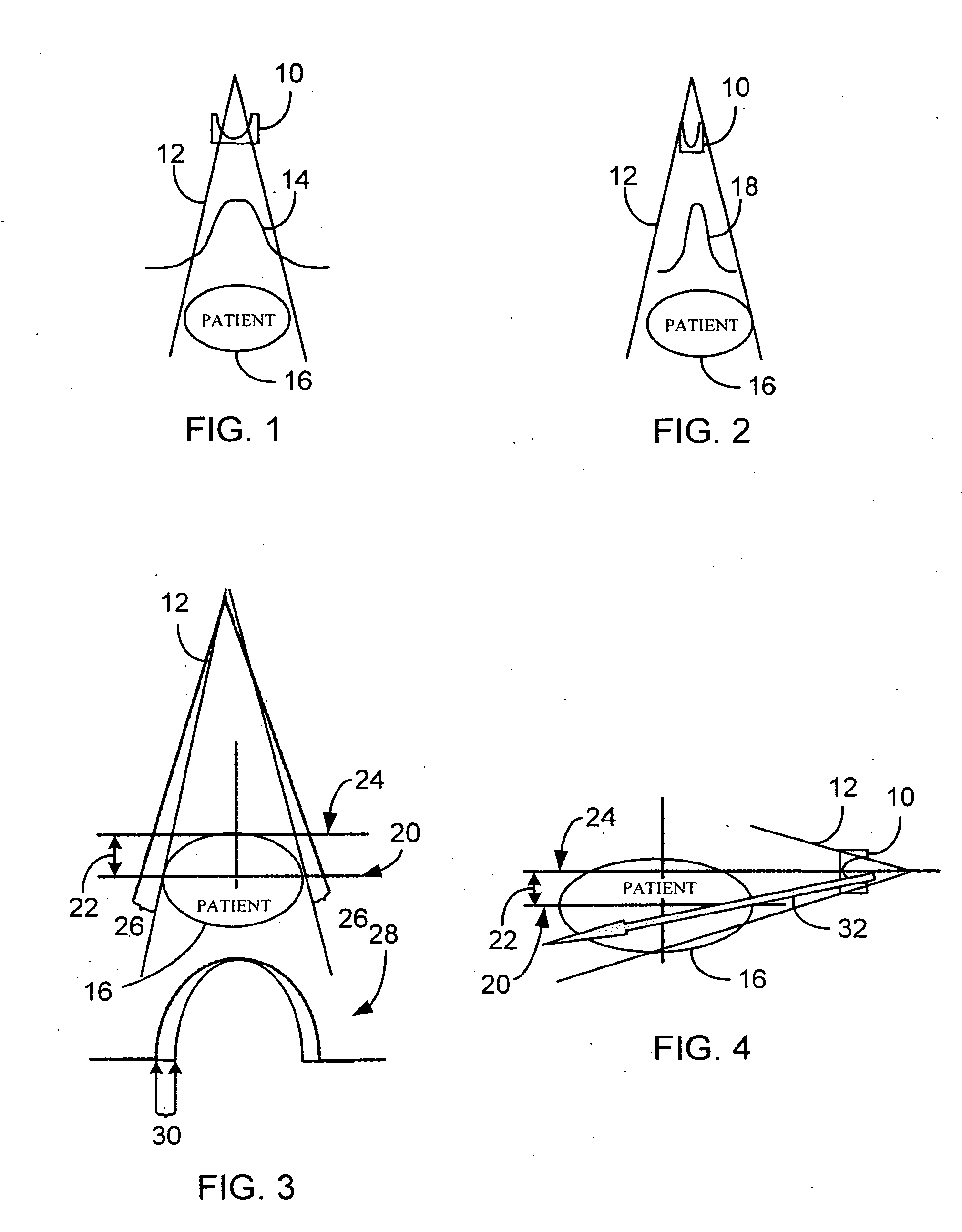
A system and method of diagnostic imaging is provided that includes positioning a subject in a scanning bay, comparing a center of mass of the subject to a reference point, and repositioning the subject in the scanning bay to reduce a difference in position between the center of mass of the subject and the reference point. The present invention automatically selects a proper attenuation filter configuration, corrects patient centering, and corrects noise prediction errors, thereby increasing dose efficiency and tube output.
Owner:GENERAL ELECTRIC CO
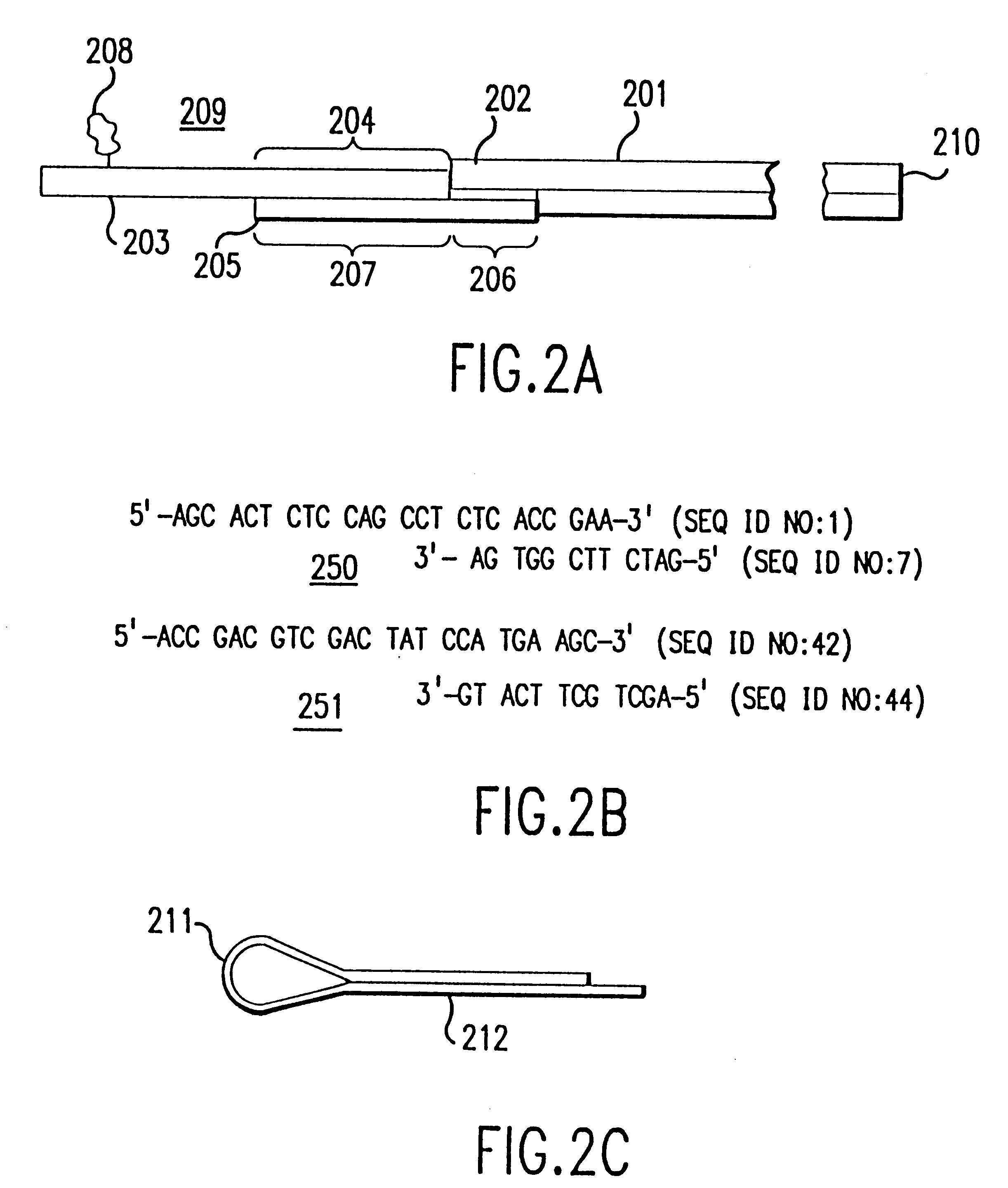
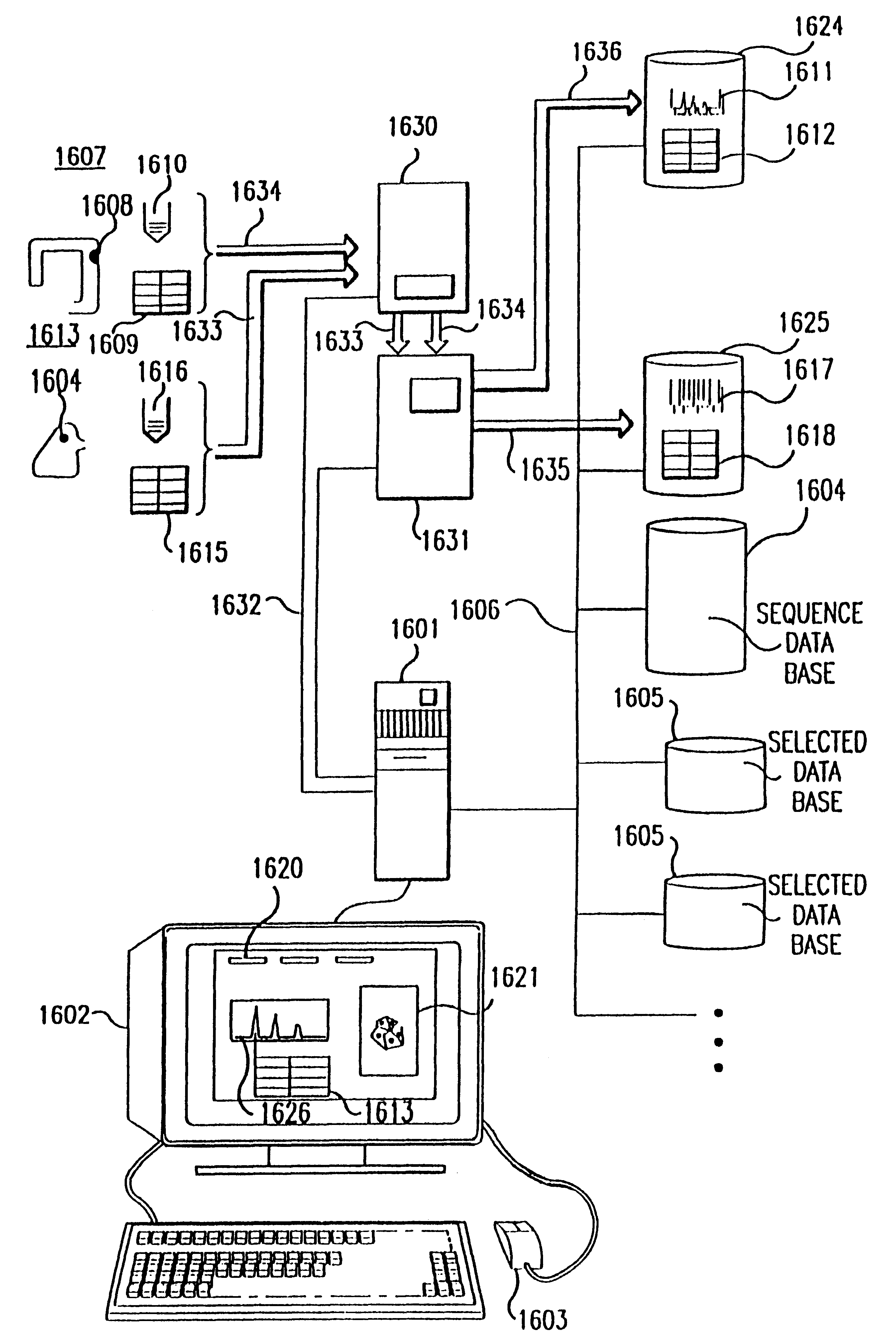
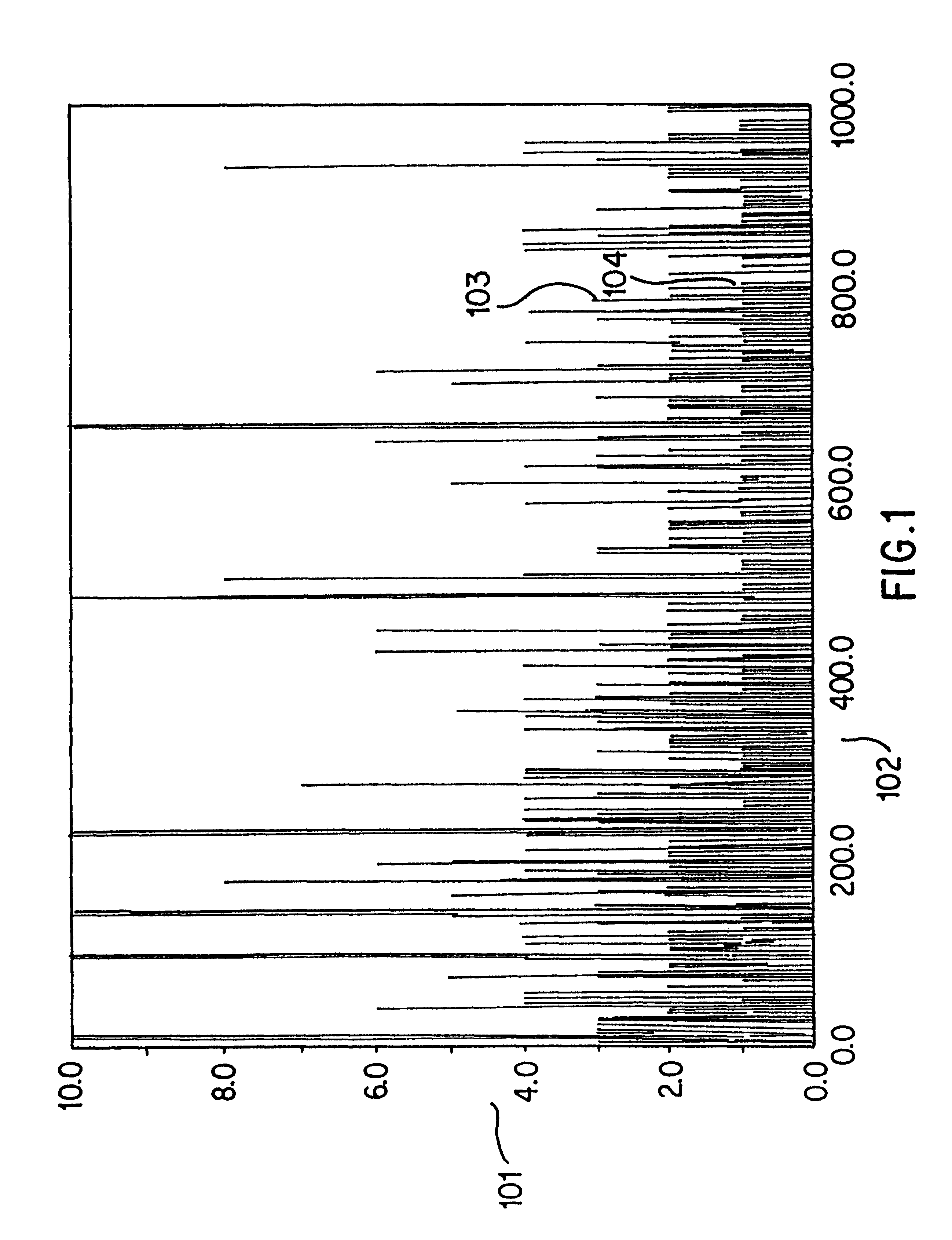
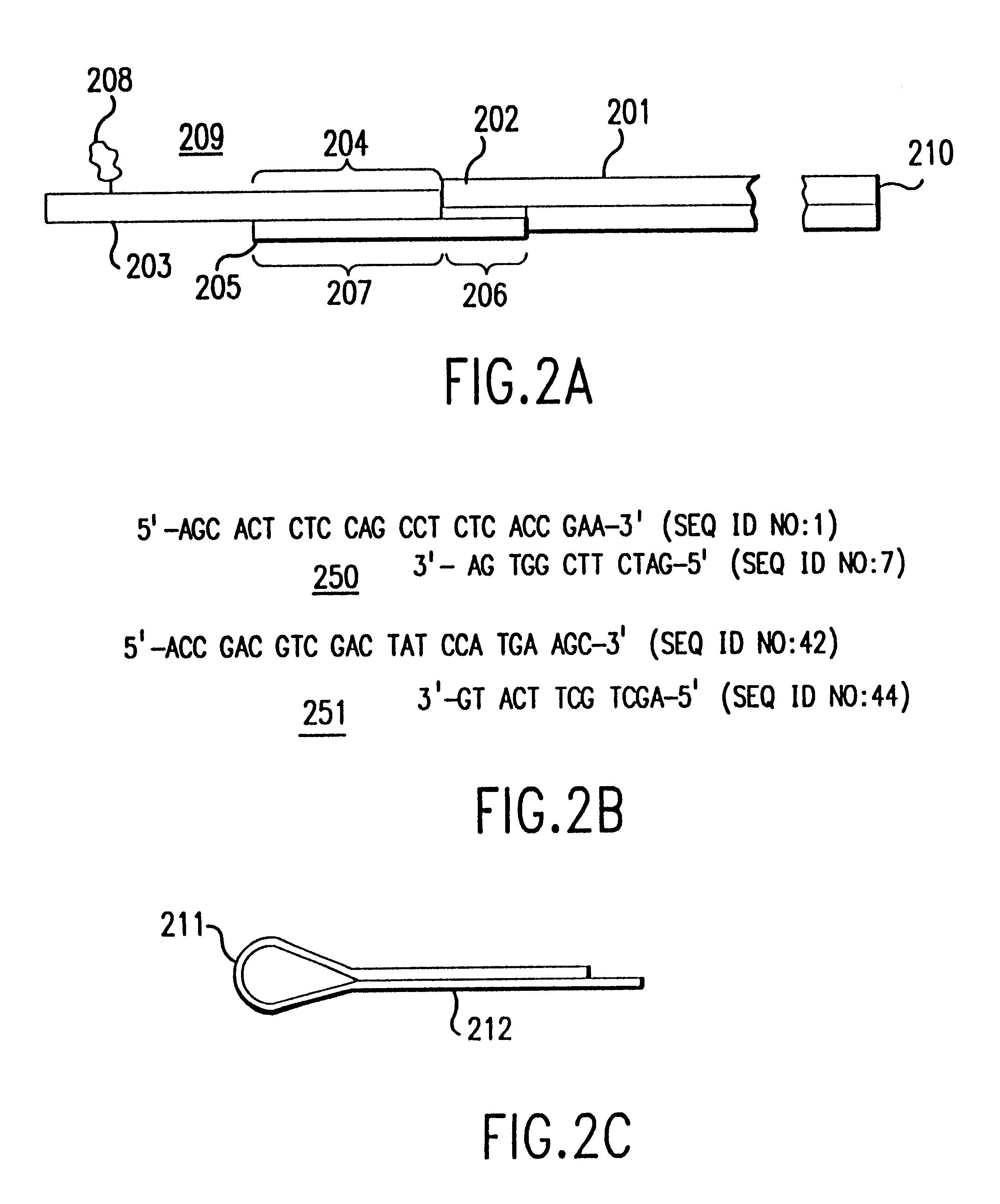
Method and apparatus for identifying classifying or quantifying DNA sequences in a sample without sequencing
InactiveUS6141657ARapid and economical and quantitative and precise determination and classificationSufficient discrimination and resolutionData processing applicationsDigital data processing detailsSequence databaseDNA Sequence Databases
This invention provides methods by which biologically derived DNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information.
Owner:CURAGEN CORP
Method and apparatus for identifying, classifying, or quantifying DNA sequences in a sample without sequencing
InactiveUS6418382B2Rapid and economical and quantitative and precise determination and classificationSufficient discrimination and resolutionData processing applicationsMicrobiological testing/measurementSample sequenceSingle sequence
This invention provides methods by which biologically derived DNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information.
Owner:CURAGEN CORP
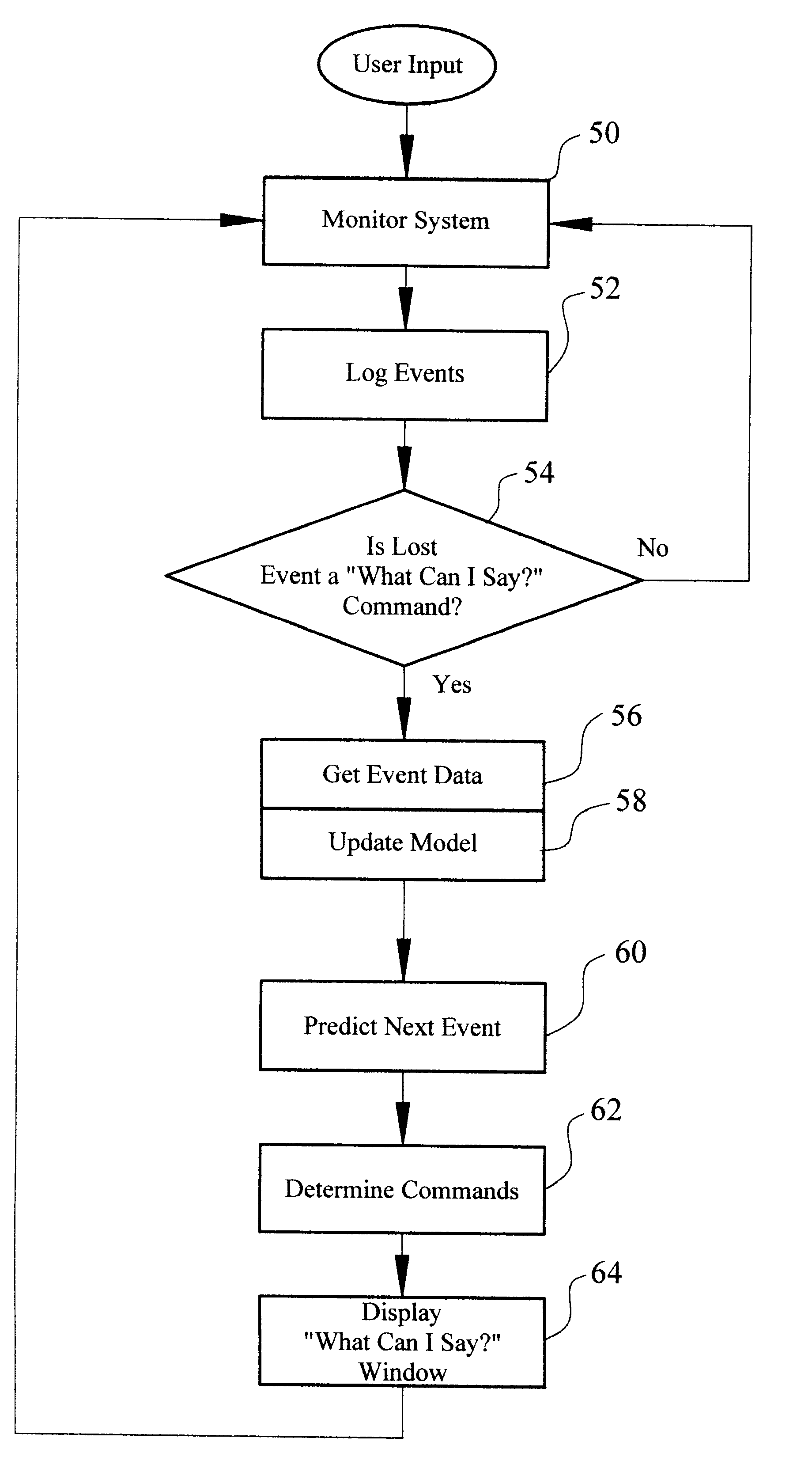
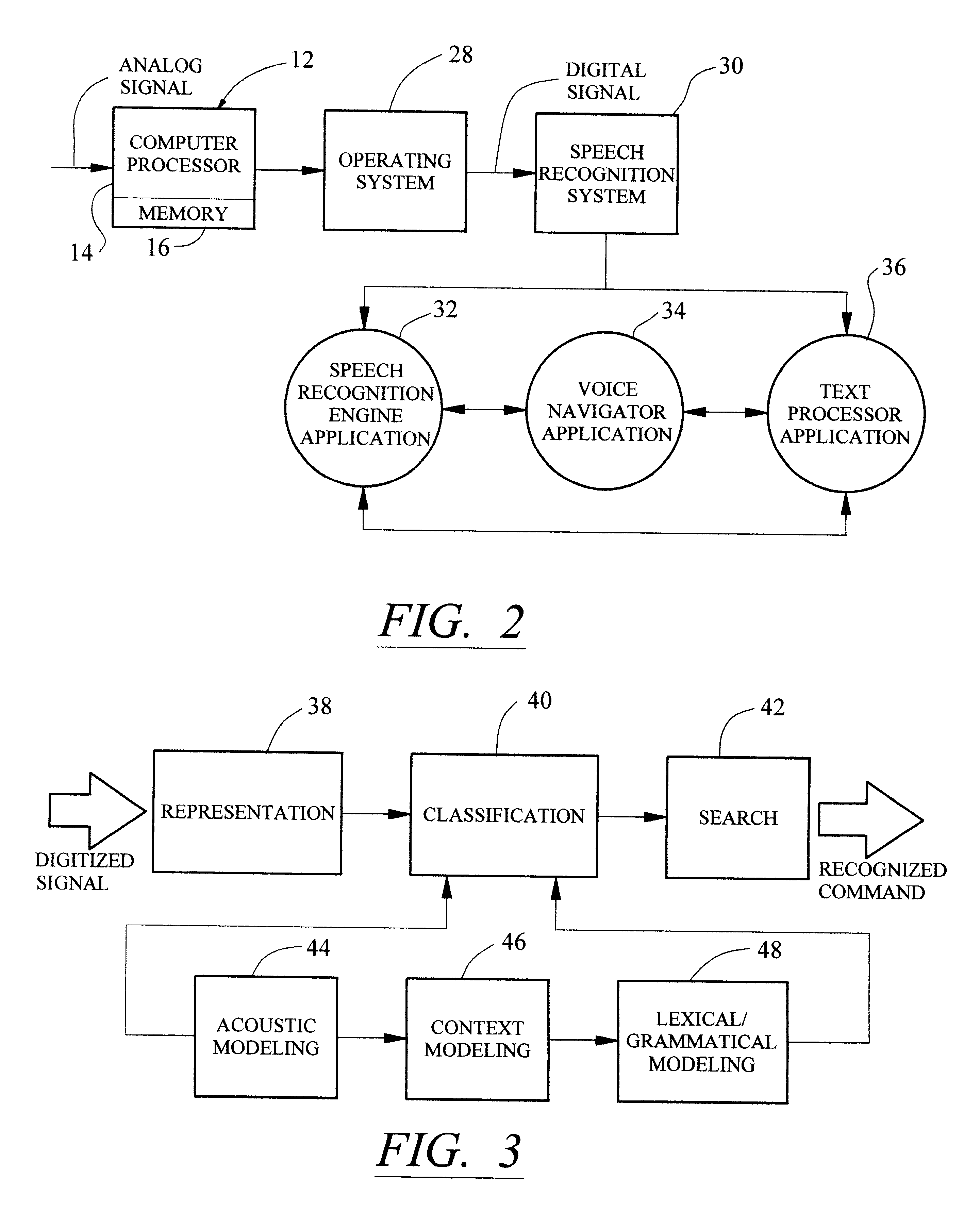
Method and apparatus for providing an event-based "What-Can-I-Say?" window
InactiveUS6308157B1Efficiently and intelligently selectingReduce in quantitySpeech recognitionStatistical analysisComputerized system
A method and system efficiently identifies voice commands for a user of a speech recognition system. The method involves a series of steps including: receiving input from a user; monitoring the computer system to log system events and ascertain a current system state; predicting a probable next event according to the current system state and logged events; and identifying acceptable voice commands to perform the next event. The system events include commands, system control activities, timed activities, and application activation. These events are statistically analyzed in light of the current system state to determine the probable next event. The voice commands for performing the probable next event are displayed to the user.
Owner:NUANCE COMM INC
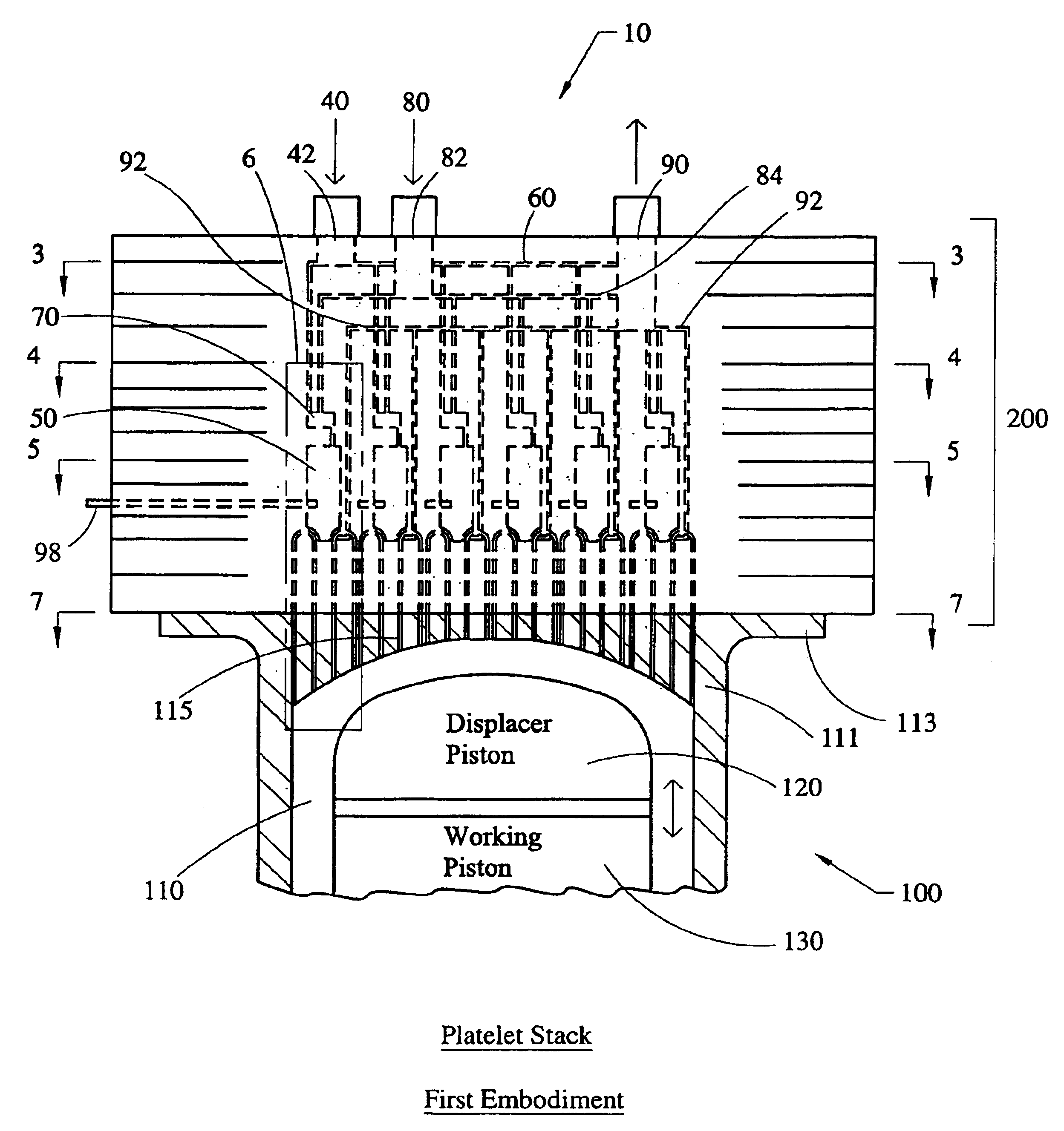
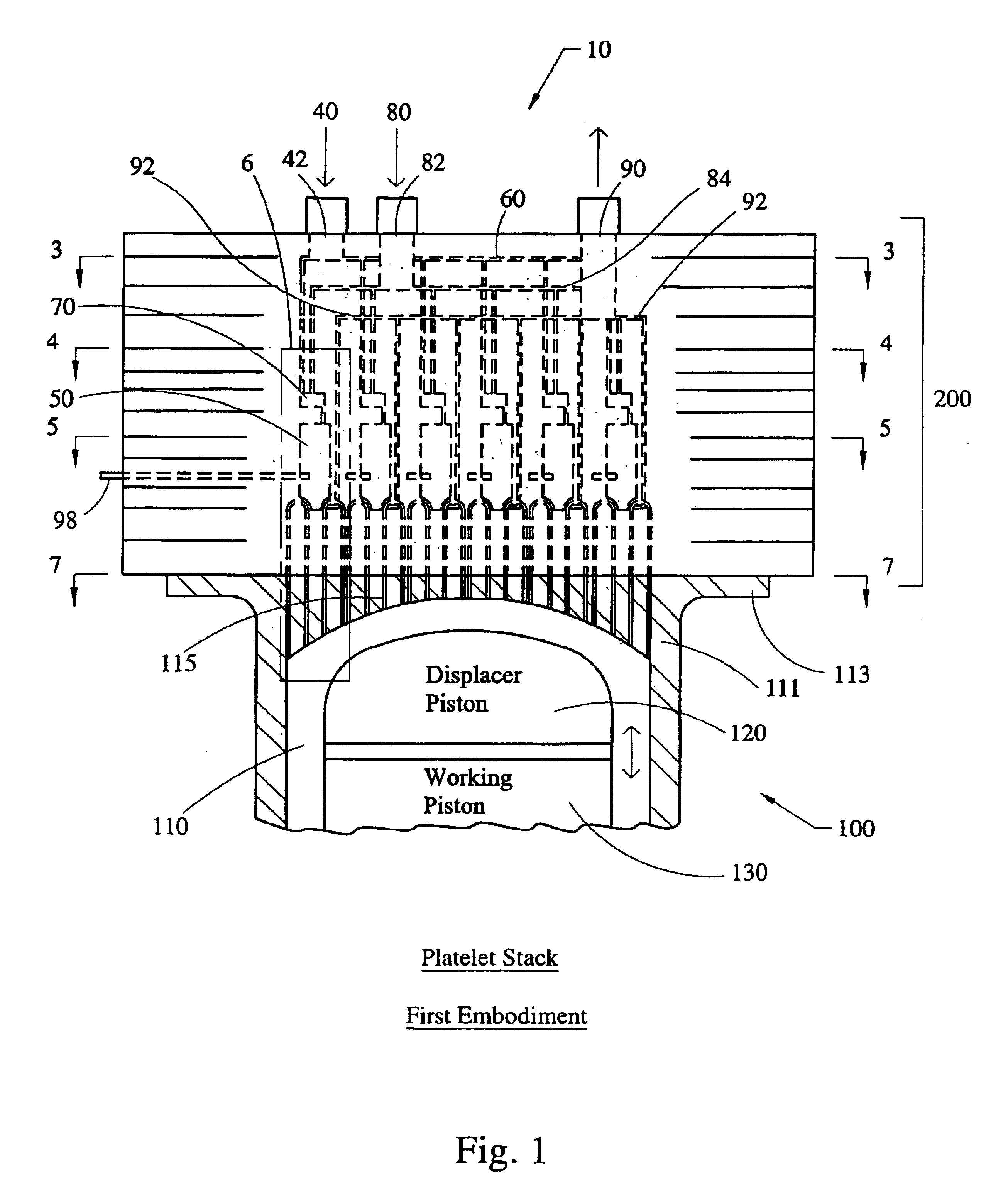
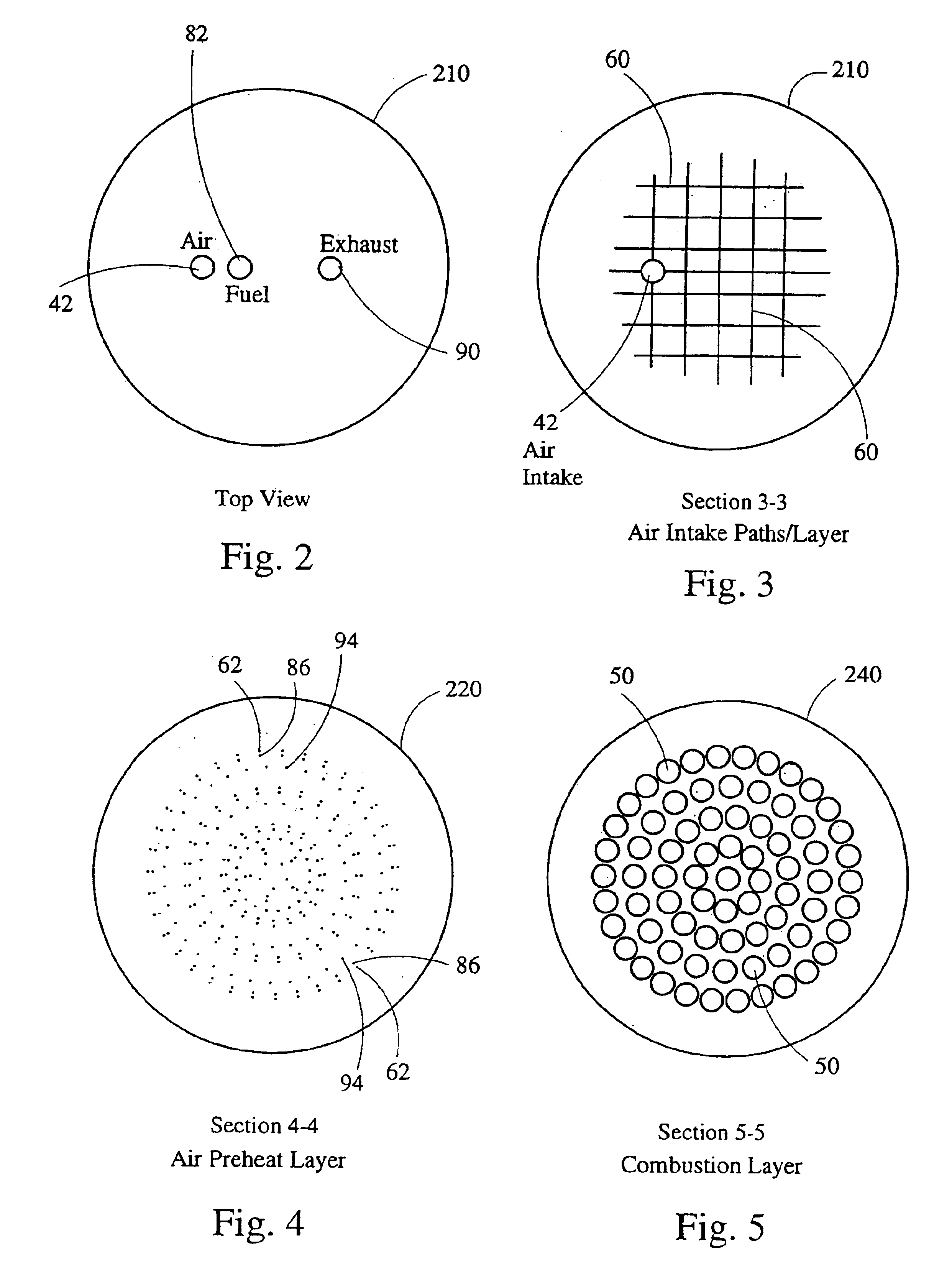
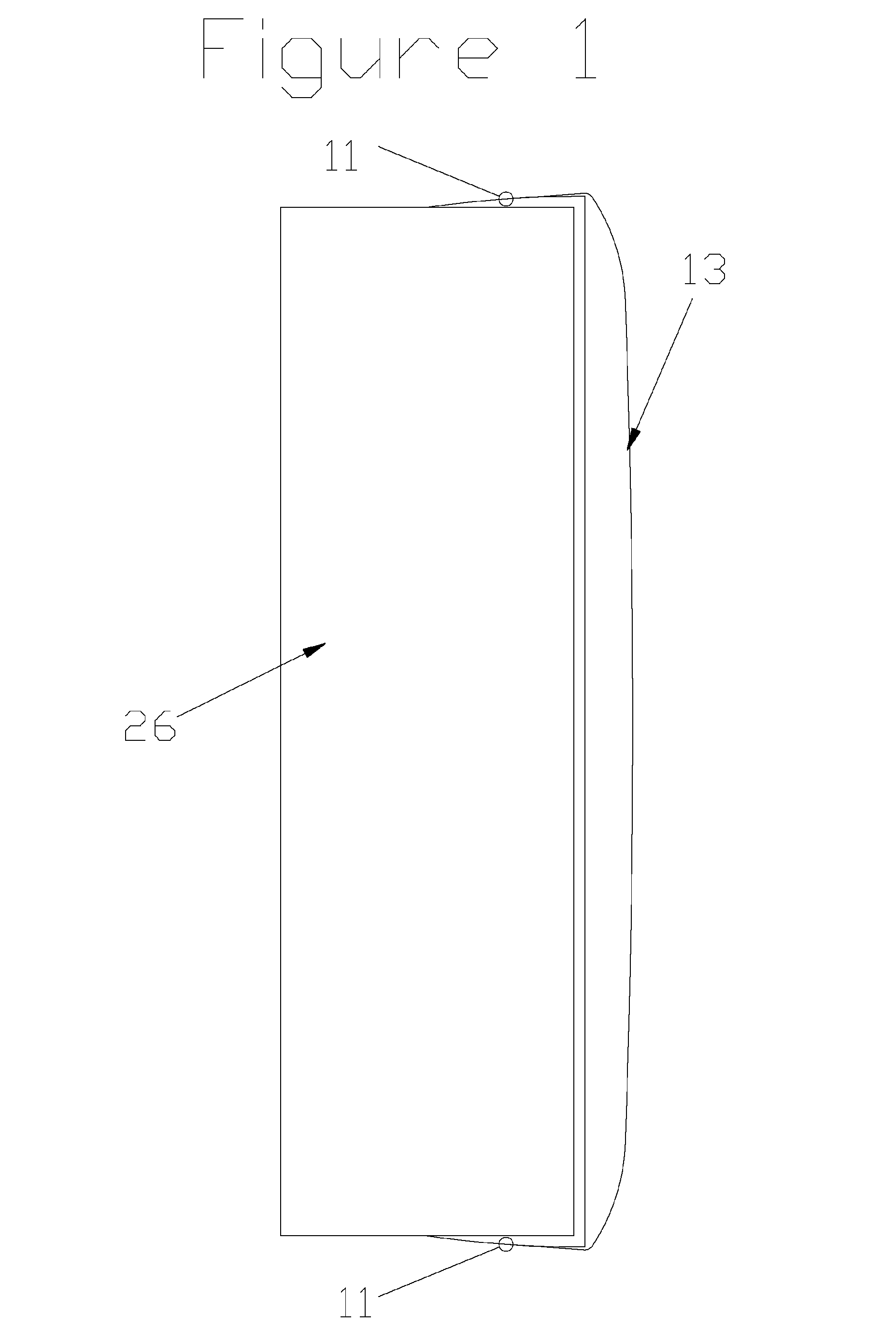
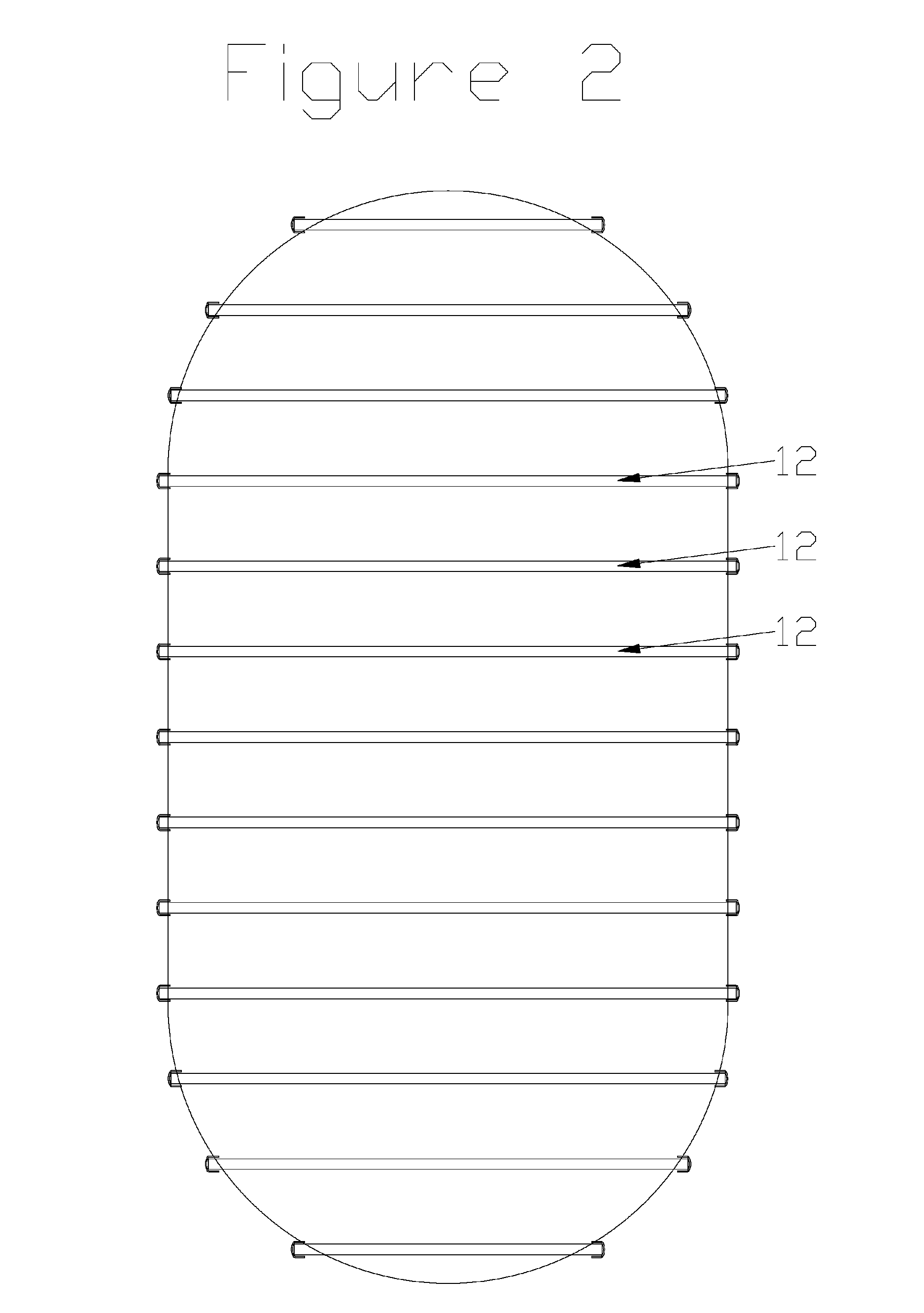
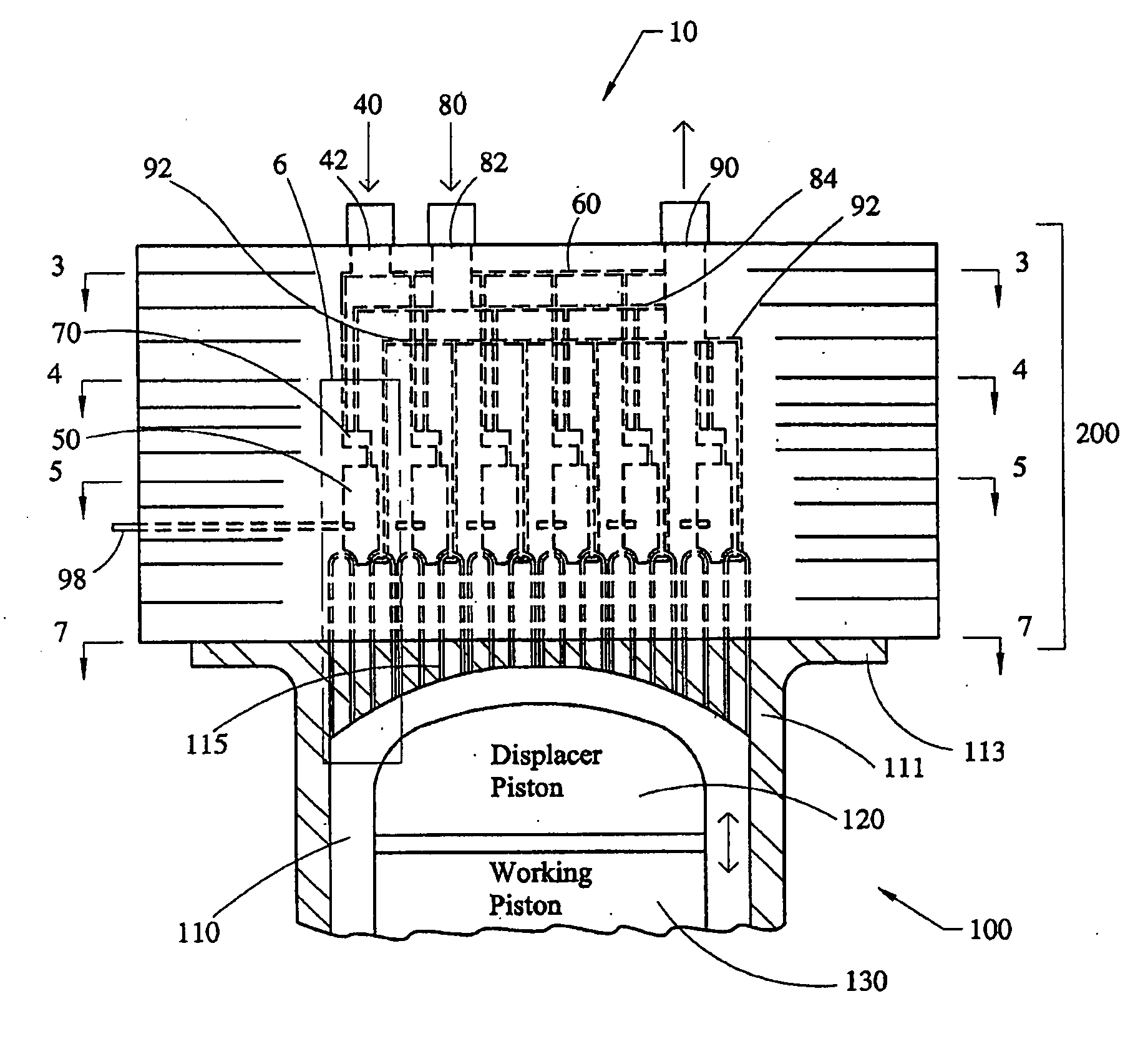
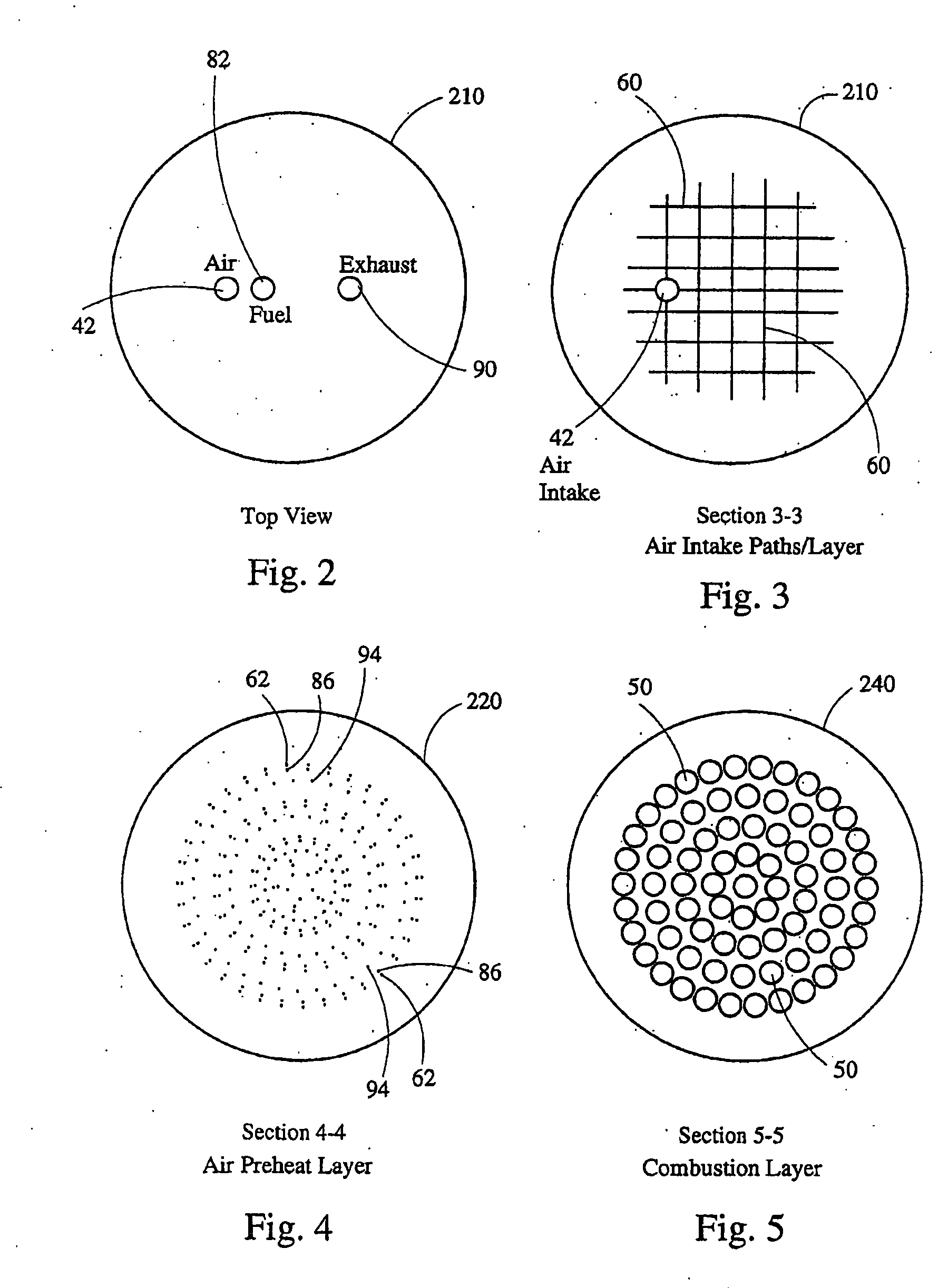
Stirling engine having platelet heat exchanging elements
InactiveUS6931848B2Guaranteed uptimeEfficient heat transferSolar heating energySolar heat devicesEngineeringStaged combustion
The present invention provides heat exchanging elements for use in Stirling engines. According to the present invention, the heat exchanging elements are made from muliple platelets that are stacked and joined together. The use of platelets to make heat exchanging elements permits Stirling engines to run more effiecient because the heat transfer and combustion processes are improved. In one embodiment, multi-stage combustion can be introduced with platlets, along with the flexibility to use different types of fuels. In another embodiment, a single component constructed from platelets can provide the heat transfer rquirements betweeen the combustion gas / working gas, working gas in the regenerator and the working gas / coolant fluid of a Stirling engine. In another embodiment, the platelet heat exchanging element can recieve solar energy to heat the Stirling engine's working gas. Also, this invention provides a heat exchanging method that allows for multiple fluids to flow in opposing or same direction.
Owner:DISENCO +1
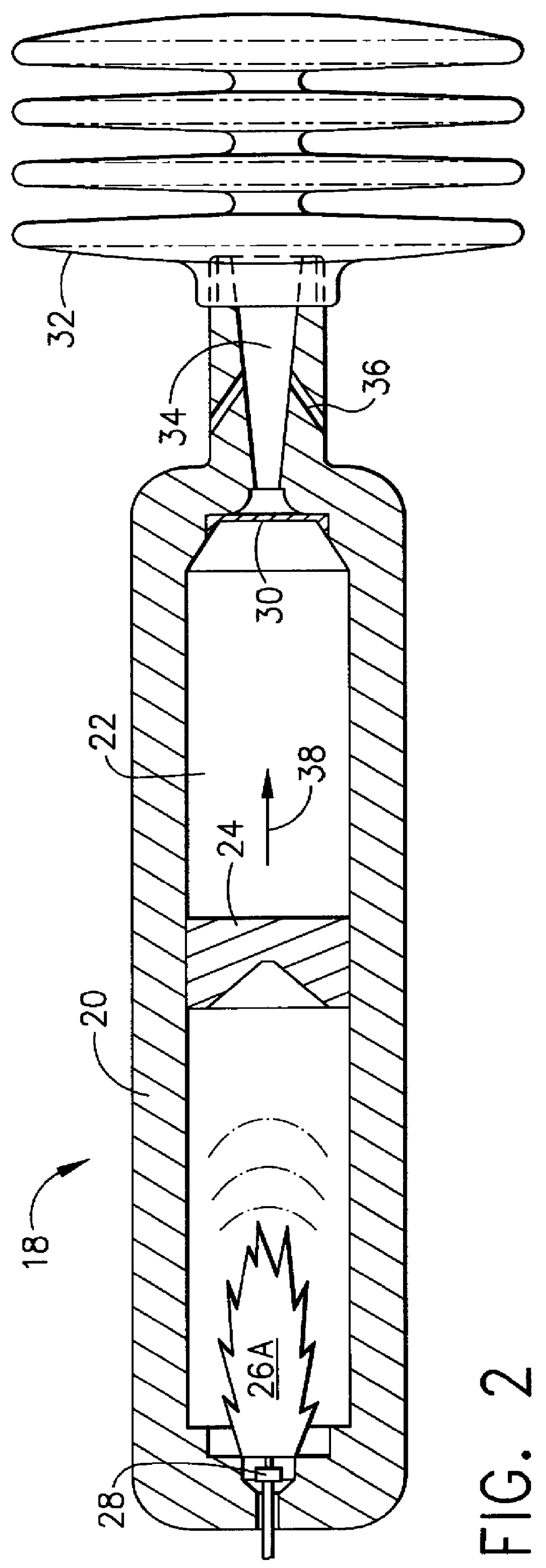
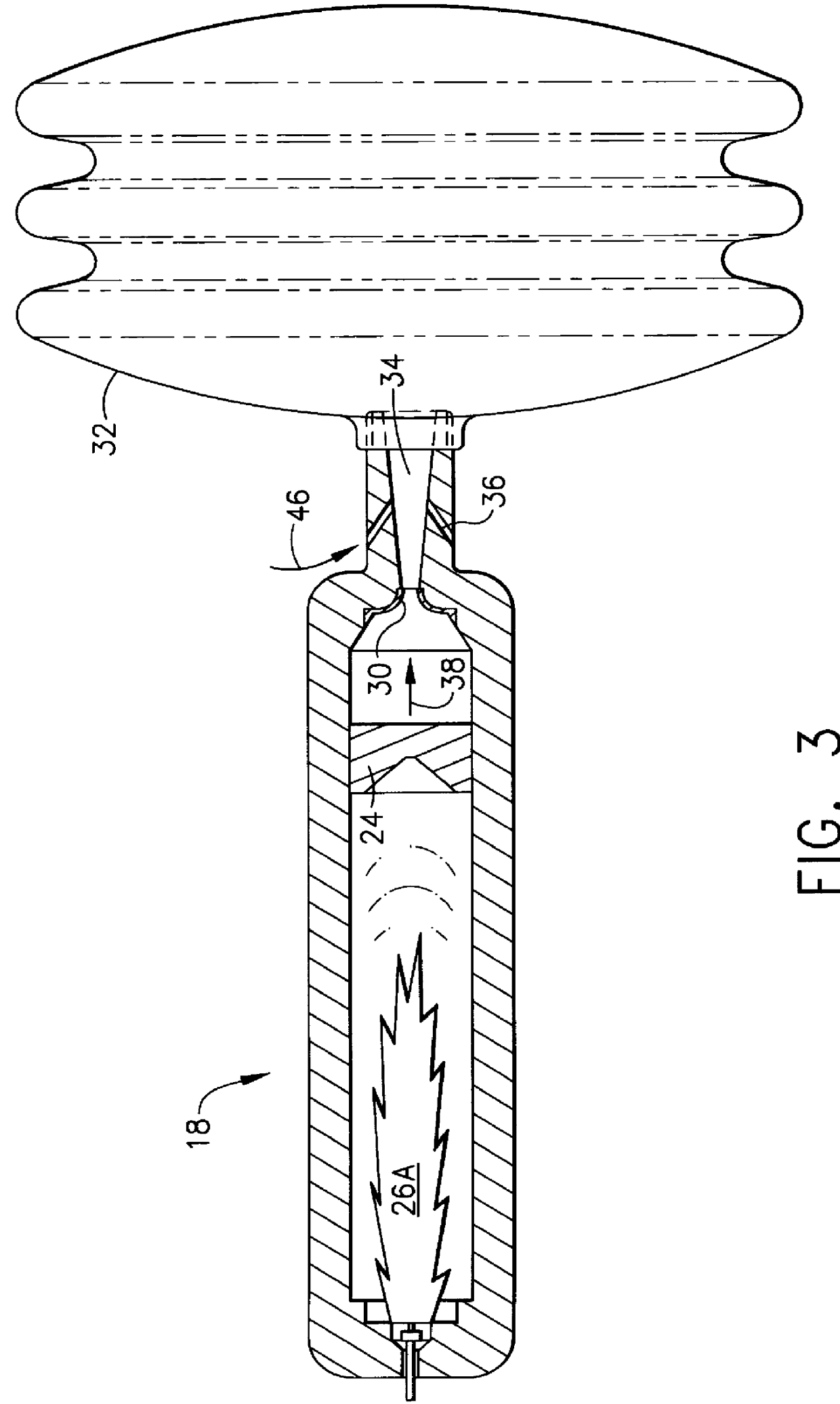
Safety air bag inflation device
InactiveUS6155600AFailure of controlSpeed up the flowPedestrian/occupant safety arrangementLiquid transferring devicesReaction rateGas passing
An air bag inflator providing a source of gas, releasable upon command, to inflate a supplemental inflation restraint (SIR) system commonly known as an automobile air bag. Provided is a pressure vessel containing one or more separate chambers for the purpose of storing gaseous fuel(s) and gaseous oxidizer(s) or liquid fuel(s) and liquid oxidizer(s) under pressure with helium as the primary filler gas. The primary function of the helium gas is to serve as a kinetic damper to modulate and control the reaction rate of the fuel(s) and oxidizer(s). Because of its low mass, high thermal conductivity and high heat capacity for its mass, helium is an excellent filler gas. In the case of the gaseous fuel(s) and oxidizer(s) a single chamber is provided. In the case of liquid fuel(s) and oxidizer(s), two or more separate housings are provided for storing the liquid fuel(s) and oxidizer(s). Along with the liquid fuel(s) and oxidizer(s) housings, two separate chambers containing pressurized helium are provided within the pressure vessel. The first helium chamber rapidly pressurizes upon initiation of a gas producing pyrotechnic igniter. This pressure acts on thin membranes on the first chamber side of the separate liquid storage housings to force the liquid fuel(s) and oxidizer(s) into the second chamber where the materials are atomized and mixed. The mixture is ignited by the arrival of hot gases from the igniter directed into the second chamber via the small diameter tube or orifice. The fuel(s) oxidizer(s) mixture burns to produce gaseous reaction products that are released into the air bag by bursting a controlled rupture burst disc.
Owner:AUTOLIV DEV AB
Fused carbon nanotube-nanocrystal heterostructures and methods of making the same
ActiveUS20060024503A1Easy to controlGood physical propertiesMaterial nanotechnologyPolycrystalline material growthNanocrystalOxygen
The present invention provides a fused heterostructure comprising a carbon nanotube and at least one nanocrystal, wherein the nanocrystal is attached by a covalent linkage to at least one oxygen moiety on the nanotube, and wherein there is no intermediary linker between the nanotube and the nanocrystal.
Owner:THE RES FOUND OF STATE UNIV OF NEW YORK
Method of fabricating a multilayer dielectric tunnel barrier structure
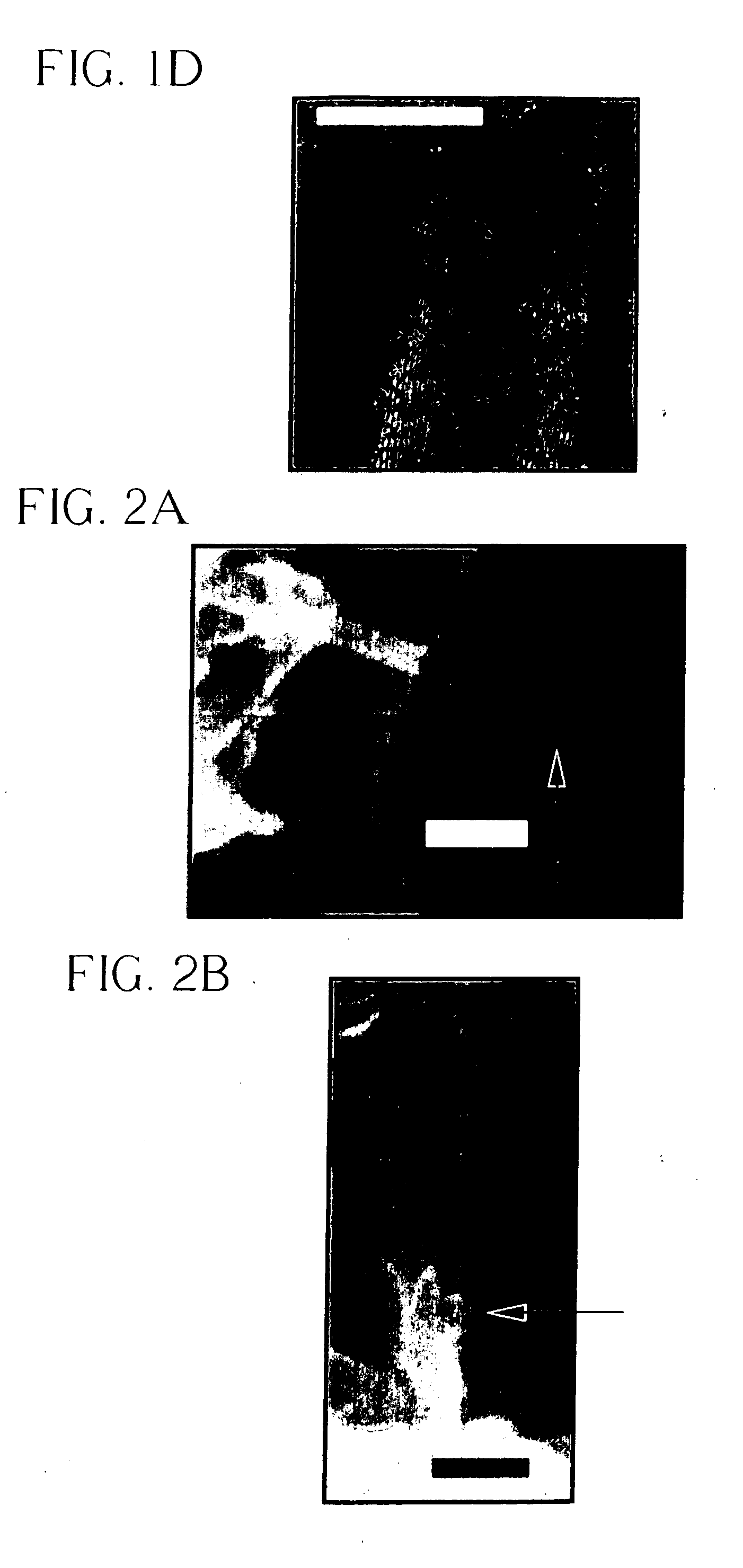
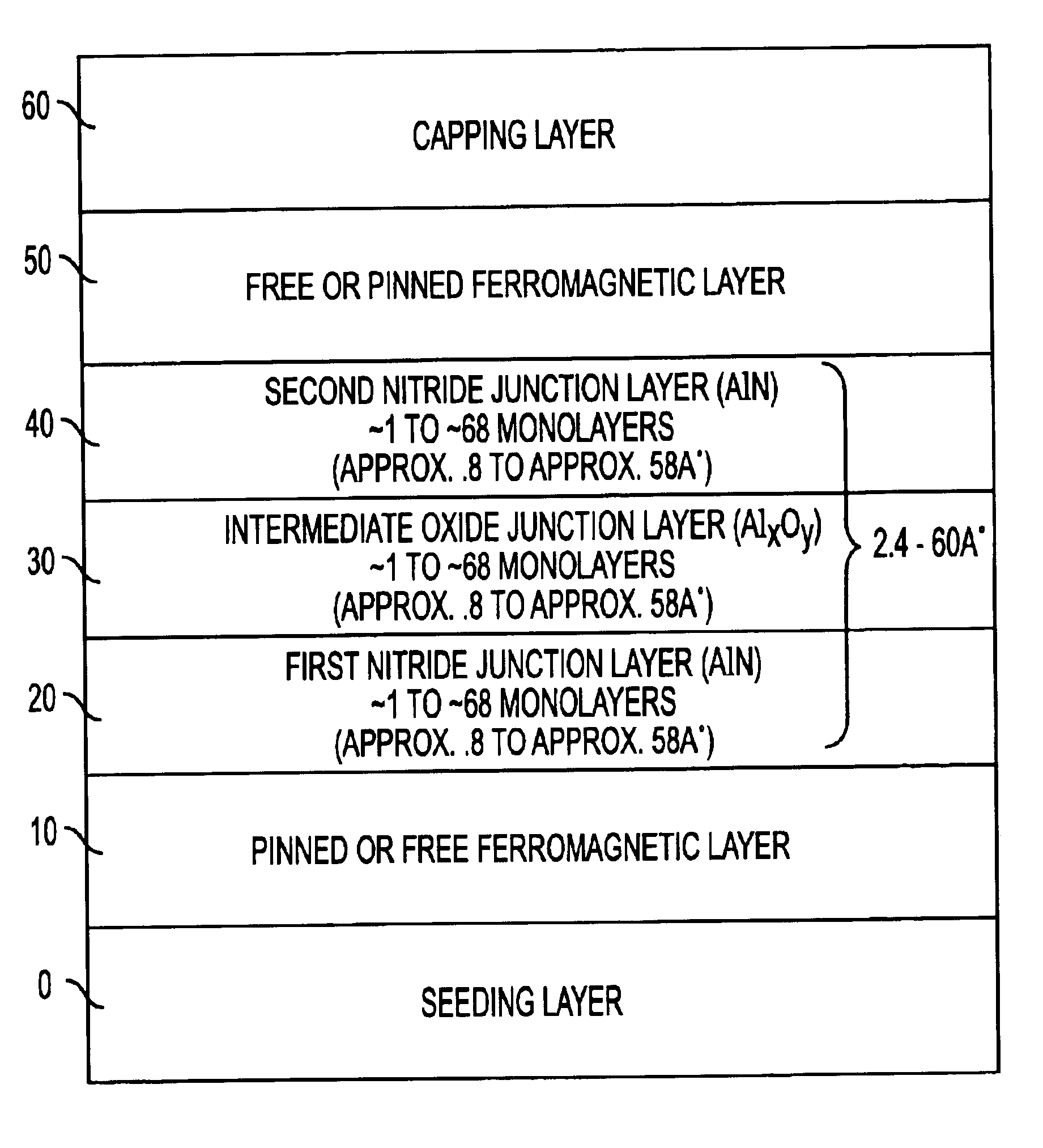
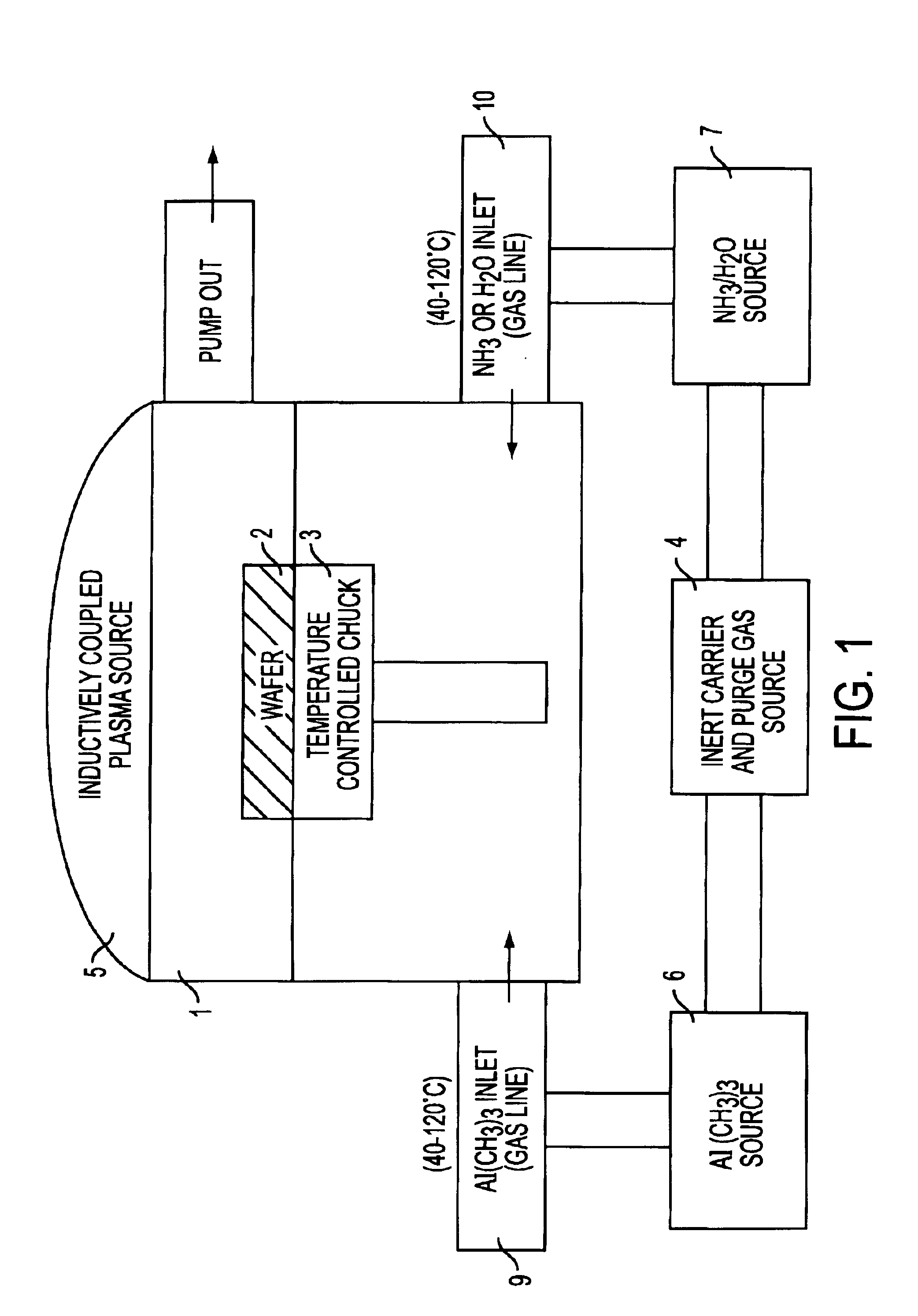
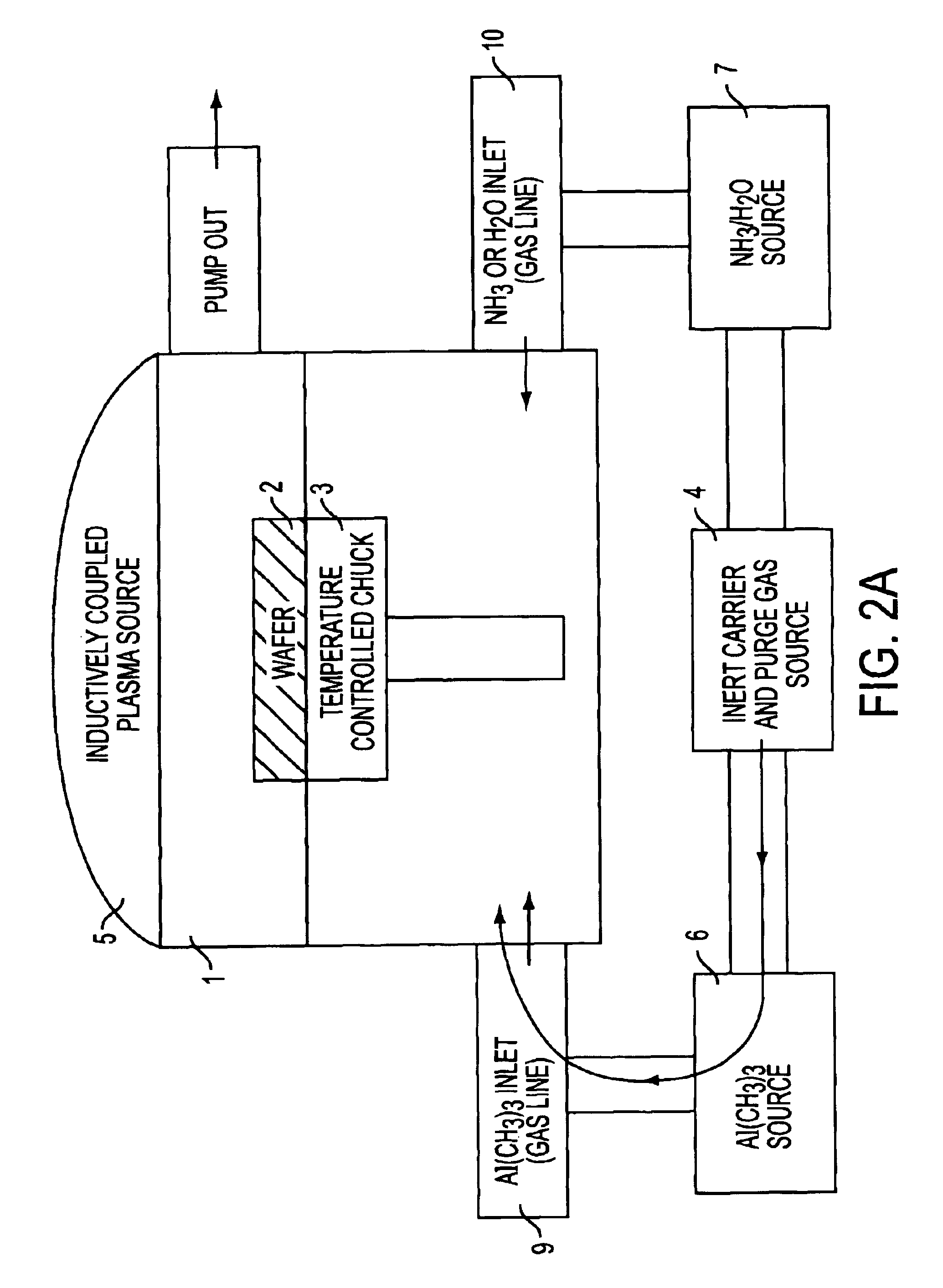
InactiveUS6849464B2Reduce resistanceGood shape retentionNanomagnetismChemical vapor deposition applicationMagnetic memoryNitride
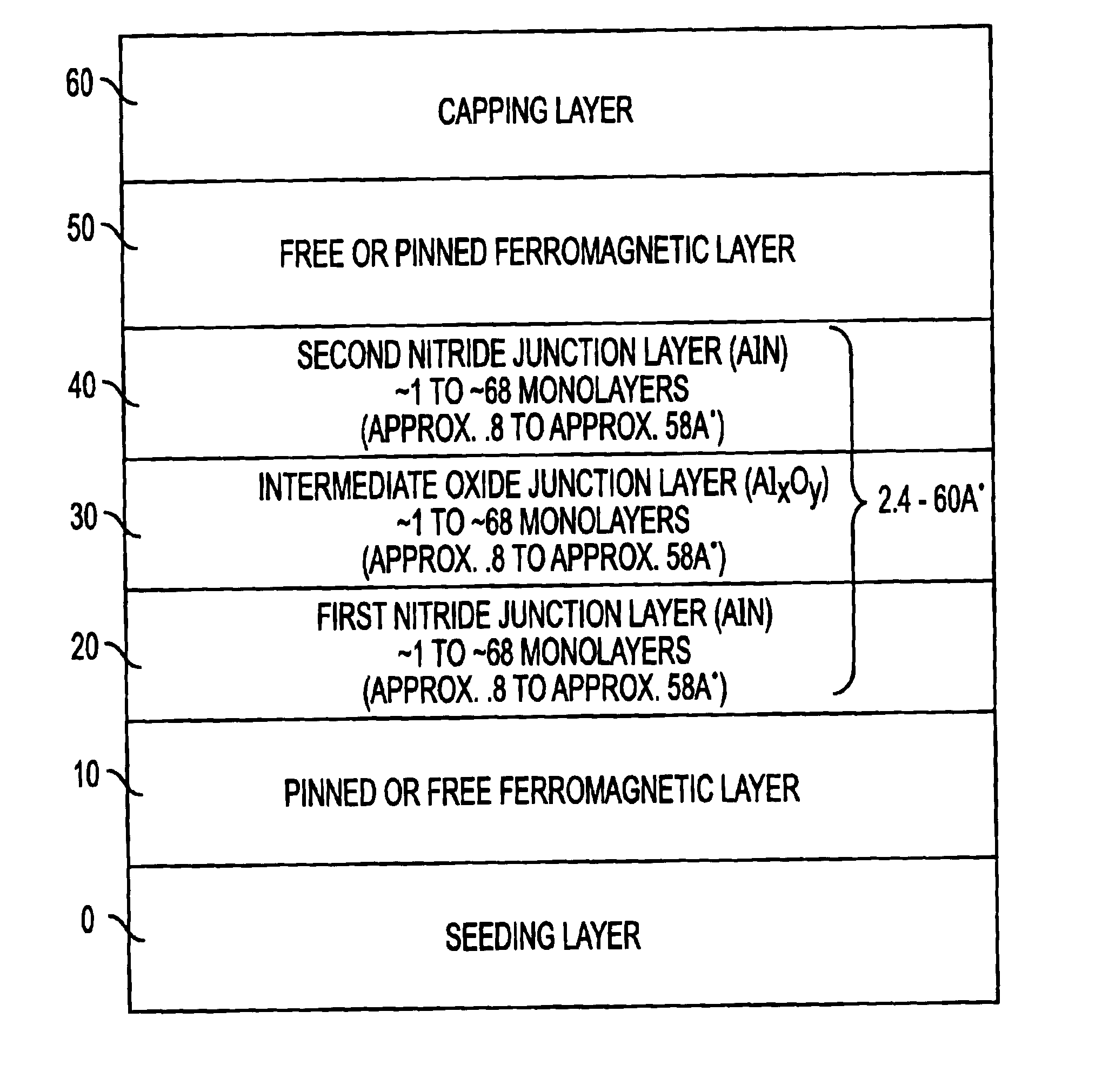
A multilayer dielectric tunnel barrier structure and a method for its formation which may be used in non-volatile magnetic memory elements comprises an ALD deposited first nitride junction layer formed from one or more nitride monolayers i.e., AlN, an ALD deposited intermediate oxide junction layer formed from one or more oxide monolayers i.e., AlxOy, disposed on the first nitride junction layer, and an ALD deposited second nitride junction layer formed from one or more nitride monolayers i.e., AlN, disposed on top of the intermediate oxide junction layer. The multilayer tunnel barrier structure is formed by using atomic layer deposition techniques to provide improved tunneling characteristics while also providing anatomically smooth barrier interfaces.
Owner:MICRON TECH INC
Multilayer dielectric tunnel barrier used in magnetic tunnel junction devices, and its method of fabrication
InactiveUS6900455B2Reduce resistanceGood shape retentionNanomagnetismChemical vapor deposition applicationDielectricMagnetic memory
Owner:MICRON TECH INC
Trailing arm suspension with optimized i-beam
InactiveUS20050051986A1Economizing costEconomizing weightRigid suspensionsResilient suspensionsTrailing armEngineering
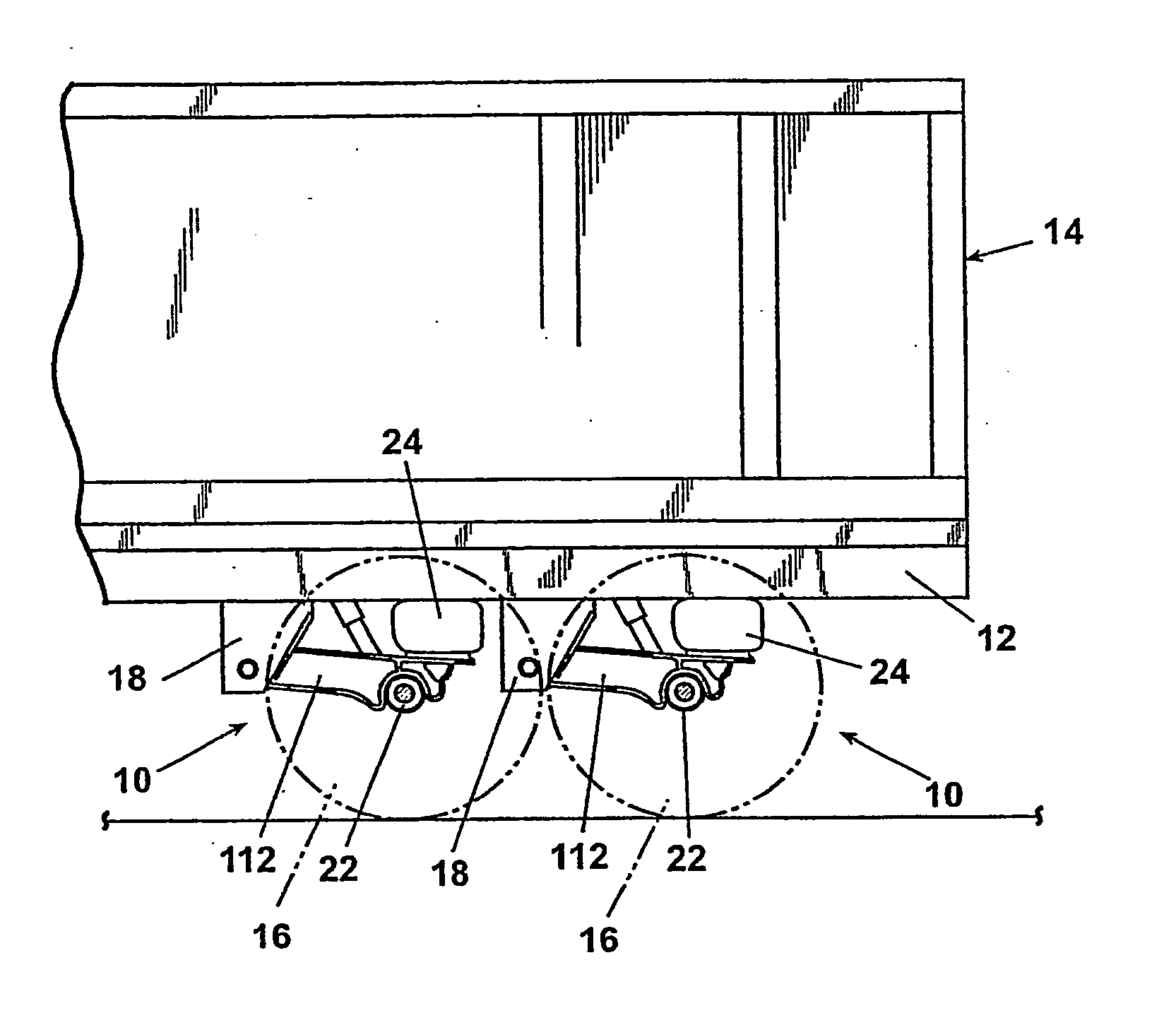
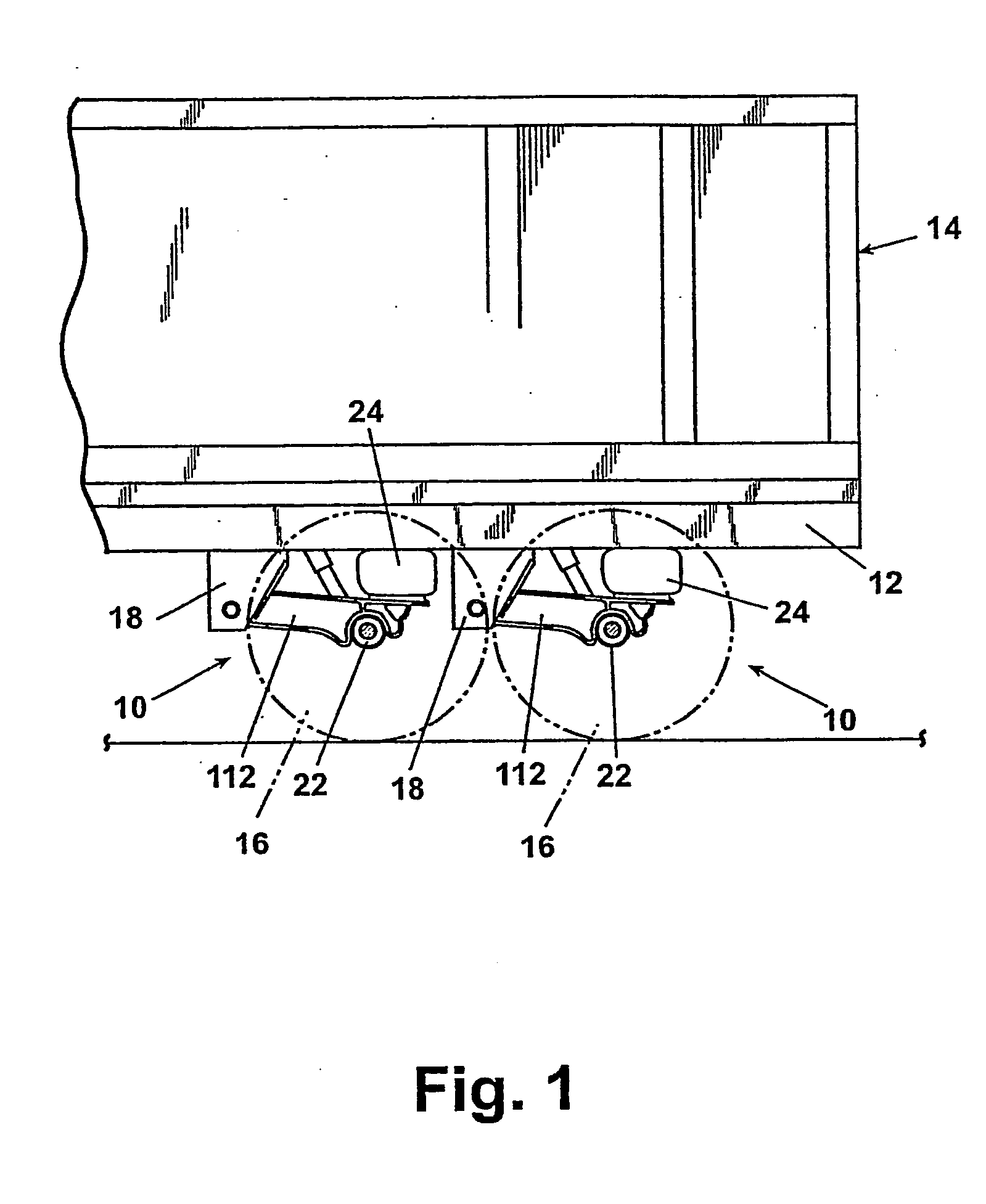
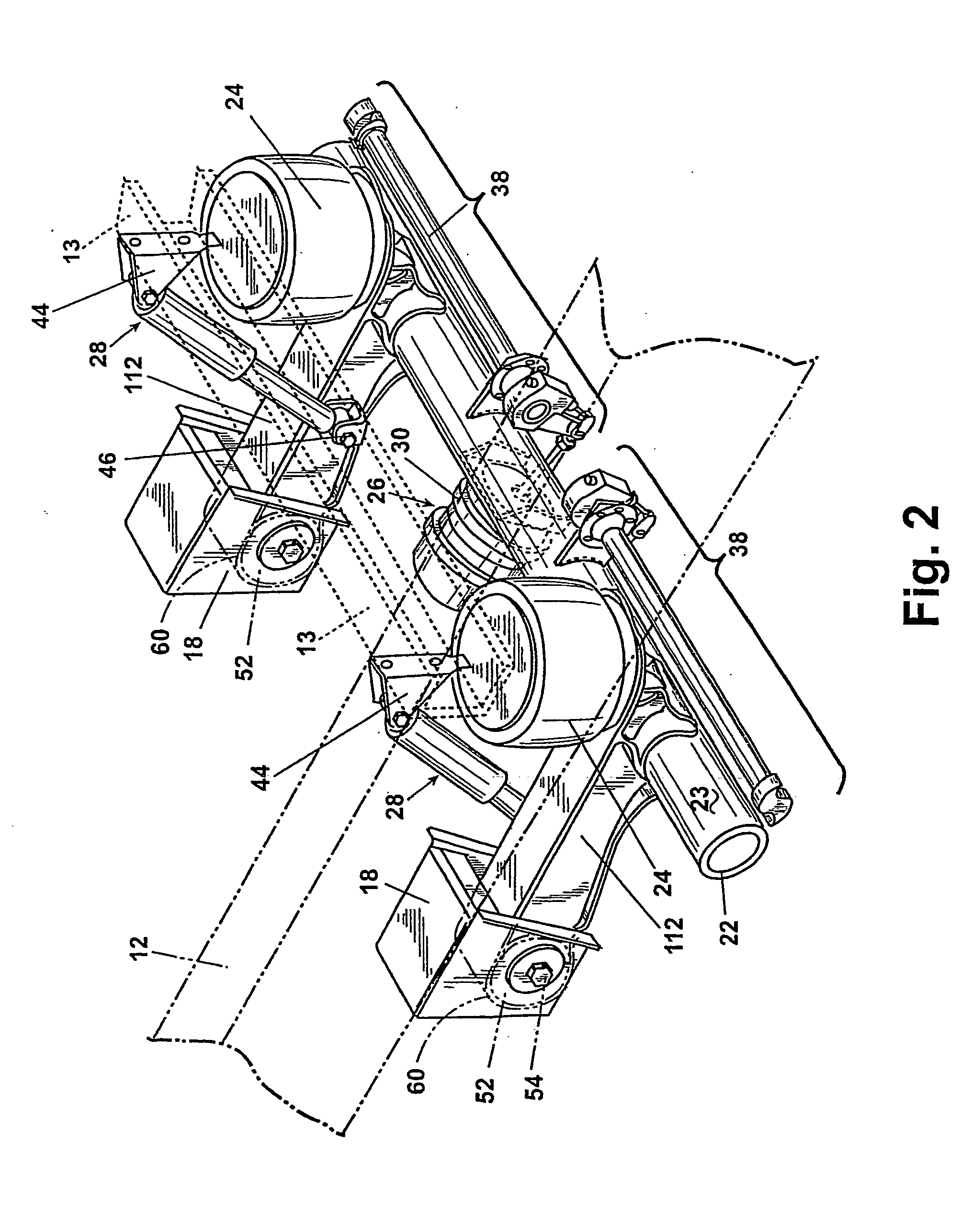
A suspension system includes a pair of trailing arms (112) extending between frame brackets (18) and a wheel-carrying axle (22). Each trailing arm (112) comrises an I-beam portion having a web section and first and second flanges, wherein the thickness of at least one of the flanges vaires along a length thereof. The trailing arms (112) are welded directly to the axle (22).
Owner:THE HOLLAND GROUP
System and method of determining a center of mass of an imaging subject for x-ray flux management control
InactiveUS20050089138A1Reduce the valueOptimize radiation exposureMaterial analysis using wave/particle radiationRadiation/particle handlingNuclear medicineUltrasound attenuation
A system and method of diagnostic imaging is provided that includes positioning a subject in a scanning bay, comparing a center of mass of the subject to a reference point, and repositioning the subject in the scanning bay to reduce a difference in position between the center of mass of the subject and the reference point. The present invention automatically selects a proper attenuation filter configuration, corrects patient centering, and corrects noise prediction errors, thereby increasing dose efficiency and tube output.
Owner:GENERAL ELECTRIC CO
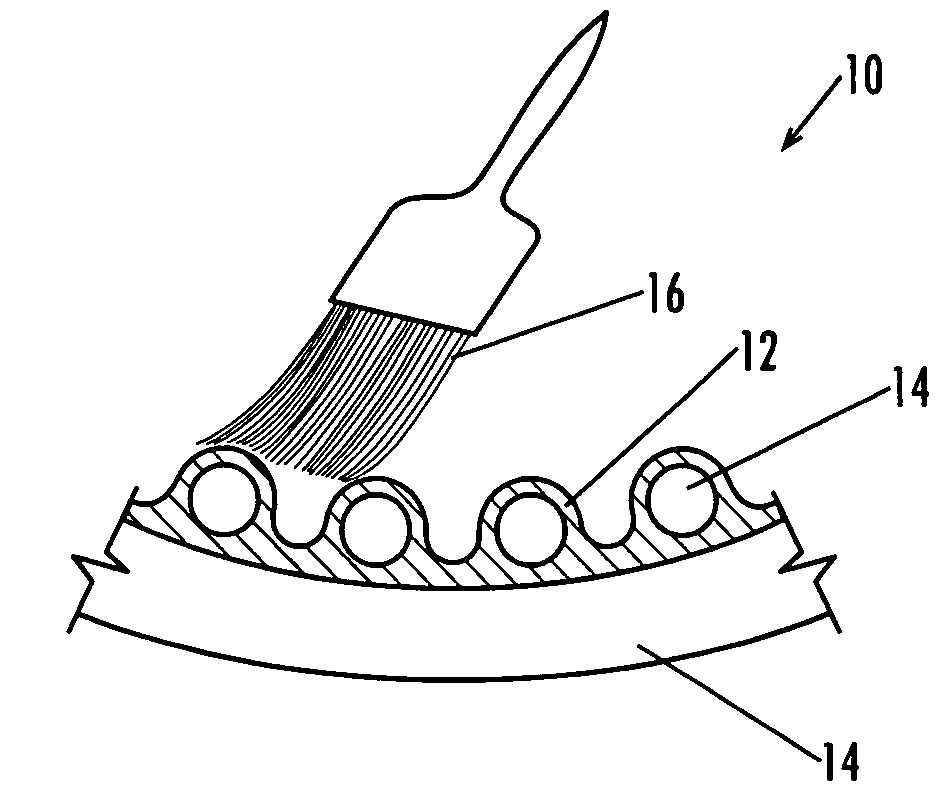
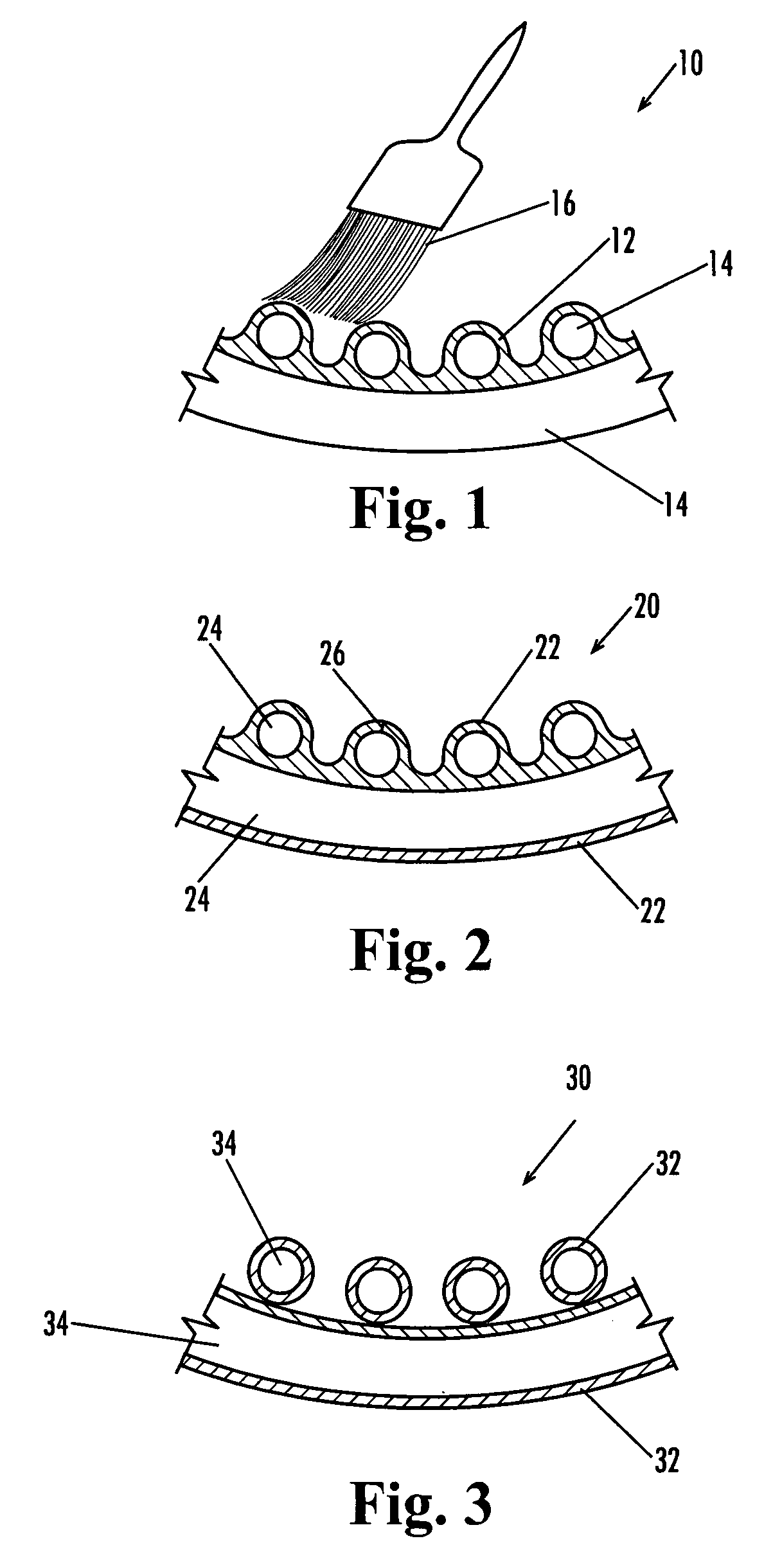
Lacrosse net
InactiveUS20080146385A1Improve adhesionImprove integrityThrow gamesCoatingsElastomerThermoplastic elastomer
A modified form of a lacrosse net provides increased adherence, integrity, and “give” as a result of a coating applied to the fibers. In one form, the fibers are manually coated by brushing them with a natural latex mixture followed by evaporation of the volatile components, which gives the coated fibers a tacky and energy absorbent nature. The soft nature of the coating may also aid in dissipating energy during impact with the ball, and this could contribute to the sense of increased “feel.” There are many natural and synthetic polymers that might be capable of delivering similar benefits, including conventional synthetic elastomers, e.g., butyl rubber. Thermoplastic elastomers are a growing field of interest in coating technologies, and with the appropriate tuning, they can be made with all of the same properties as the natural rubber coating. A brush-on method delivers the desired coating thickness and properties, but other methods, such as dip coating and spray coating, may increase feasibility for scaling-up the process. In one arrangement, the entire fiber is composed of a rubbery material. The resulting properties may not be exactly what is desired, however, because the underlying integrity of the nylon fiber would be absent, and this might result in netting with a “springy” feel.
Owner:LUNDBERG STEPHEN F
Fused carbon nanotube-nanocrystal heterostructures and methods of making the same
ActiveUS7189455B2Easy to controlGood physical propertiesMaterial nanotechnologyPolycrystalline material growthOxygenNanocrystal
The present invention provides a fused heterostructure comprising a carbon nanotube and at least one nanocrystal, wherein the nanocrystal is attached by a covalent linkage to at least one oxygen moiety on the nanotube, and wherein there is no intermediary linker between the nanotube and the nanocrystal.
Owner:THE RES FOUND OF STATE UNIV OF NEW YORK
Process to create buried heavy metal at selected depth
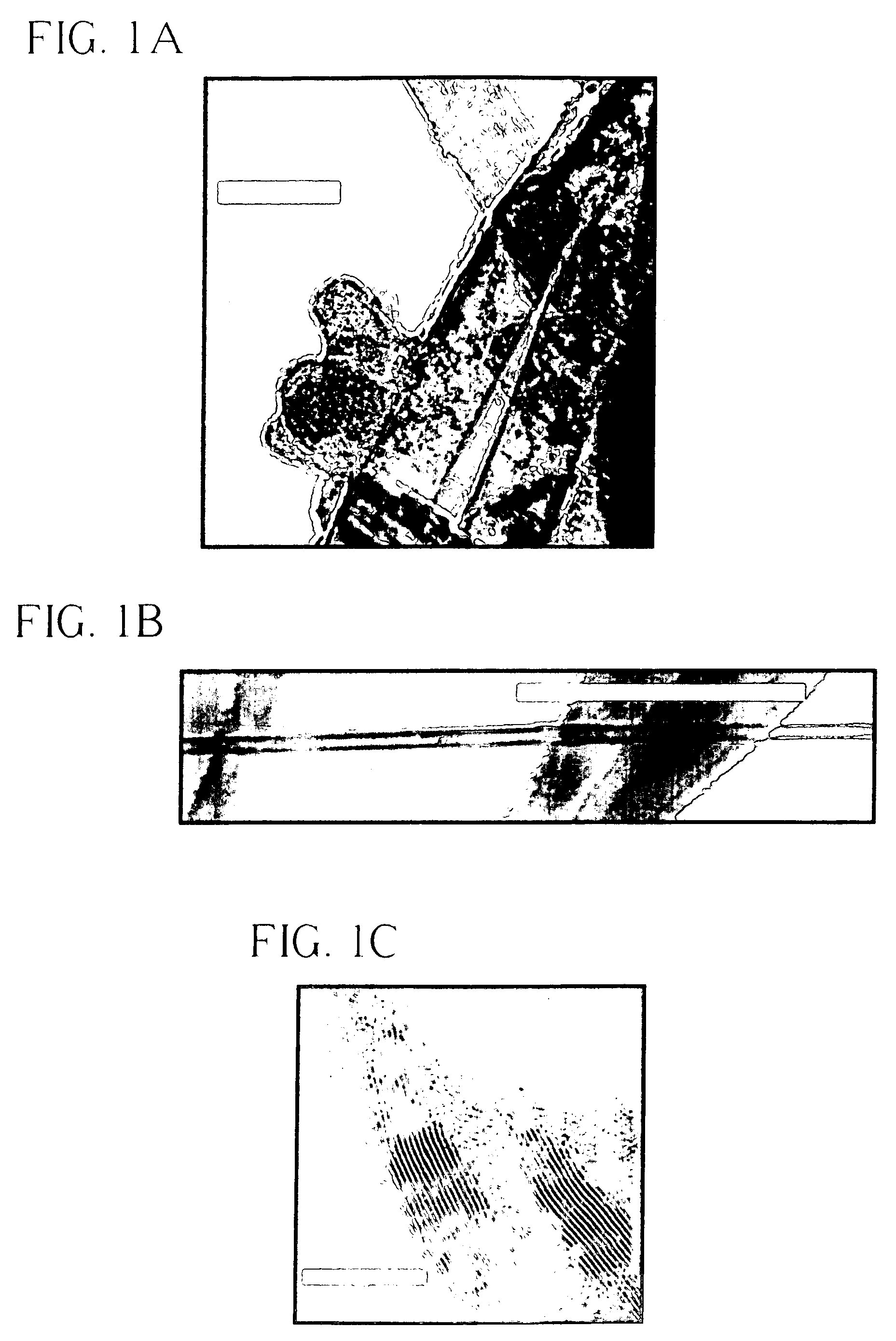
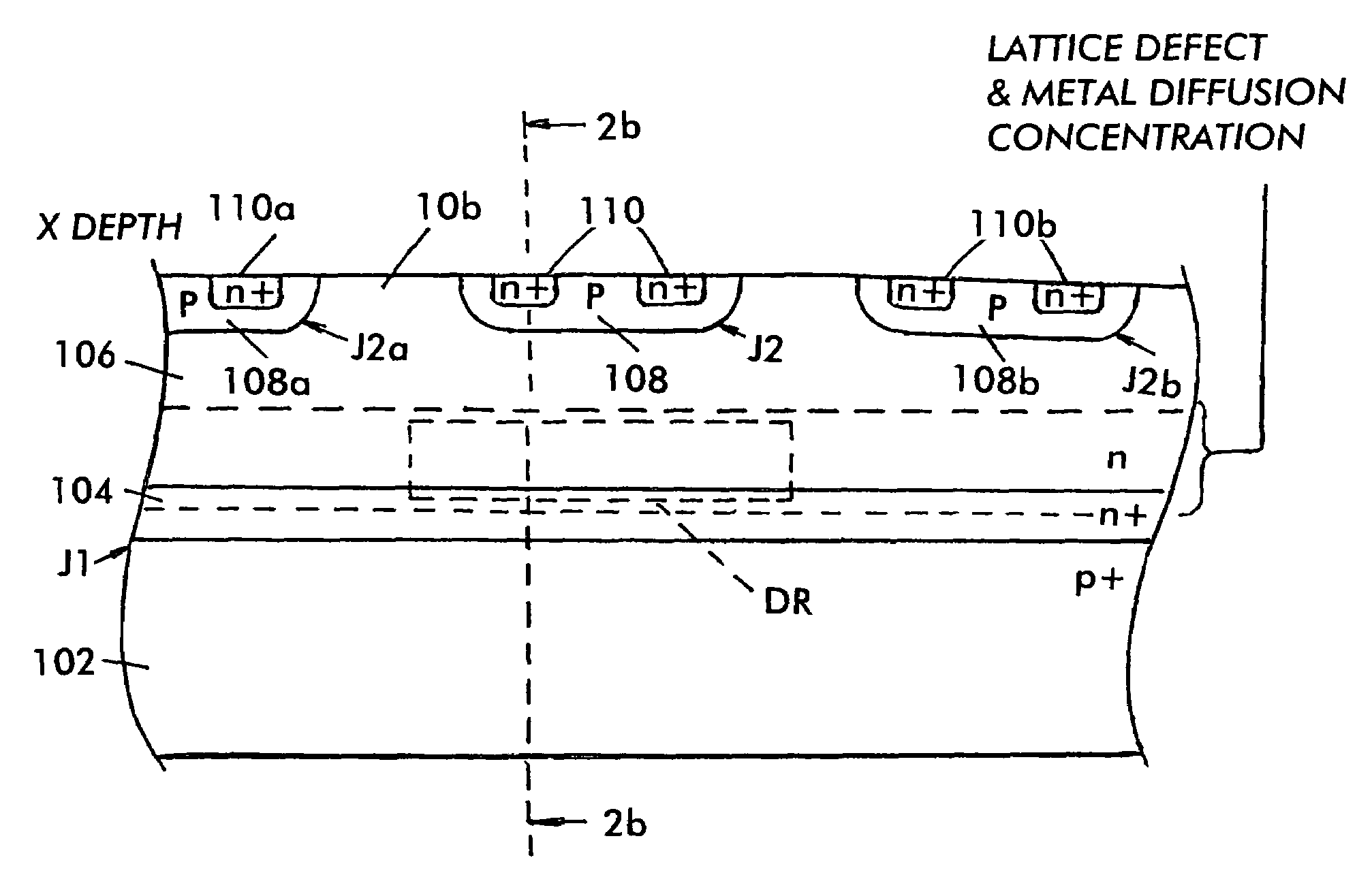
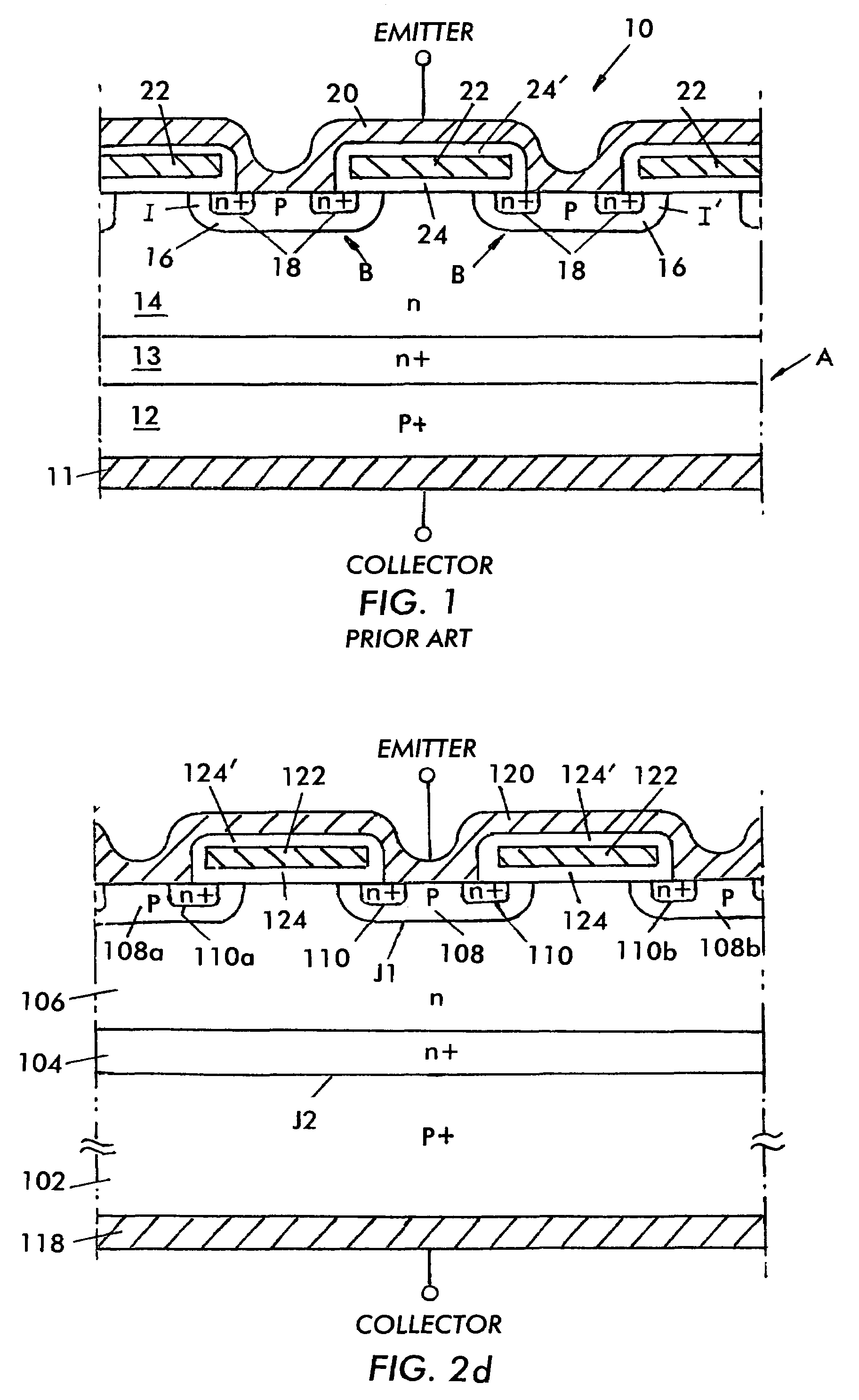
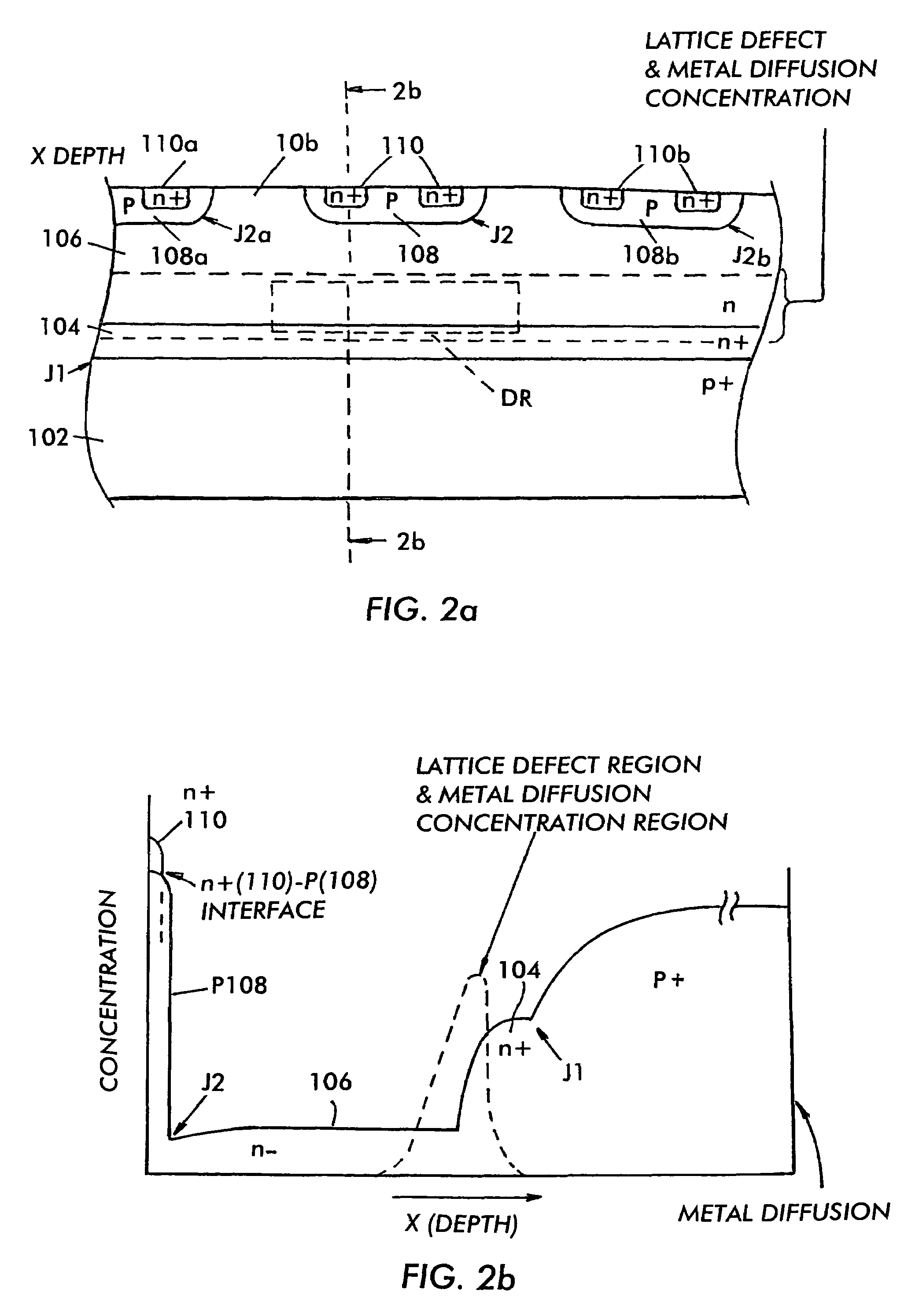
InactiveUS7485920B2Reduced carrier lifetimeReduce disadvantagesTransistorThyristorLattice defectsParticle beam
Semiconductor devices having recombination centers comprised of well-positioned heavy metals. At least one lattice defect region within the semiconductor device is first created using particle beam implantation. Use of particle beam implantation positions the lattice defect region(s) with high accuracy in the semiconductor device. A heavy metal implantation treatment of the device is applied. The lattice defects created by the particle beam implantation act as gettering sites for the heavy metal implantation. Thus, after the creation of lattice defects and heavy metal diffusion, the heavy metal atoms are concentrated in the well-positioned lattice defect region(s).
Owner:INFINEON TECH AMERICAS CORP
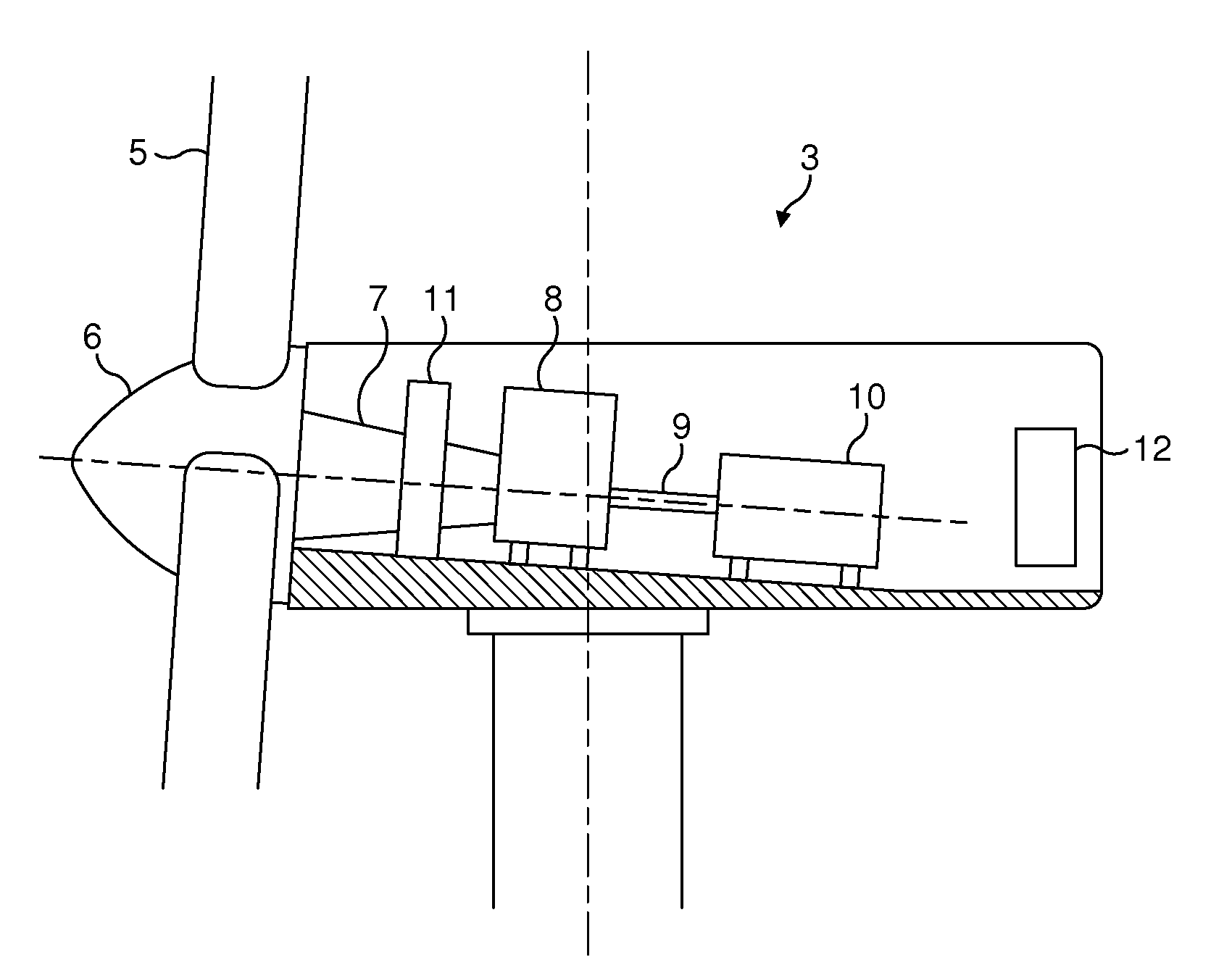
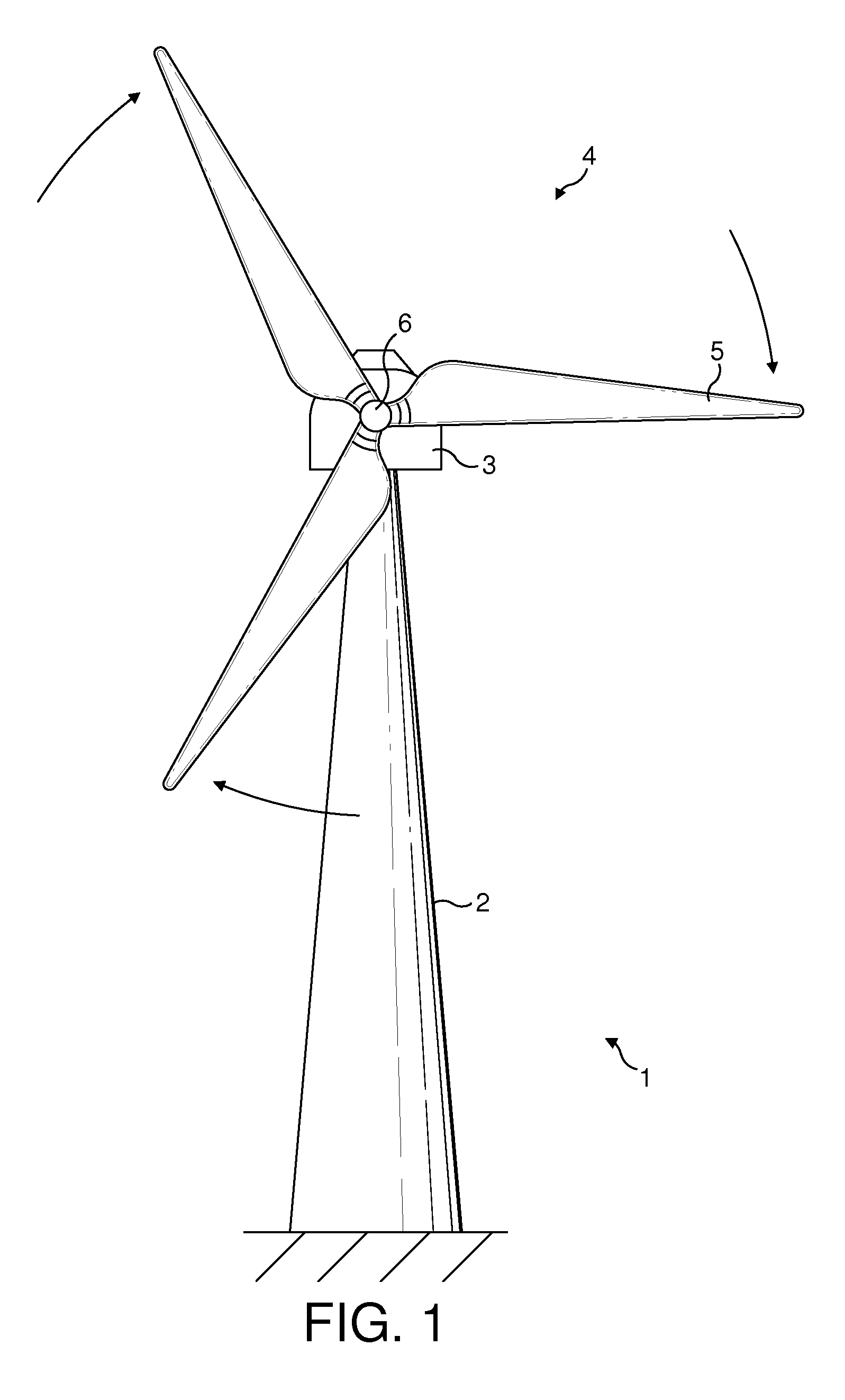
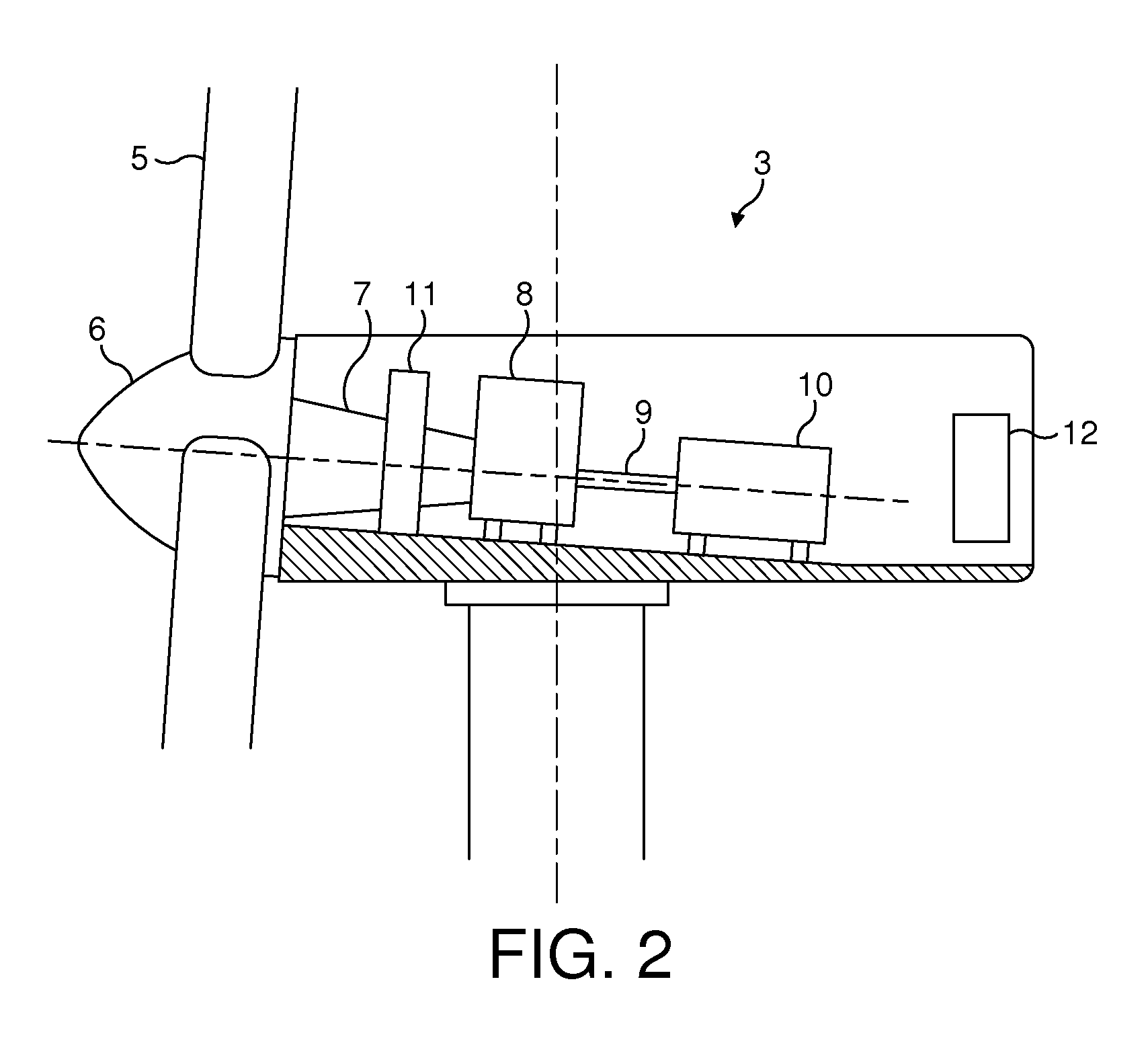
Wind energy power plant equipped with an optical vibration sensor
ActiveUS20130078095A1Reduce spacingImproved electricity generation regimePropellersWind motor controlPower stationWind power
A wind energy power plant optical vibration sensor is described, using two light sources 15, 16 that emit light at different respective frequencies. The light from the first light source falls on a surface 44 of the wind energy power plant at a detection site. Movements in the surface result in changes to the phase of the light reflected back from the surface which can be detected by mixing the first light with the light emitted from the second light source. The difference in frequencies between the two light sources results in a beating of the resulting interference signal, whereas movements in the sensor surface result in changes in the phase timing and frequency of the beats.
Owner:VESTAS WIND SYST AS
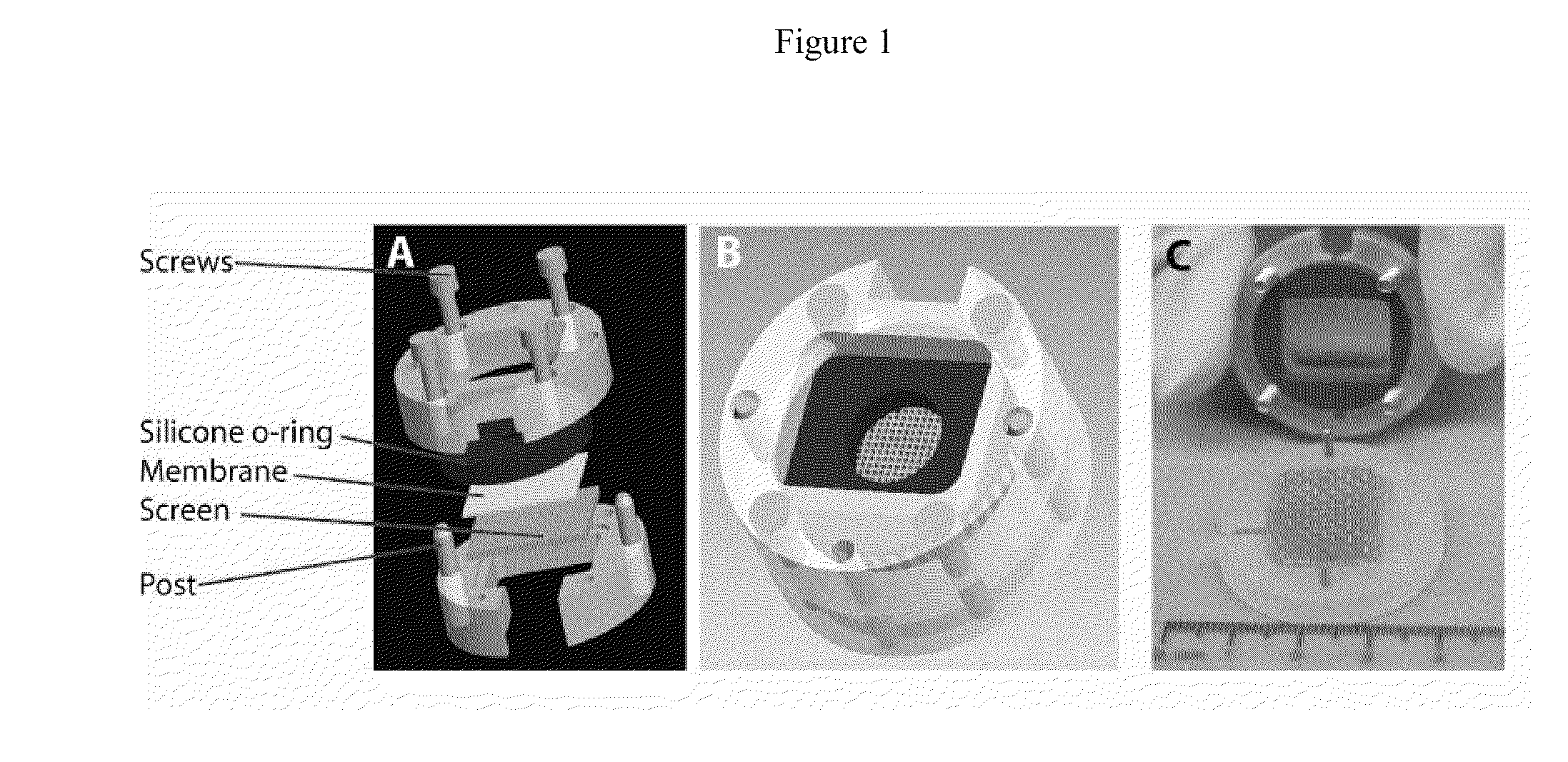
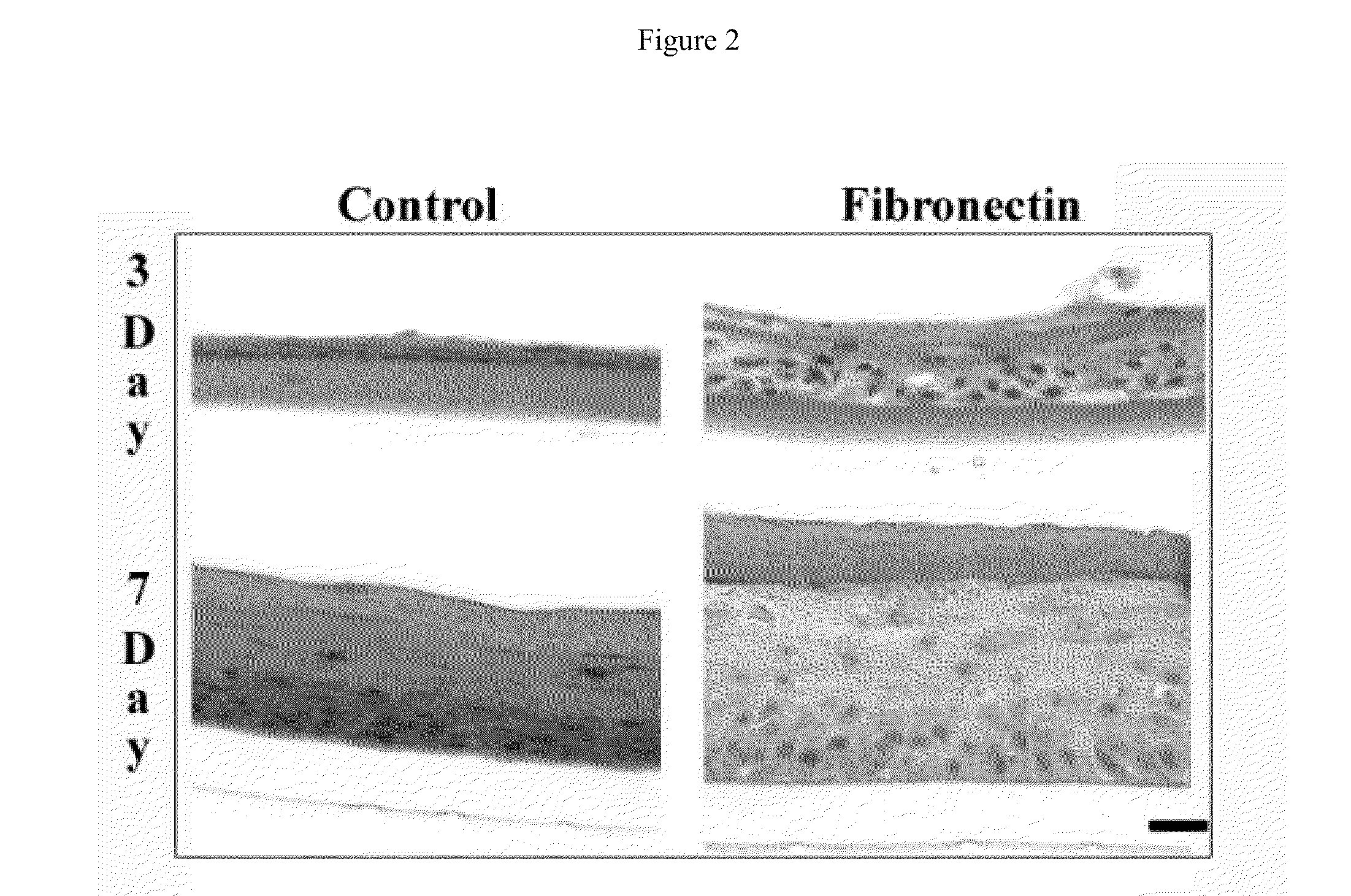
Bioengineered skin substitutes
InactiveUS20110171180A1Reduce frictionImprove regenerative abilityBiocideEpidermal cells/skin cellsDermisBiomedical engineering
A bioengineered skin substitute was developed that contains a microfabricated basal lamina analog that recapitulates the native microenvironment found at the dermal-epidermal junction (DEJ).
Owner:WORCESTER POLYTECHNIC INSTITUTE
Magnetic method and apparatus for increasing foot traction on sports boards
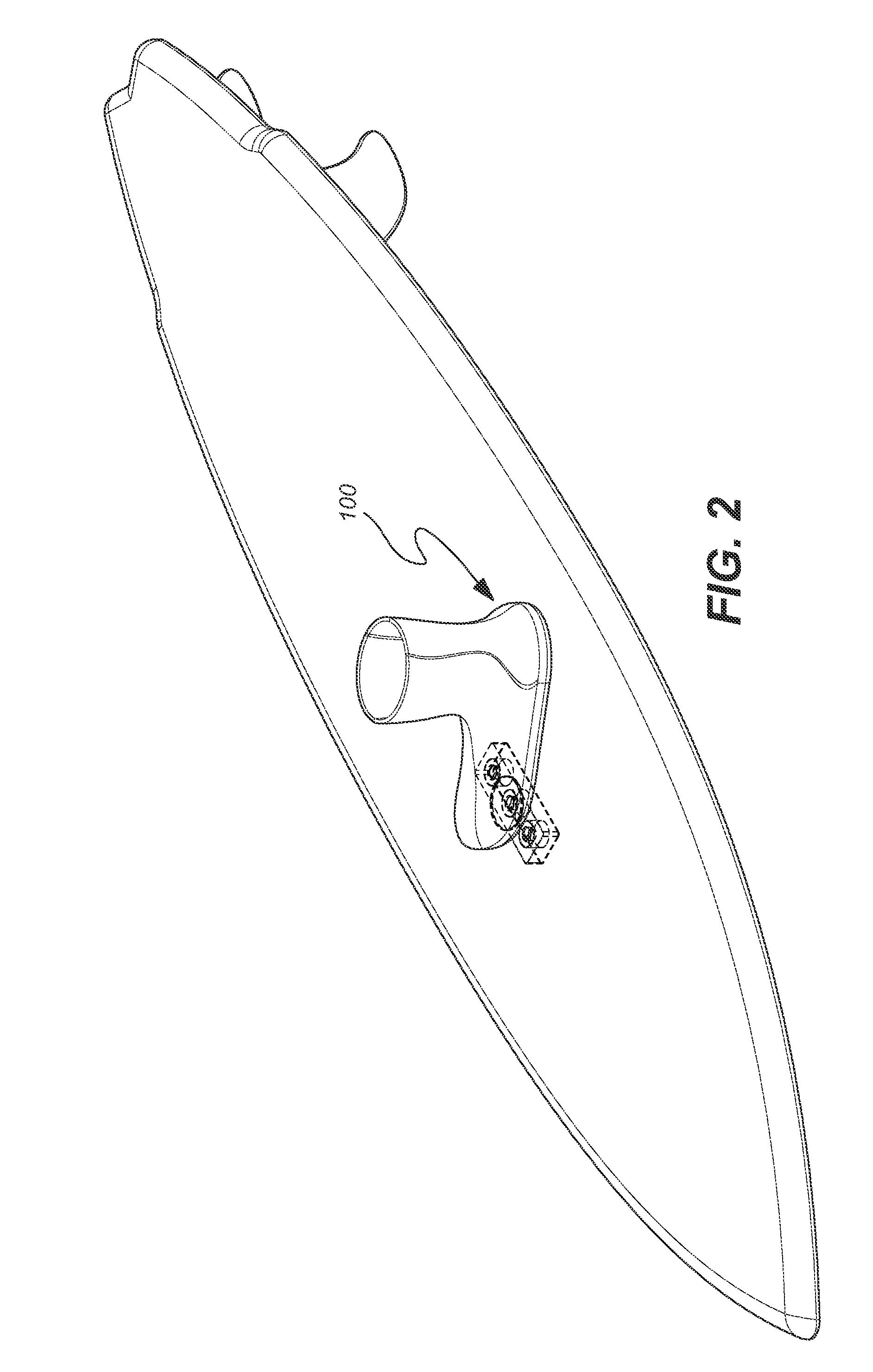
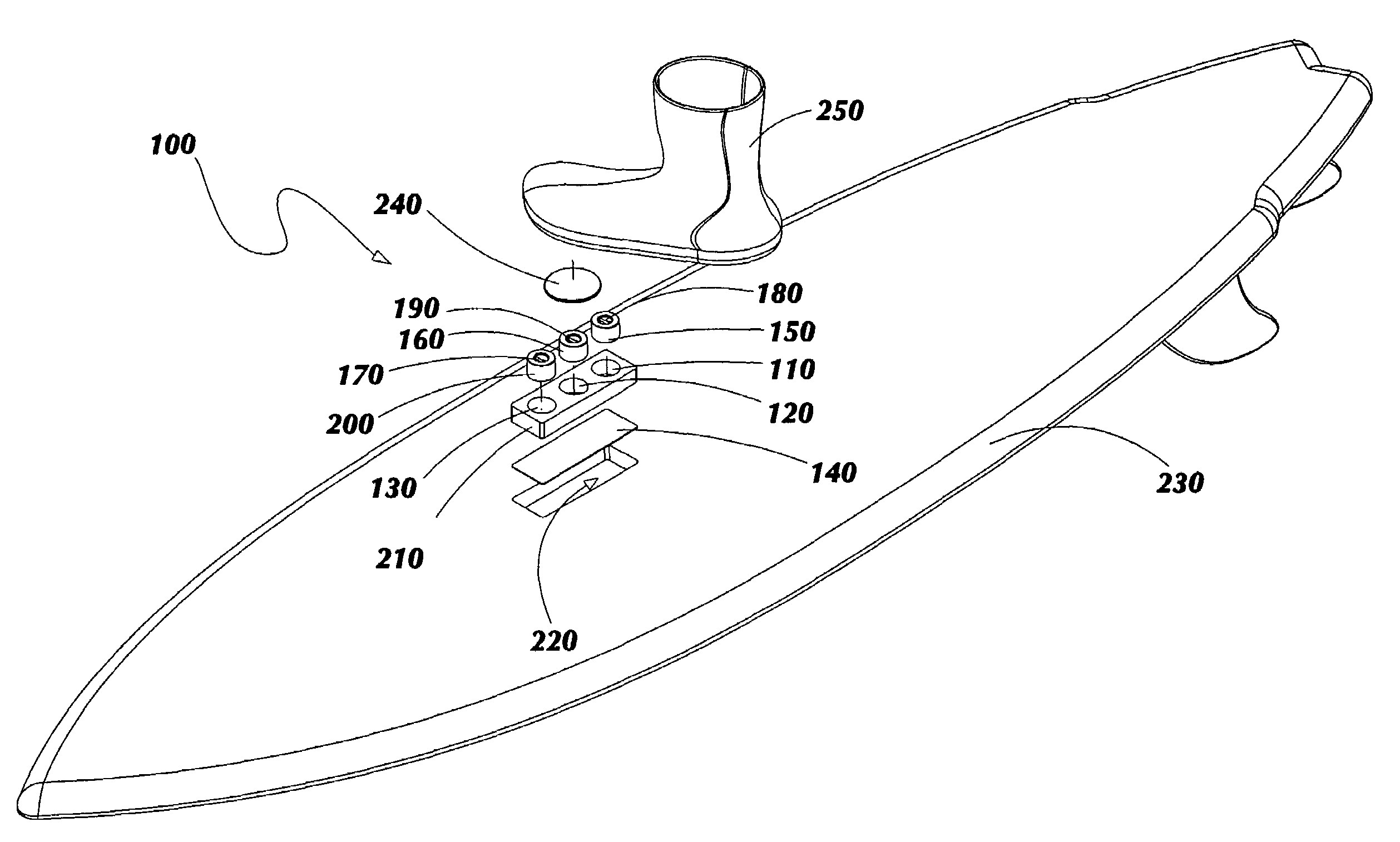
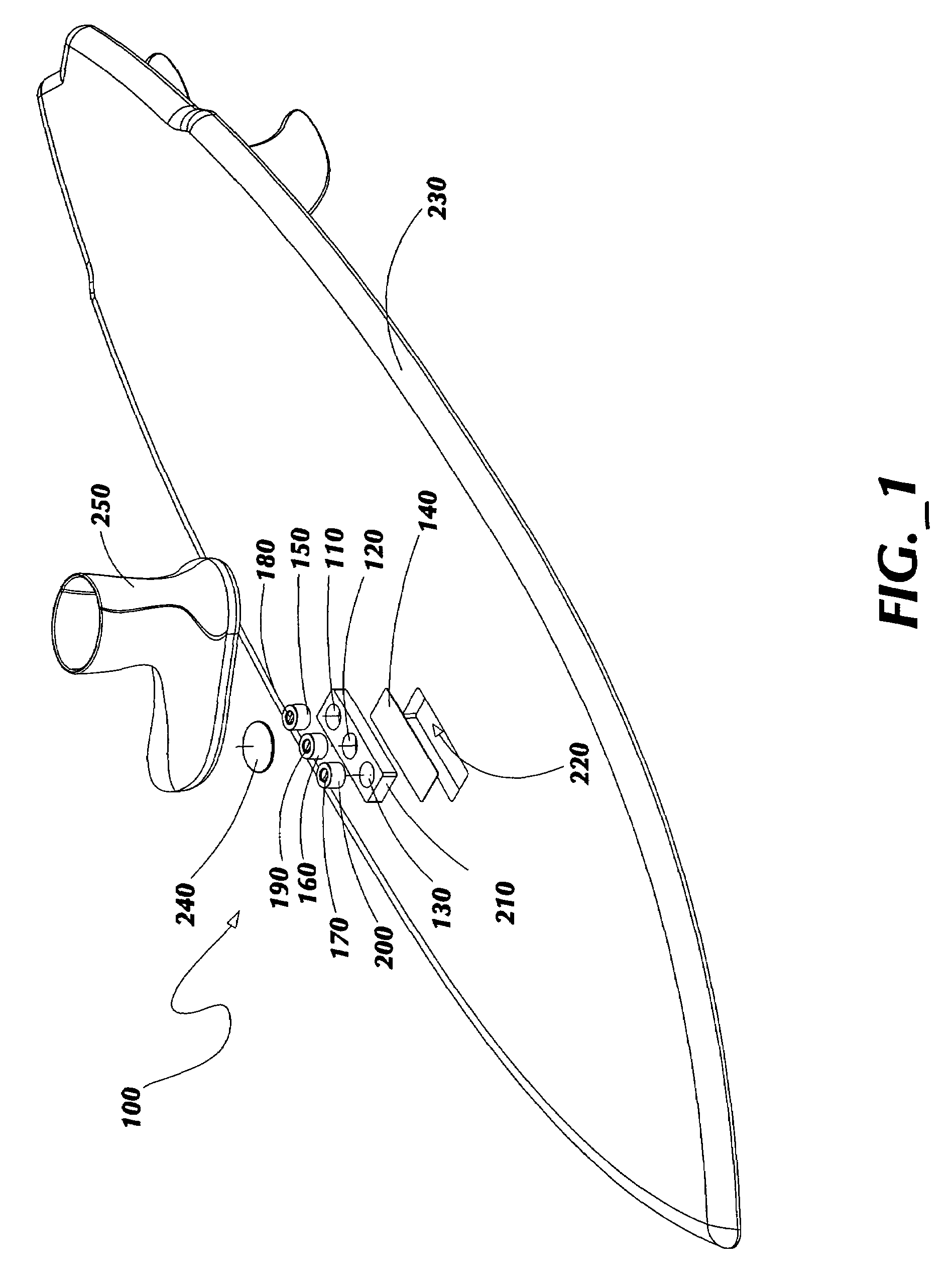
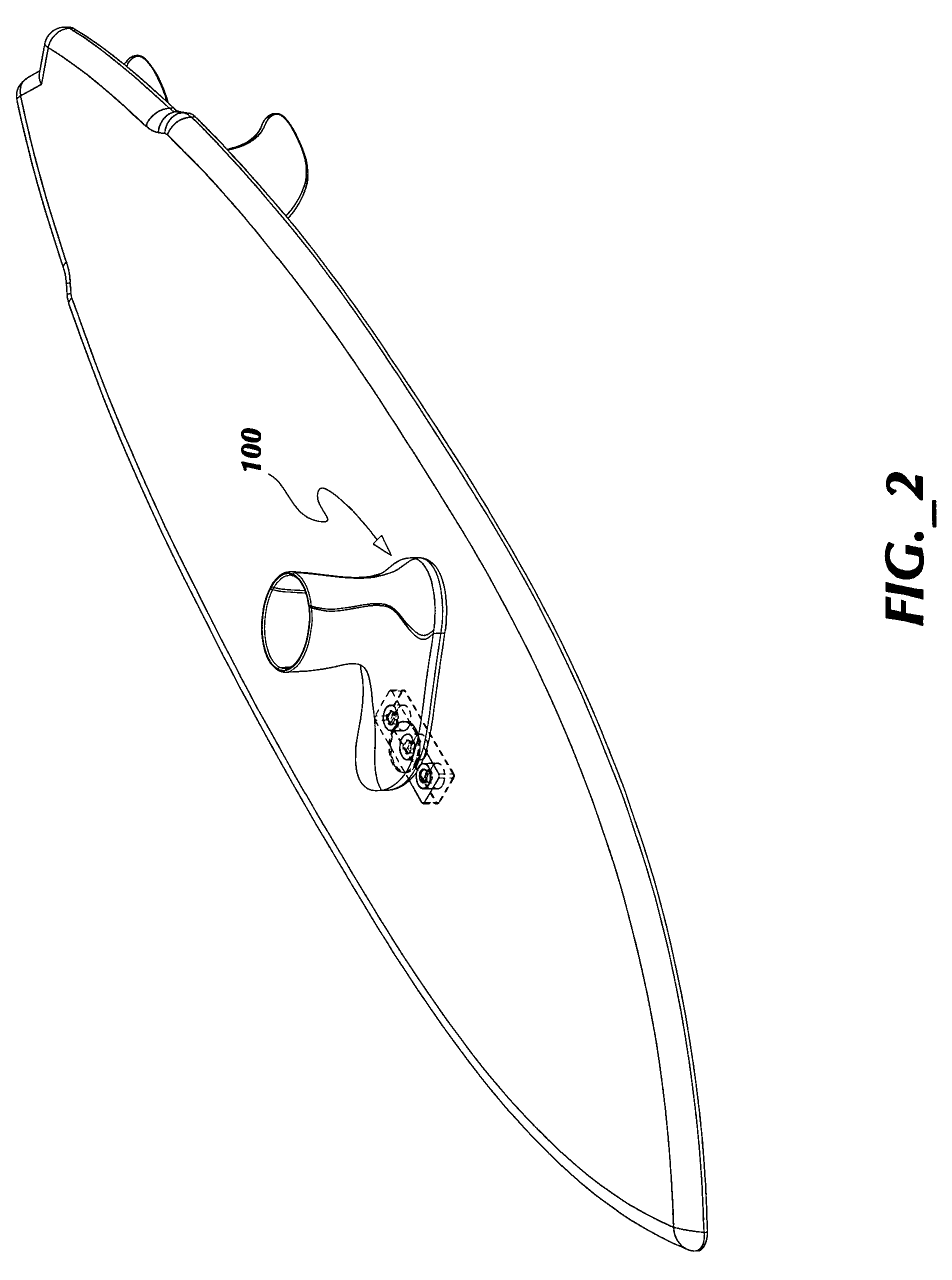
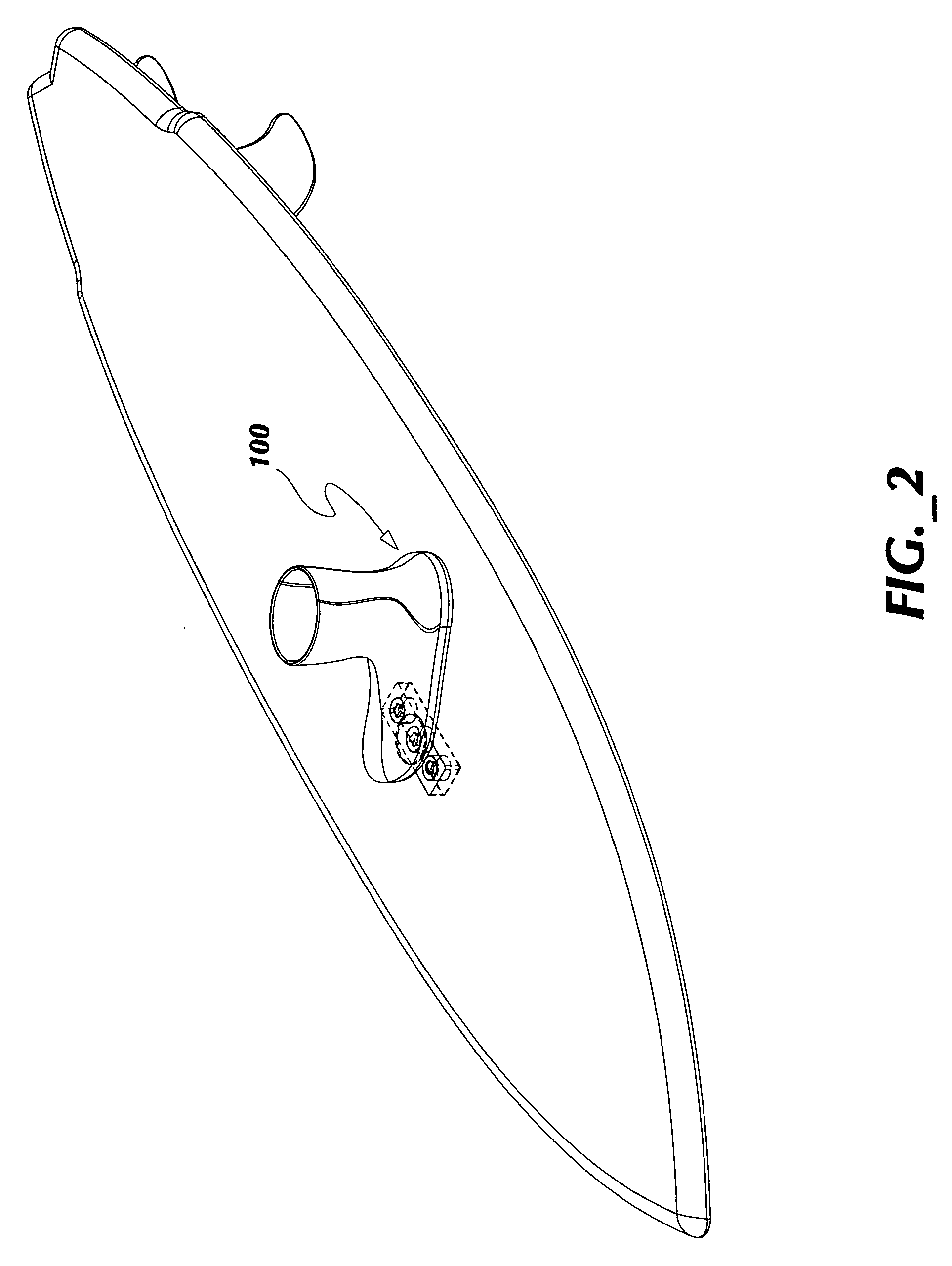
ActiveUS7837218B2Great tractionPlace stableCarriage/perambulator accessoriesSki bindingsEngineeringBinding force
A sports board binding and foot traction apparatus for use in sports boards such as surfboard, skateboard, wakeboards, and the like. The apparatus includes a sports board having a deck and a bottom and at least one cutout box disposed in one or both of the deck and bottom. One or more permanent magnets is disposed and retained in one or more of the cutout boxes to form a magnetic region on the deck of the board. An article of sports footwear having a sole is provided, and a ferromagnetic plate is disposed in at lest a portion of each footwear sole. When the footwear is brought into proximity with the magnetic field, a binding force is applied to the footwear and the wearer is thereby enabled to manipulate the sports board in ways not possible without the inventive apparatus.
Owner:FLAIG THEODORE J
Magnetic method and apparatus for increasing foot traction on sports boards
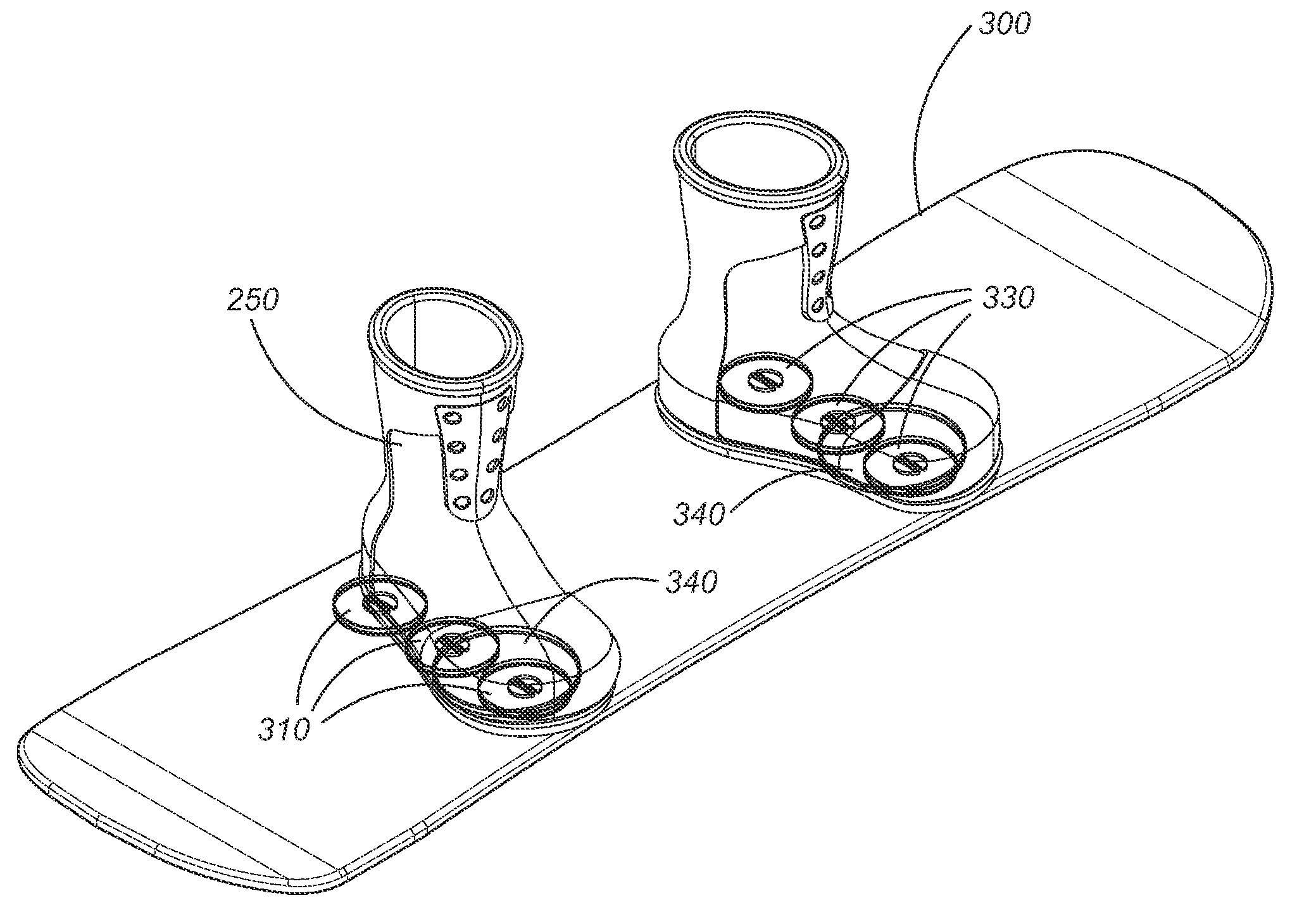
ActiveUS7338067B2Great tractionPlace stableSki bindingsCarriage/perambulator accessoriesEngineeringBinding force
A sports board binding and foot traction apparatus for use in sports boards such as surfboard, skateboard, wakeboards, and the like. The apparatus includes a sports board having an upper surface and at least one magnet holding receptable disposed in the upper surface; A a ferromagnetic bar is disposed under the bottom of the magnet holding receptacles, and permanent magnets are placed in the receptacles to form a magnetic region on the surface of the board. An article of sports footwear having a sole is provided, and a ferromagnetic plate is disposed in at lest a portion of the footwear soles. When the footwear is brought into proximity with the magnetic field, a binding force is applied to the footwear.
Owner:FLAIG THEODORE J
Method and apparatus for identifying, classifying, or quantifying protein sequences in a sample without sequencing
InactiveUS6432361B1Rapid and economical and quantitative and precise determination and classificationSufficient discrimination and resolutionData processing applicationsMicrobiological testing/measurementSequence databaseDNA Sequence Databases
This invention provides methods by which biologically derived DNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information.
Owner:CURAGEN CORP
Optimized multi-layer optical waveguiding system
InactiveUS20030026569A1Low enough refractive indexAvoid lostCladded optical fibreCoupling light guidesRefractive indexWaveguide
A single mode optical waveguide which is lithographically formed and employs polymeric materials having low propagation loss. The optical waveguide has a substrate, a polymeric, buffer layer having an index of refraction nb disposed on a surface of the substrate, a patterned, light-transmissive core layer having an index of refraction nc disposed directly on a surface of the cladding layer, and an overcladding layer having an index of refraction no on a top surface of the core, side walls of the core, and exposed portions of the buffer layer, with nb<no<nc and DELTAn=nc-no.
Owner:ENABLENCE TECH USA
Magnetic method and apparatus for increasing foot traction on sports boards
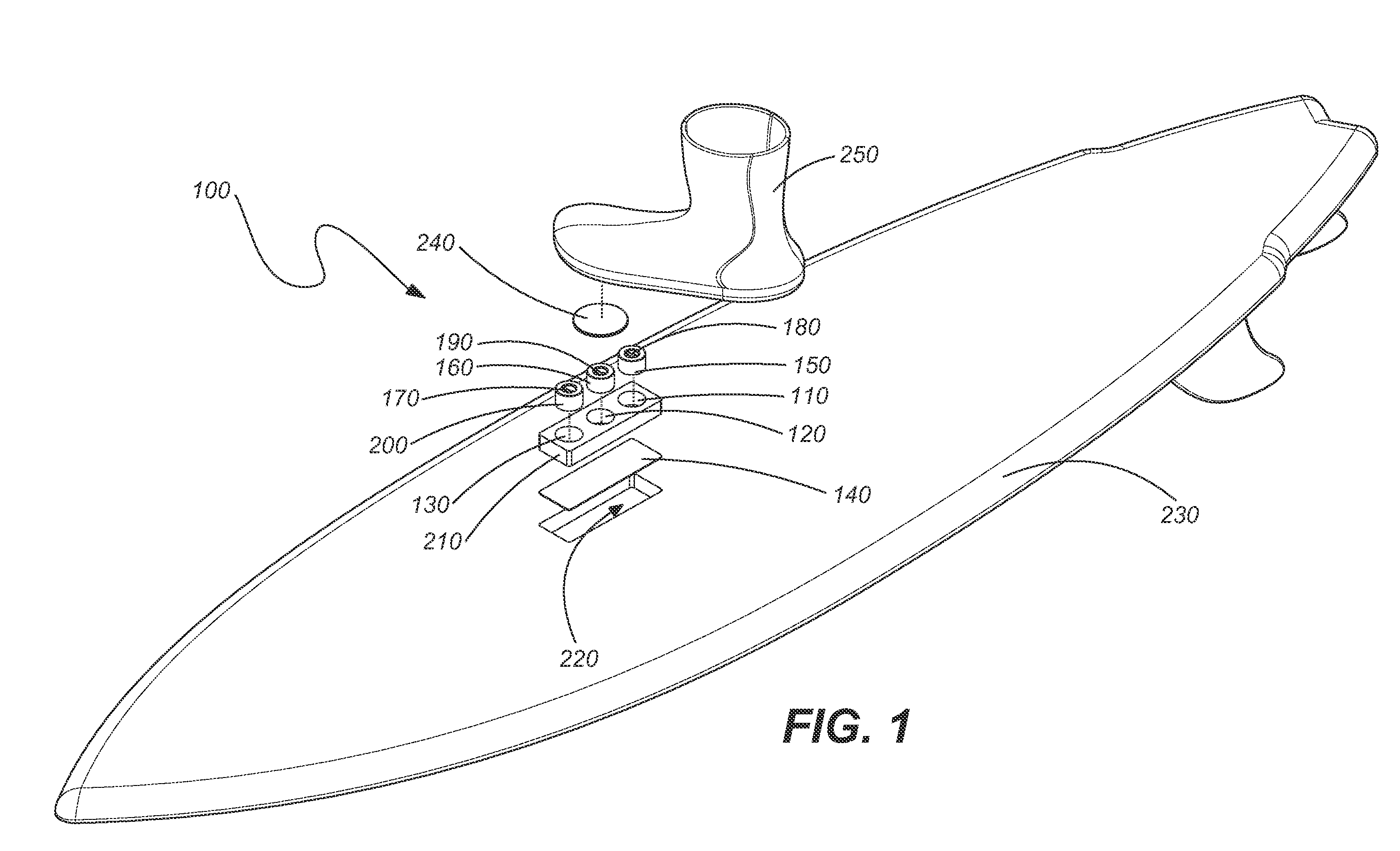
ActiveUS20060197311A1Great tractionPlace stableCarriage/perambulator accessoriesSki bindingsEngineeringBinding force
A sports board binding and foot traction apparatus for use in sports boards such as surfboard, skateboard, wakeboards, and the like. The apparatus includes a sports board having an upper surface and at least one magnet holding receptable disposed in the upper surface; A a ferromagnetic bar is disposed under the bottom of the magnet holding receptacles, and permanent magnets are placed in the receptacles to form a magnetic region on the surface of the board. An article of sports footwear having a sole is provided, and a ferromagnetic plate is disposed in at lest a portion of the footwear soles. When the footwear is brought into proximity with the magnetic field, a binding force is applied to the footwear.
Owner:FLAIG THEODORE J
Magnetic method and apparatus for increasing foot traction on sports boards
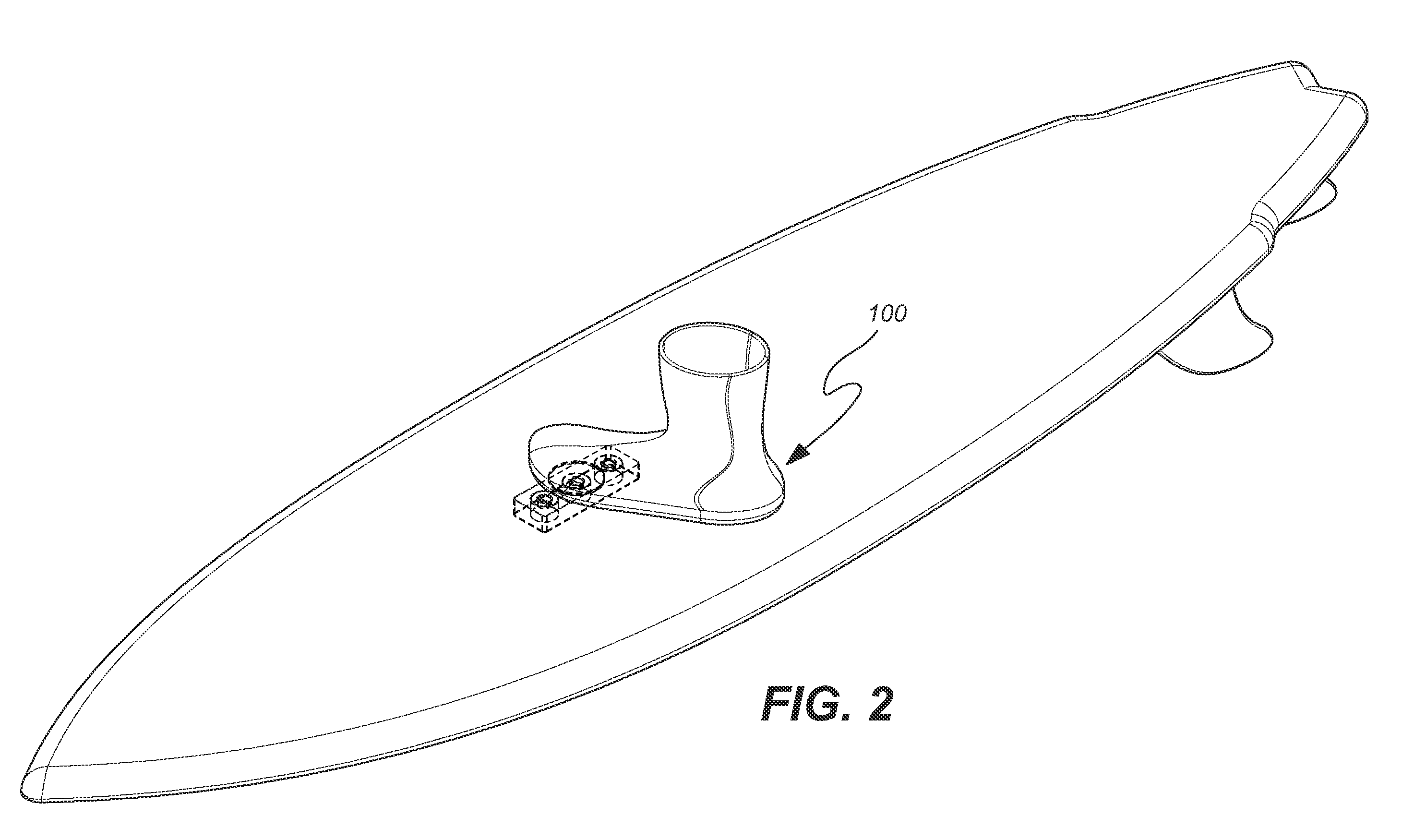
InactiveUS20100025967A1Great tractionPlace stableSki bindingsWater sport boardsEngineeringBinding force
A sports board binding and foot traction apparatus for use in sports boards such as surfboard, skateboard, wakeboards, and the like. The apparatus includes a sports board having a deck and a bottom and at least one cutout box disposed in one or both of the deck and bottom. One or more permanent magnets is disposed and retained in one or more of the cutout boxes to form a magnetic region on the deck of the board. An article of sports footwear having a sole is provided, and a ferromagnetic plate is disposed in at lest a portion of each footwear sole. When the footwear is brought into proximity with the magnetic field, a binding force is applied to the footwear and the wearer is thereby enabled to manipulate the sports board in ways not possible without the inventive apparatus.
Owner:FLAIG THEODORE J
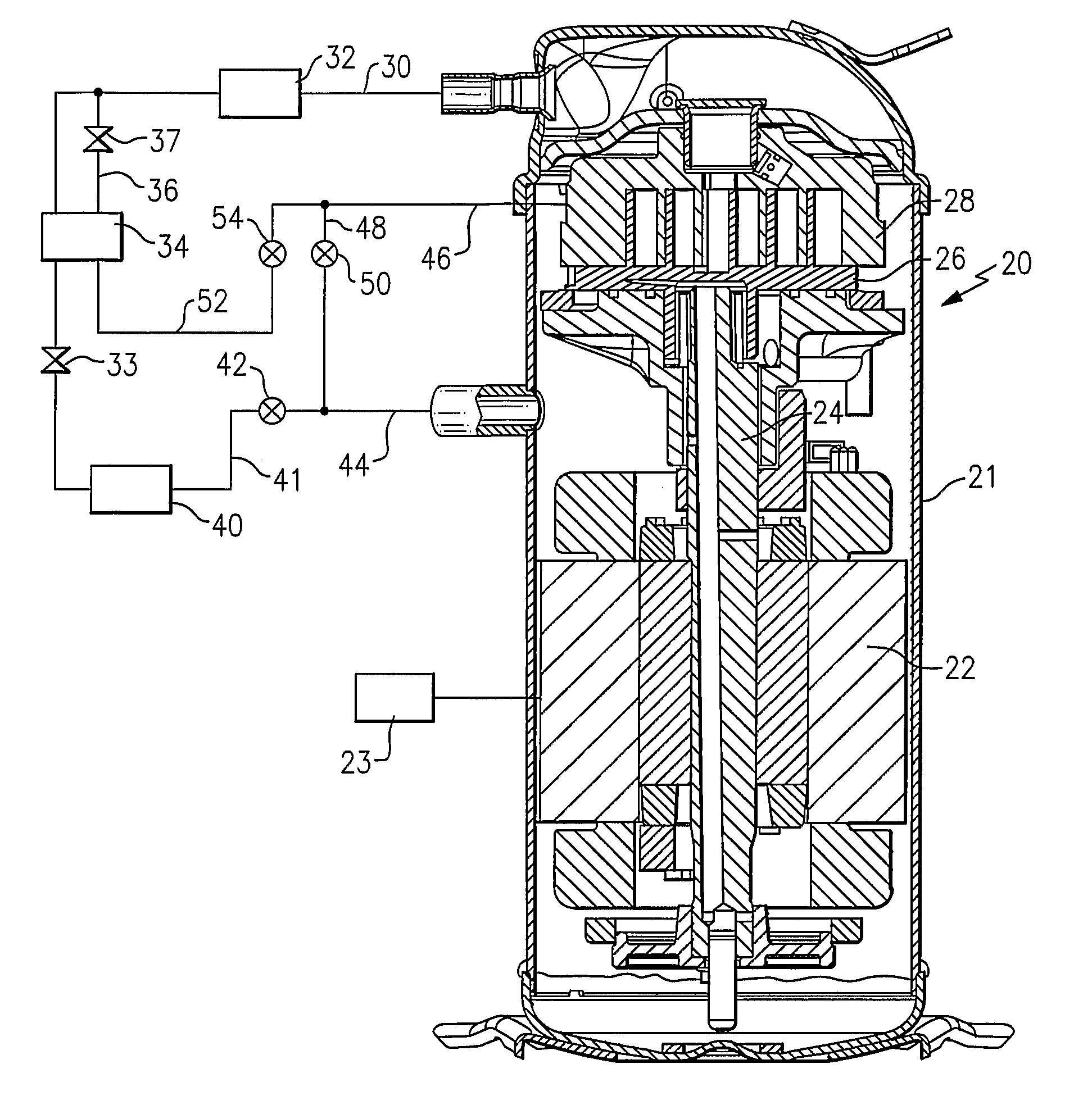
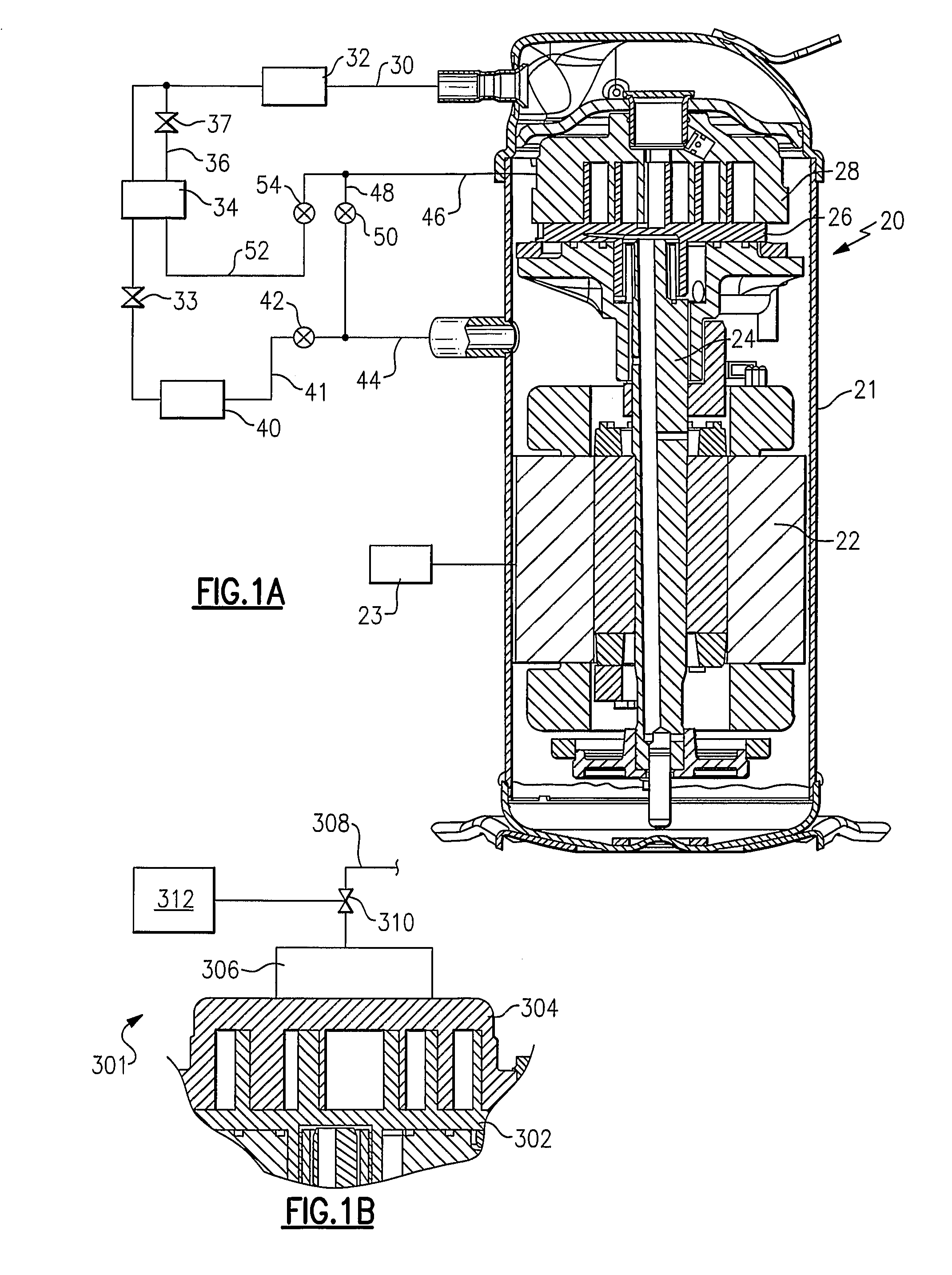
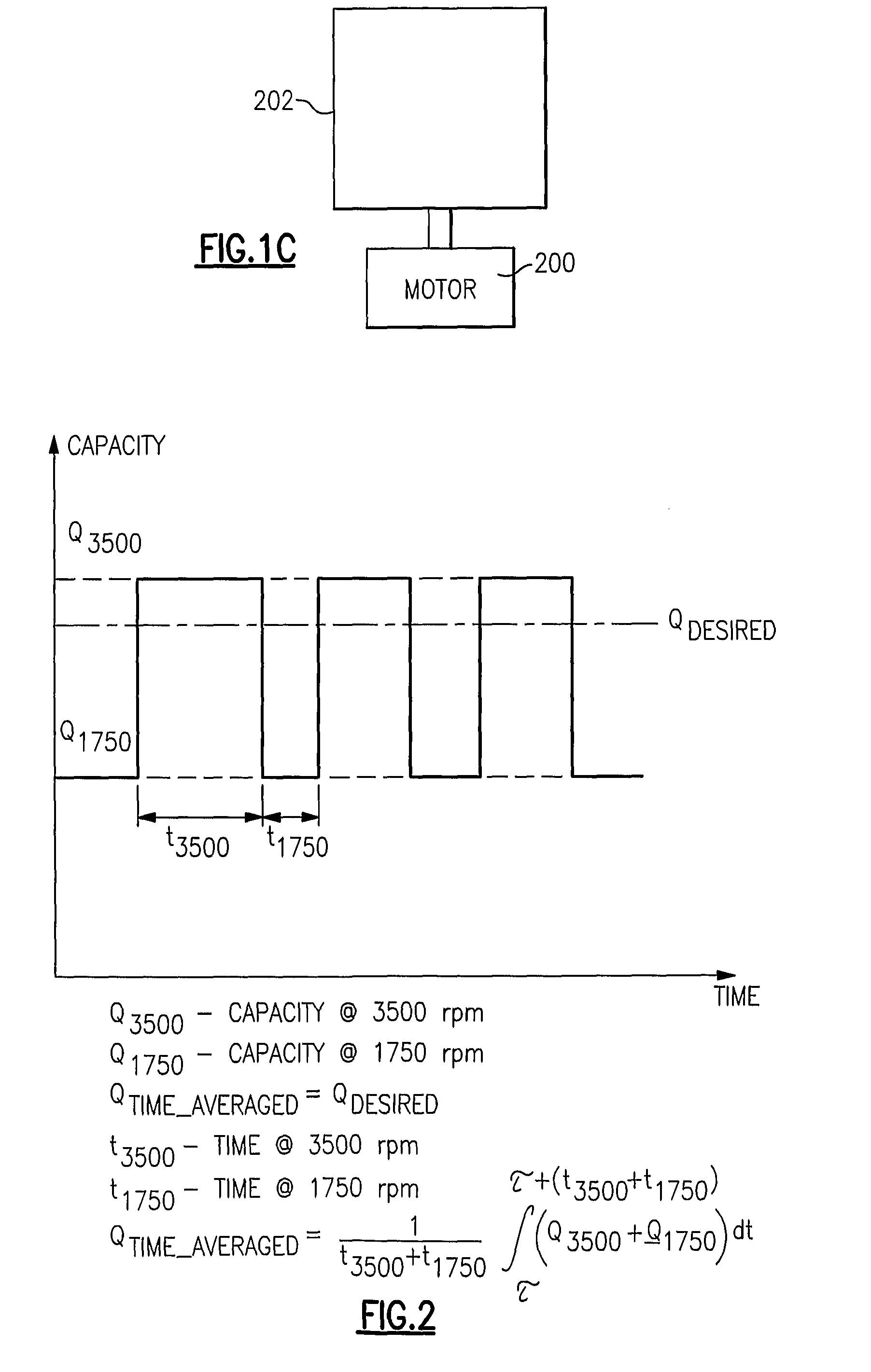
Refrigerant system with multi-speed pulse width modulated compressor
InactiveUS20090308086A1Improve reliabilityReduce capacityRotary/oscillating piston combinations for elastic fluidsEngine of arcuate-engagement typeEngineeringControl theory
A refrigerant system is provided with a compressor having a motor that is operable at least at two distinct speeds. The pulse width modulation control is provided to cycle a compressor motor operation between its at least two speeds at a specific rate to exactly match thermal load demands in a conditioned space. The present invention reduces cycling and other efficiency losses as have been experienced in the prior art, as well as minimizes cost and may improve reliability. Also, the present invention can be utilized in conjunction with other known unloading techniques.
Owner:CARRIER CORP
Roll-up spa and swim spa cover
ActiveUS8683621B1Improve structural strengthAccurately tailoredGymnasiumSwimming poolsGlass fiberHot spring
A spa cover constructed of layers of pliable insulation encapsulated within a weatherproof vinyl reinforced fabric or a rubber membrane which is attached to rigid square support tubes that run parallel to the short end of the spa. The encapsulated insulation is fastened to the bottom of the square support tubes with corrosion-free nylon anchors which allows the soft insulation assembly to span over the water and rest directly on the spa bartop surface sealing the heat in and keeping debris out. An arched fiberglass rod structure supports a weatherproof rainfly which is permanently affixed to the flat cover assembly to shed rain and debris. The entire assembly is held in place at each end with adjustable straps which stretch the cover from end to end keeping the rainfly taut. Removing the cover to access the spa involves undoing the straps on one end and rolling the cover assembly towards the other end until the desired amount of spa exposure is reached. Covering the spa after use is just the opposite procedure.
Owner:E2E LLC
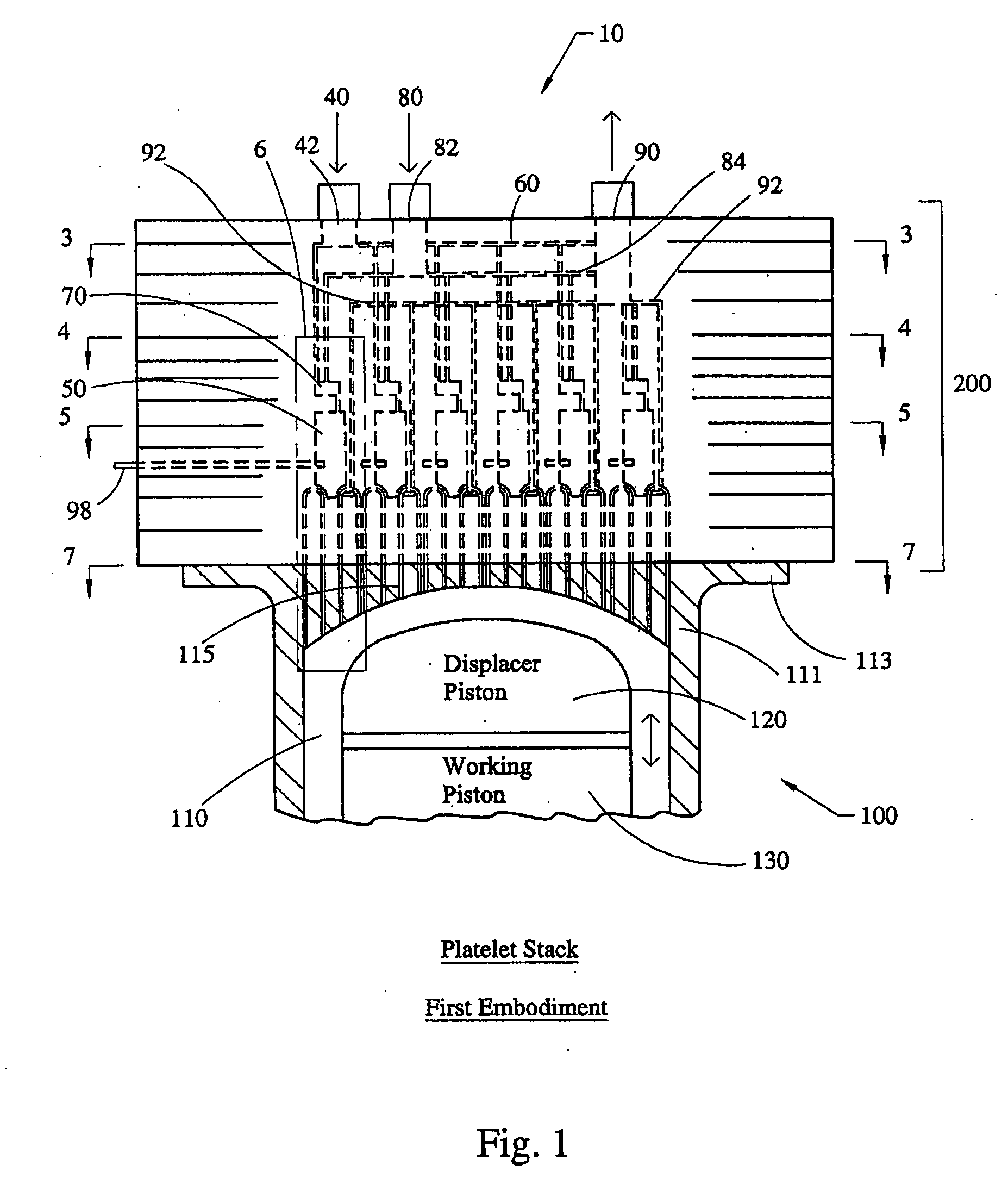
Stirling engine having platelet heat exchanging elements
InactiveUS20060021343A1Guaranteed uptimeEfficient heat transferSolar heating energySolar heat devicesEngineeringStaged combustion
The present invention provides heat exchanging elements for use in Stirling engines. According to the present invention, the heat exchanging elements are made from muliple platelets that are stacked and joined together. The use of platelets to make heat exchanging elements permits Stirling engines to run more effiecient because the heat transfer and combustion processes are improved. In one embodiment, multi-stage combustion can be introduced with platlets, along with the flexibility to use different types of fuels. In another embodiment, a single component constructed from platelets can provide the heat transfer rquirements betweeen the combustion gas / working gas, working gas in the regenerator and the working gas / coolant fluid of a Stirling engine. In another embodiment, the platelet heat exchanging element can recieve solar energy to heat the Stirling engine's working gas. Also, this invention provides a heat exchanging method that allows for multiple fuilds to flow in opposing or same direction.
Owner:DISENCO +2
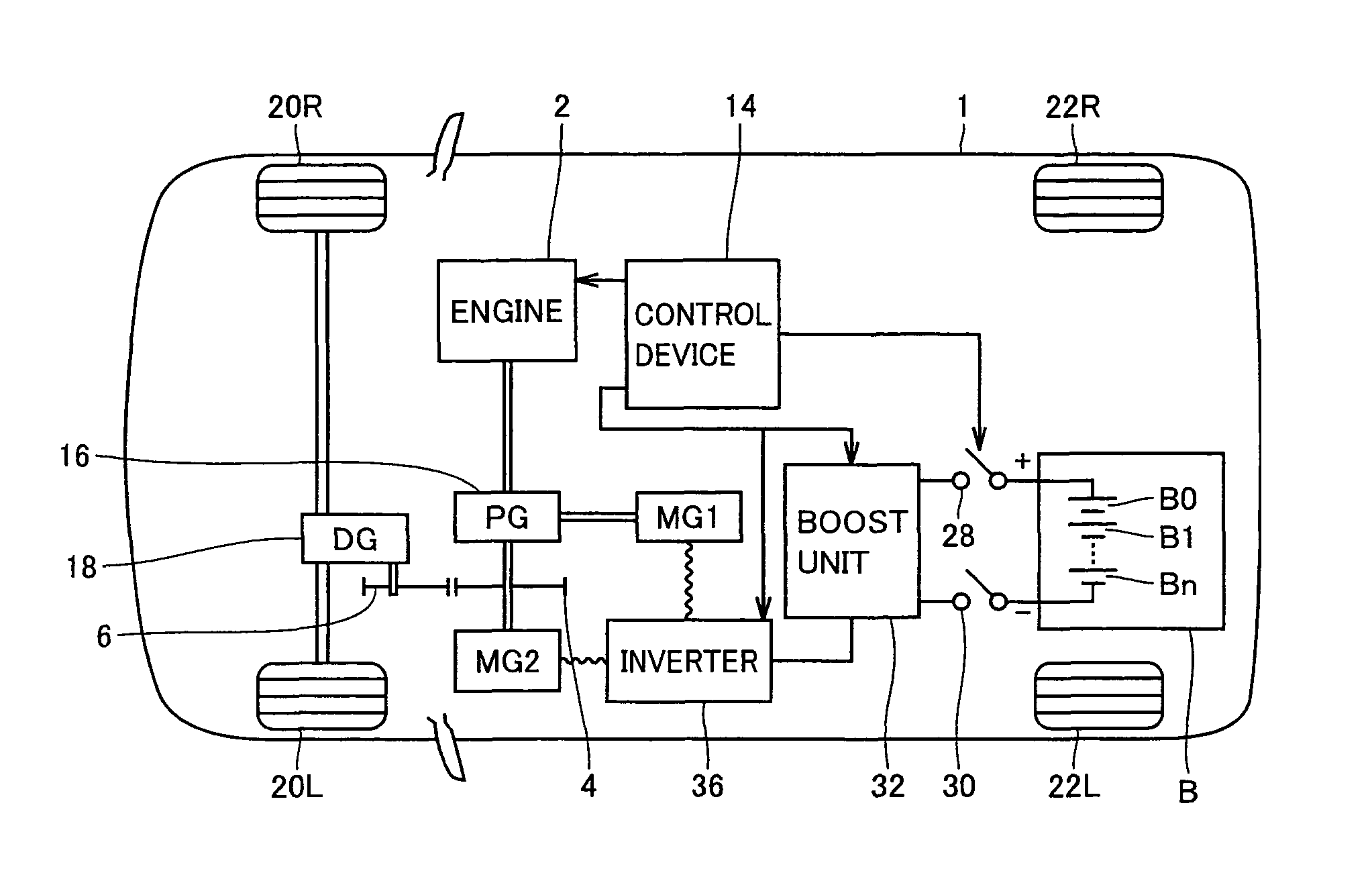
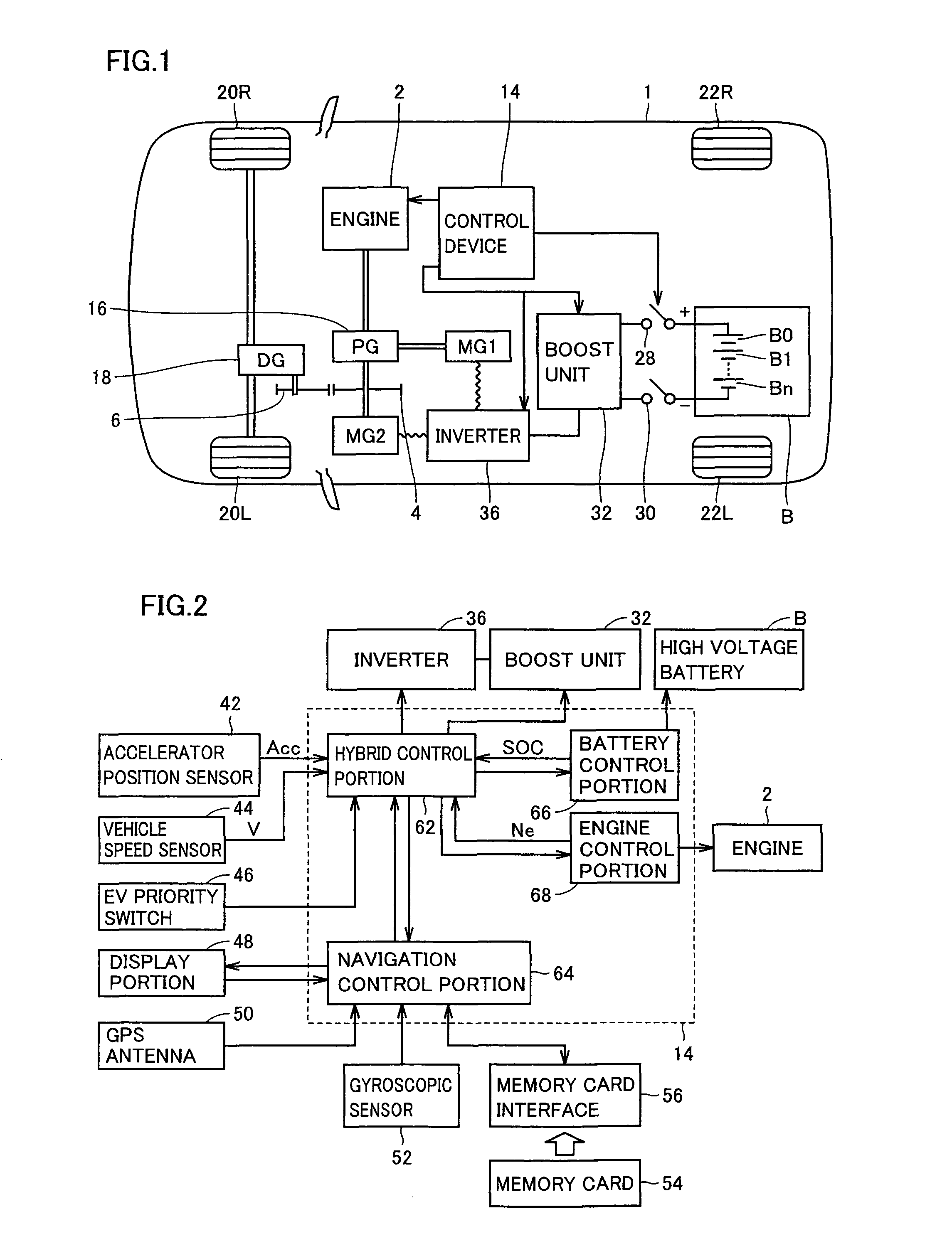
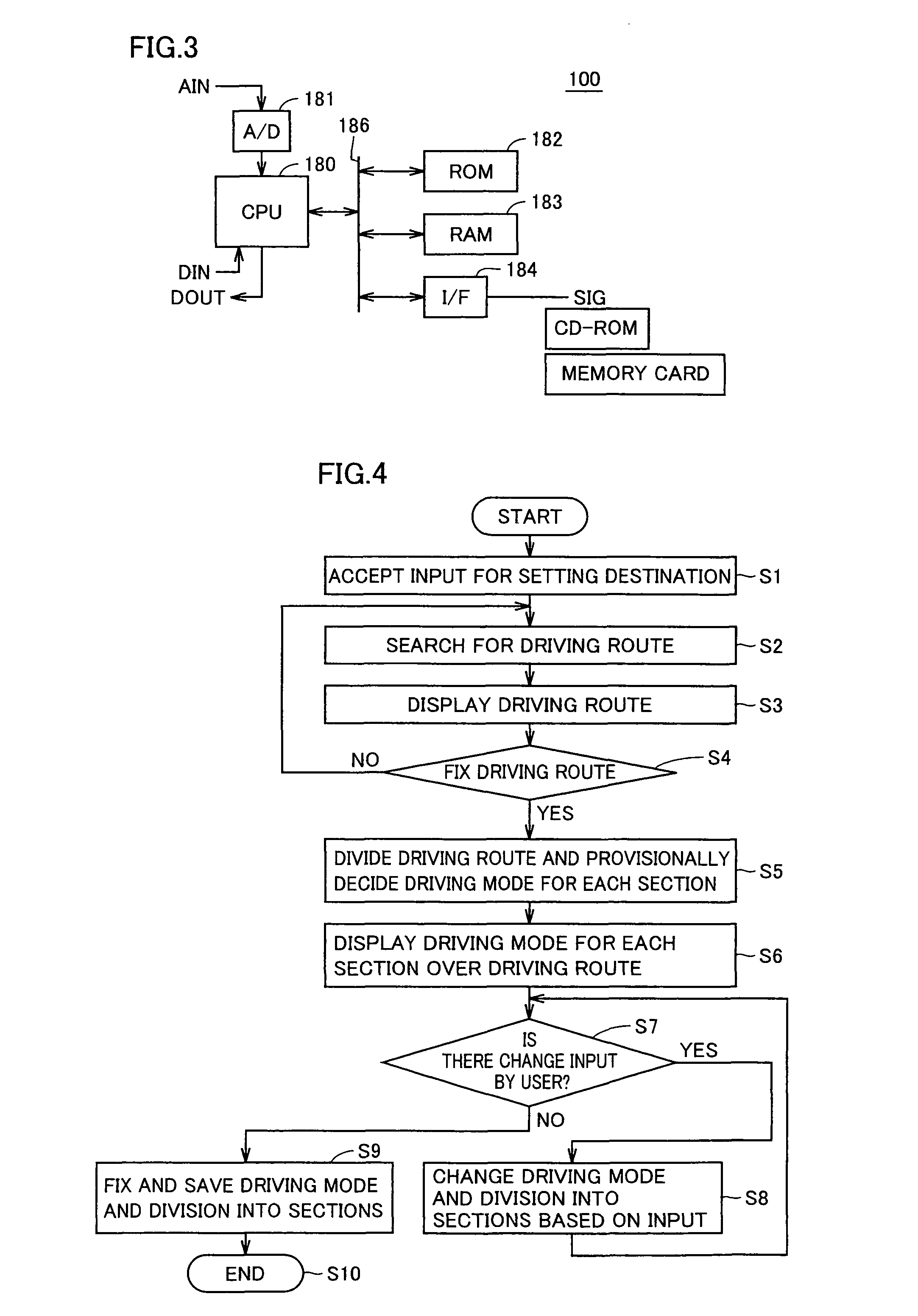
Vehicle control device, vehicle control method, and recording medium having program recorded thereon
ActiveUS8285433B2Accurately tailoredIncrease awarenessHybrid vehiclesDigital data processing detailsInternal combustion engineVehicle control
A vehicle control method having an HV driving mode in which operation of an internal combustion engine is permitted and an EV driving mode in which a vehicle is driven by using a motor with the internal combustion engine stopped includes the steps of: setting a destination; setting a driving route from a starting point to the destination; dividing the driving route and associating any of the driving modes with each section of the divided driving route; fixing the division of the driving route and the driving mode associated with each section based on an instruction from an operator; and causing the vehicle to travel each section in the associated driving mode.
Owner:TOYOTA JIDOSHA KK
Production of nano-powder based combinatorial libraries
InactiveUS20080070801A1Minimize and avoid formationAccurately tailoredSequential/parallel process reactionsLibrary screeningNanoparticleMaterials science
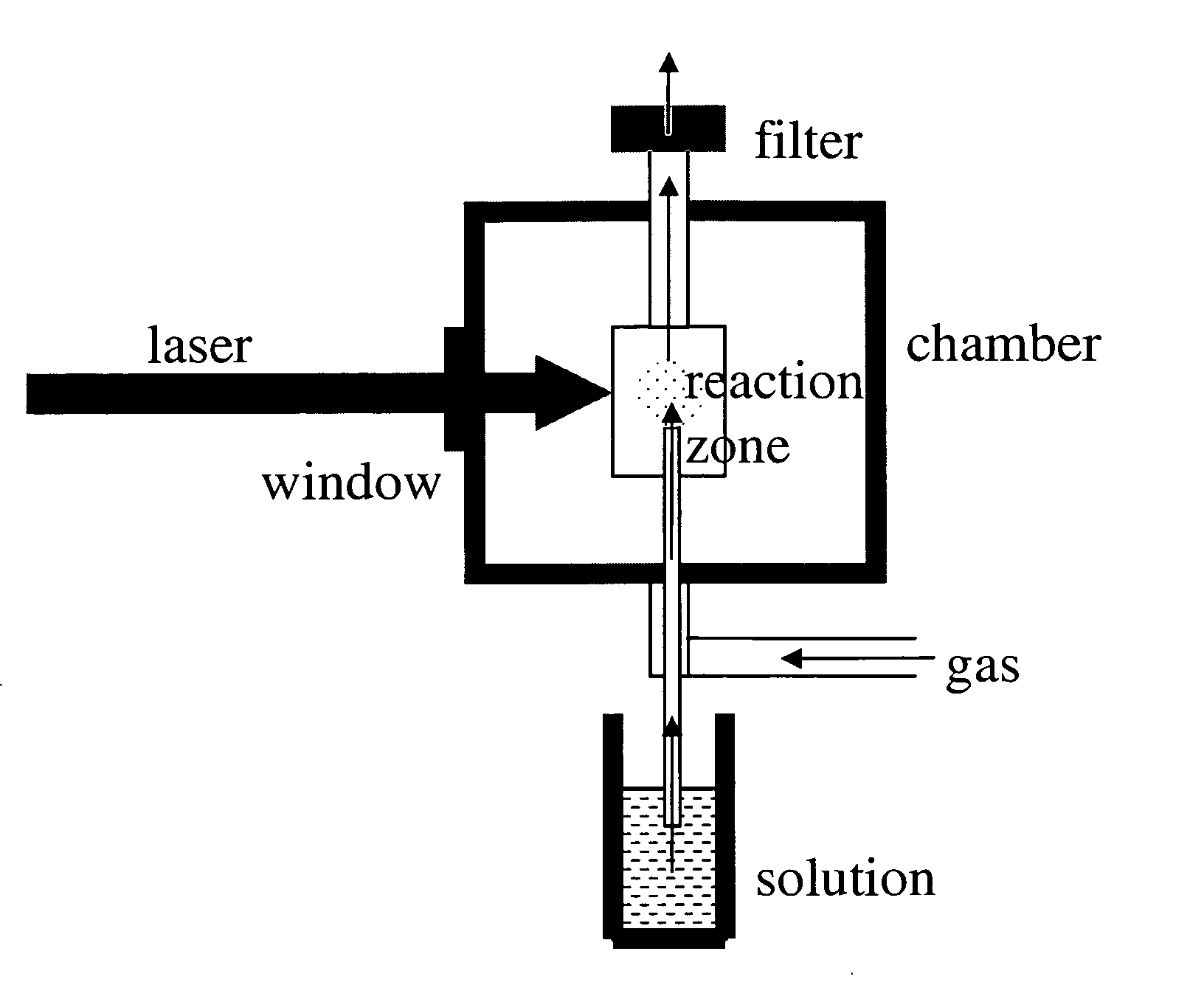
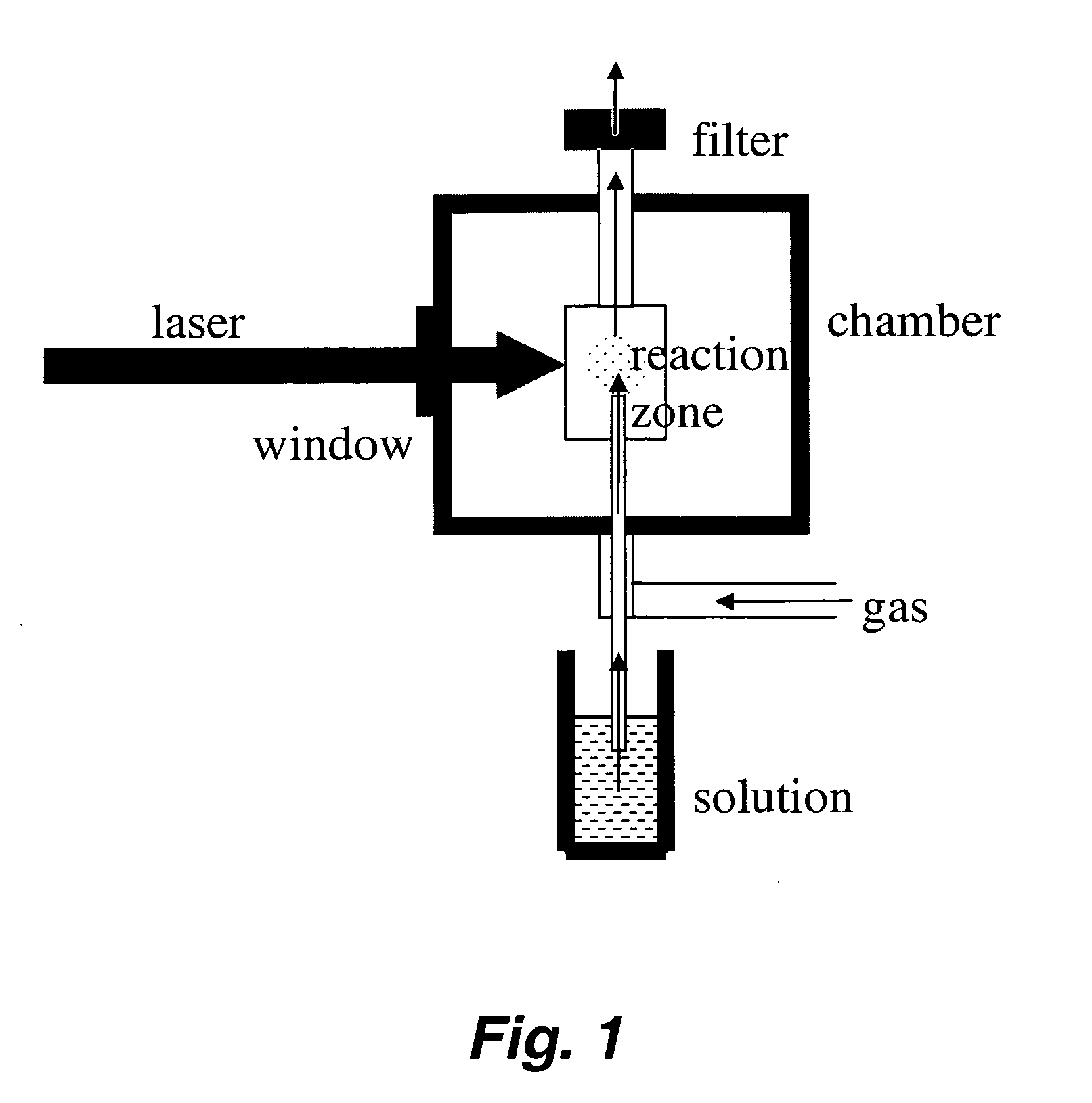
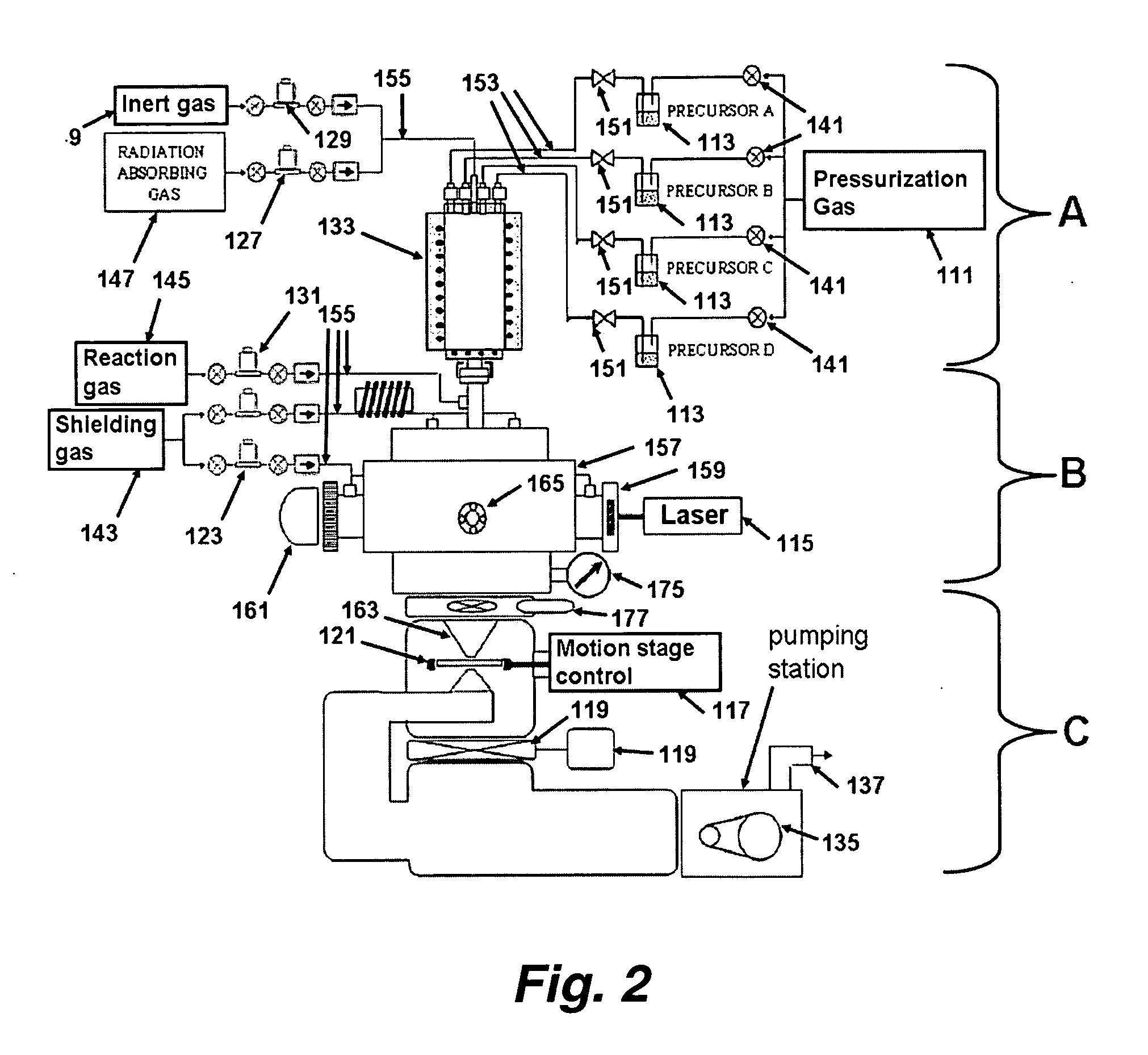
The present invention relates to methods and apparatus for synthesizing combinatorial materials libraries using pyrolysis techniques. In certain embodiments, the methods involve varying the precursors and / or reactant gases in an operating pyrolysis unit to continuously vary the resulting nanoparticle composition and collecting different nanoparticles at different locations on a substrate using a spatially addressable particle collector.
Owner:INTEMATIX
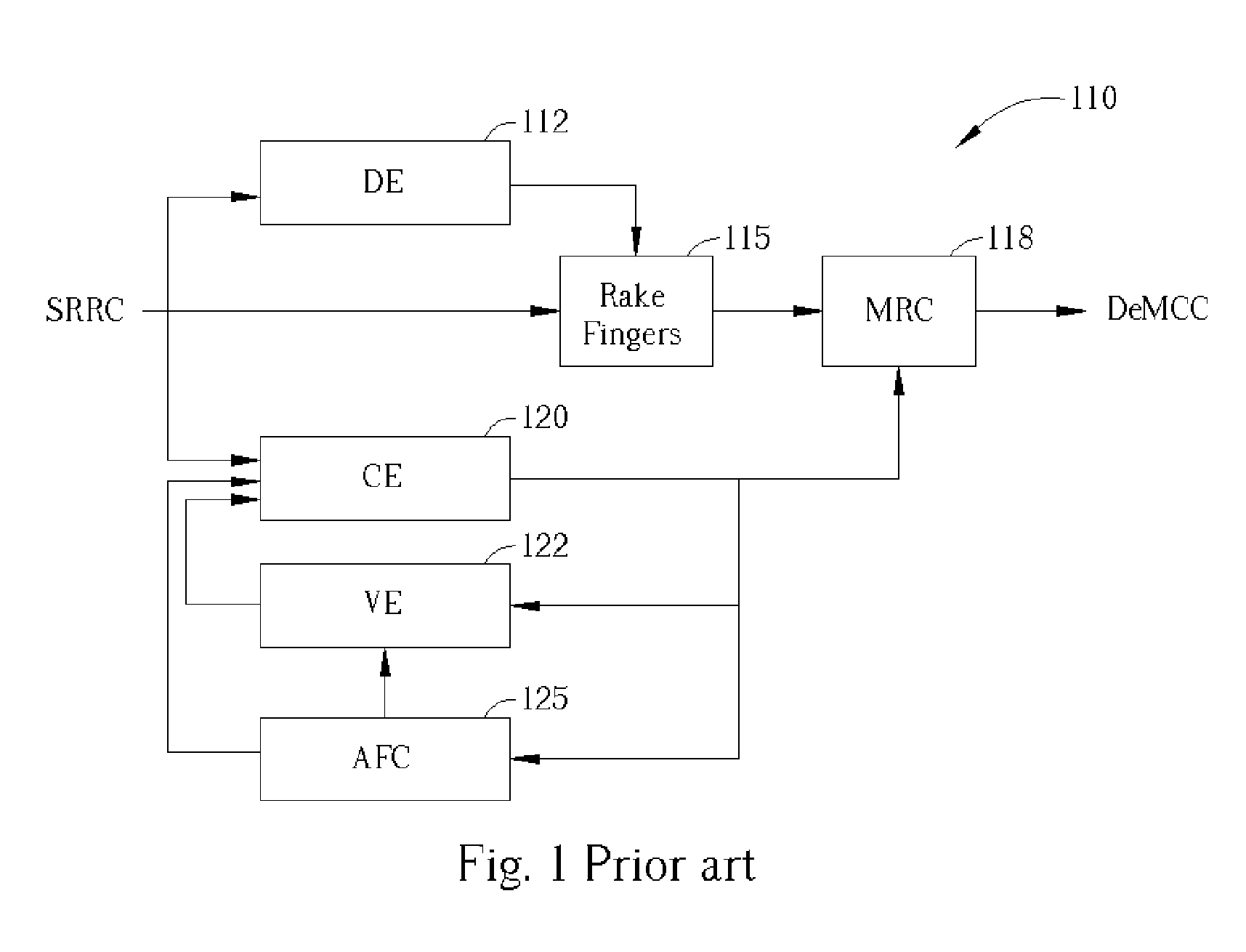
Signal demodulation in a mobile receiver
InactiveUS20050101278A1Improve performanceAccurately tailoredAmplitude-modulated carrier systemsSubstation equipmentFrequency mixerEngineering
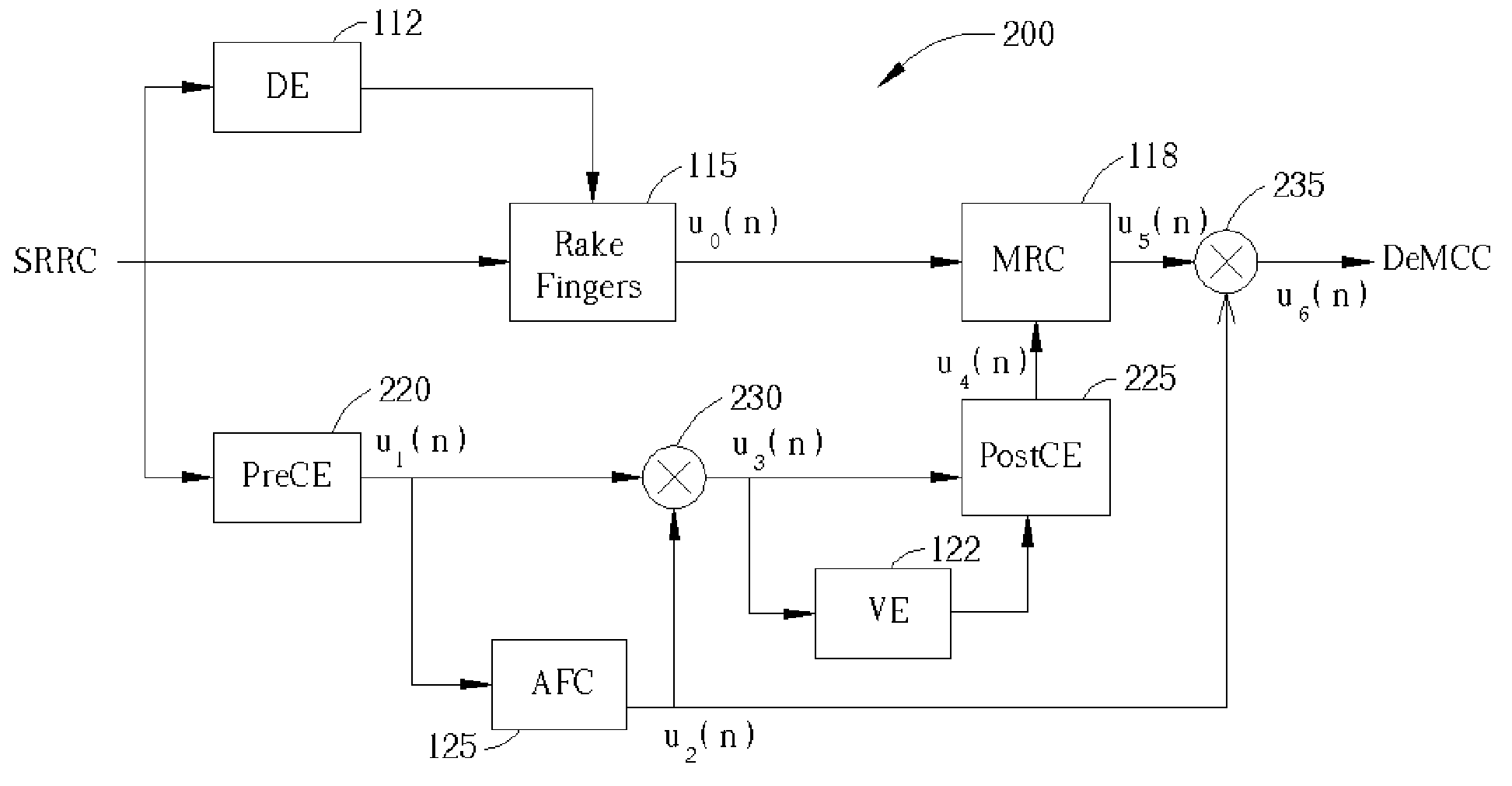
A WCDMA receiver in which the dependencies of different functional modules are arranged to allow proper tailoring of the Channel Estimation (CE) module bandwidth. A PreCE provides rough estimation results of the channel complex gain. The rough estimation results are passed to the Automatic Frequency Control (AFC). The AFC outputs a signal transmitted to two mixers. The first mixer mixes the output of the PreCe with the output of the AFC and outputs the result to a Velocity estimator (VE) and a PostCE to generate the compensating signals. The output of the VE is also transmitted to the PostCE. The output of the PostCE is sent to a Maximum Ratio Combining module, whose output is mixed by the second mixer with the output of the AFC to generate a final signal.
Owner:BENQ CORP
Multi-Mode Filter Having Aperture Arrangement with Coupling Segments
InactiveUS20150380799A1Accurately tailoredCarefully controlledResonatorsCouplingCondensed matter physics
A multi-mode filter including a resonator body including a piece of dielectric material, the resonator body being configured to support a first resonant mode and a second resonant mode, the resonator body having a covering of an electrically conductive material, wherein the covering is provided with a coupling aperture arrangement, the coupling aperture arrangement including a first coupling segment and a second coupling segment, wherein the first coupling segment is configured to couple a first magnetic field or a first electric field to the first resonant mode or the second resonant mode within the resonator body and the second coupling segment is configured to couple a second magnetic field or a second electric field to the first resonant mode or the second resonant mode within the resonator body.
Owner:MESAPLEXX
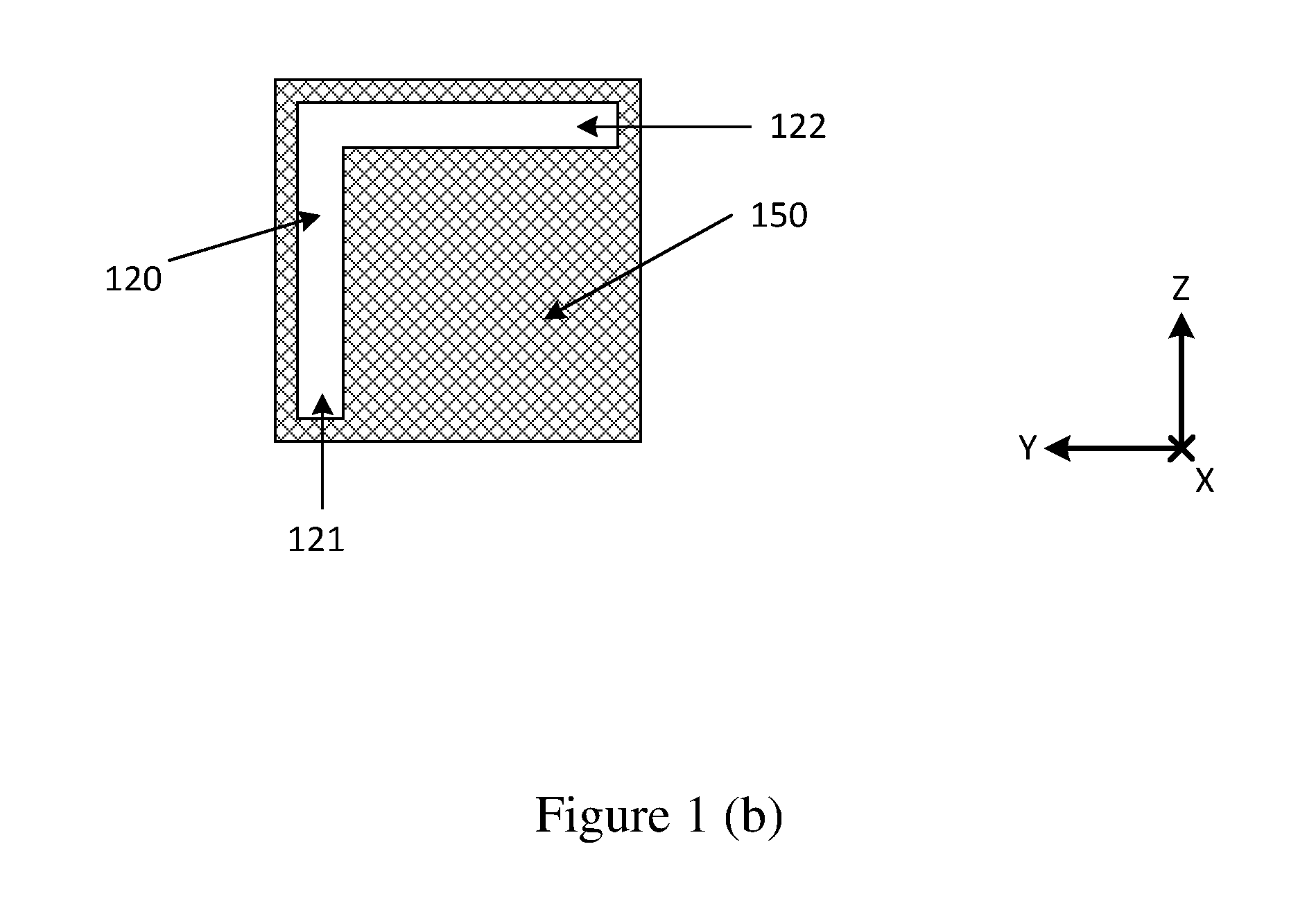
Method for manufacturing a FBG having improved performances and an annealing-trimming apparatus for making the same
ActiveUS20040161195A1Accurately tailoredImprove performanceCladded optical fibreSemiconductor/solid-state device manufacturingSingle processEngineering
A method for manufacturing a FBG having improved performances and an annealing-trimming apparatus for making the same are provided. The trimming and the annealing steps are advantageously combined into a single process in order to efficiently fabricate complex FBG filters with improved performance. The method comprises the steps of UV-writing a FBG in an optical fiber prior to annealing-trimming characteristics of the FBG by performing the sub-steps of monitoring characteristic data of the FBG while generating a controlled complex temperature profile along the FBG with a heating means according to the characteristic data for providing an accurate controlled annealing process of the FBG, thereby providing an accurate trimming thereof.
Owner:TERAXION
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com