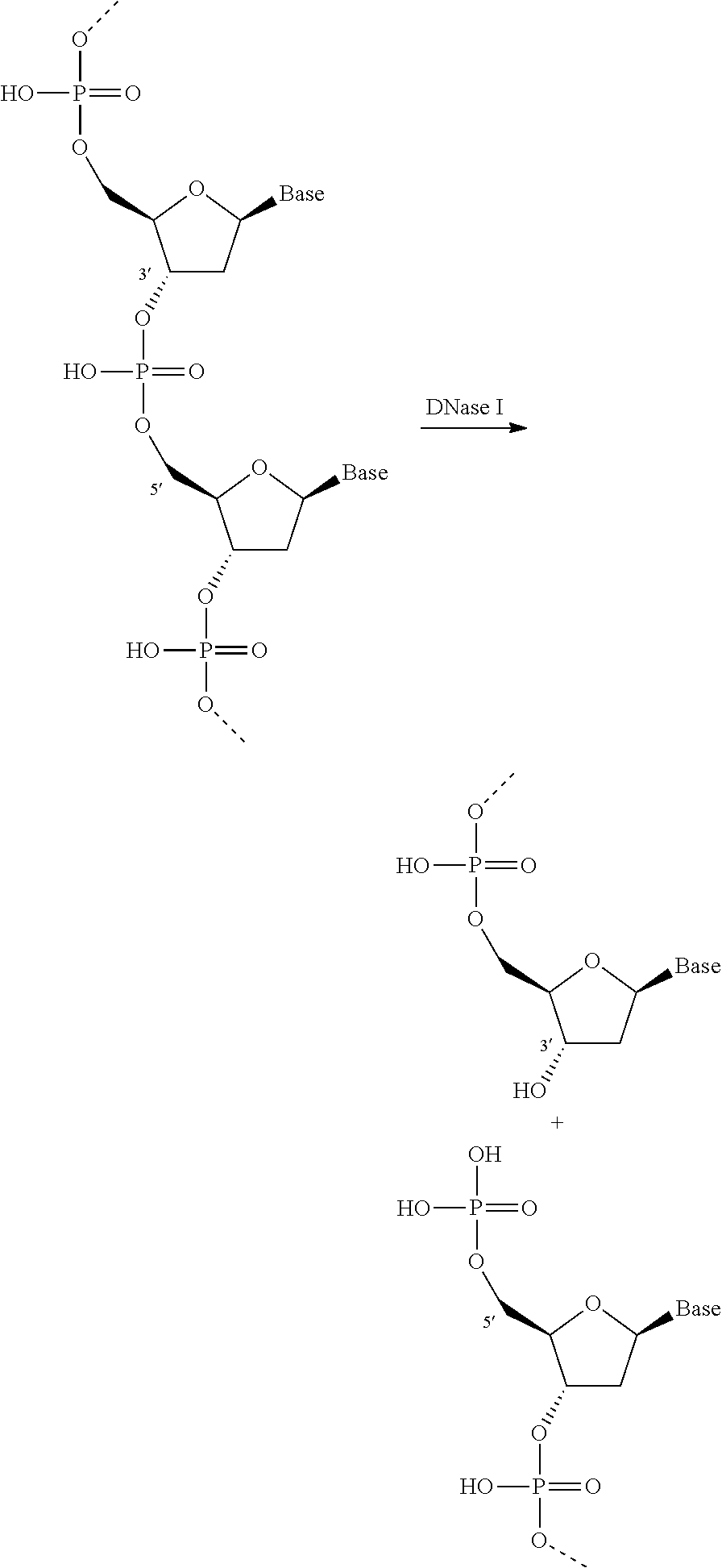
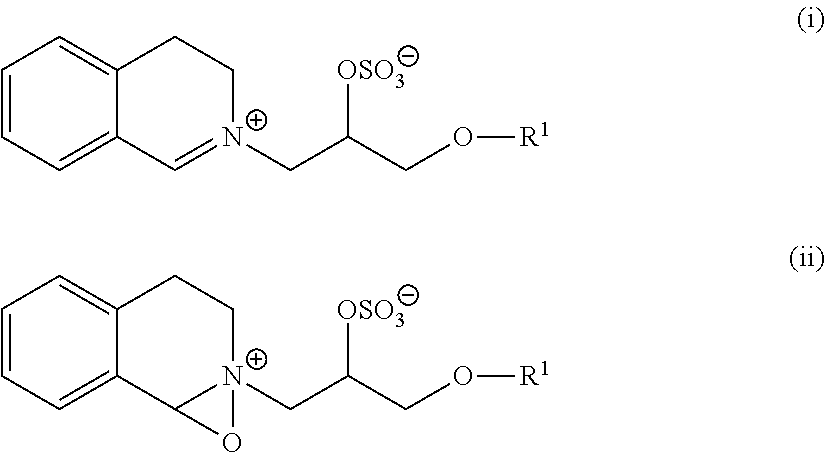
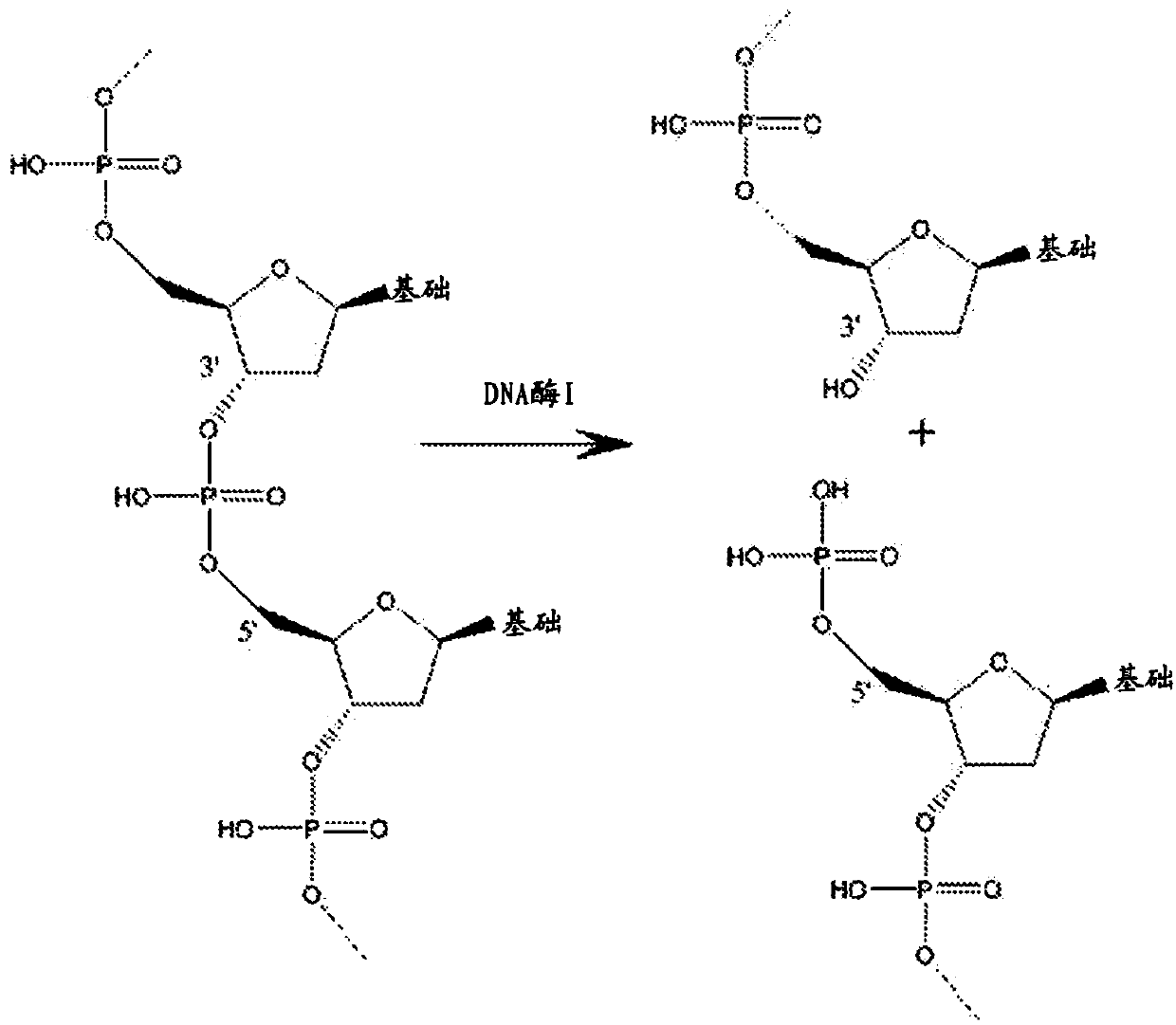
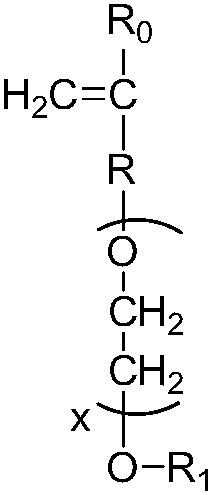
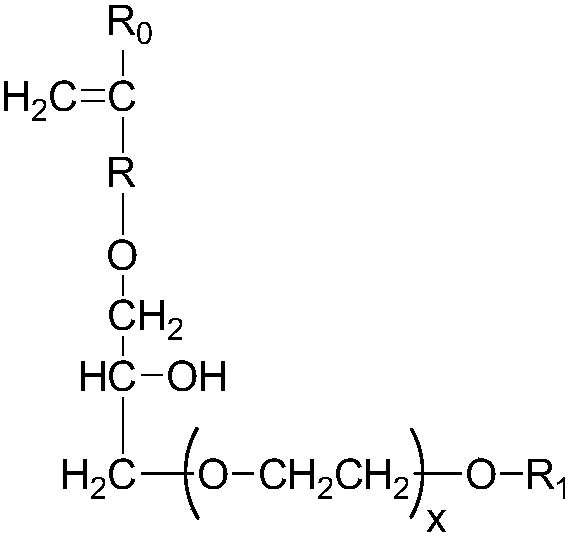
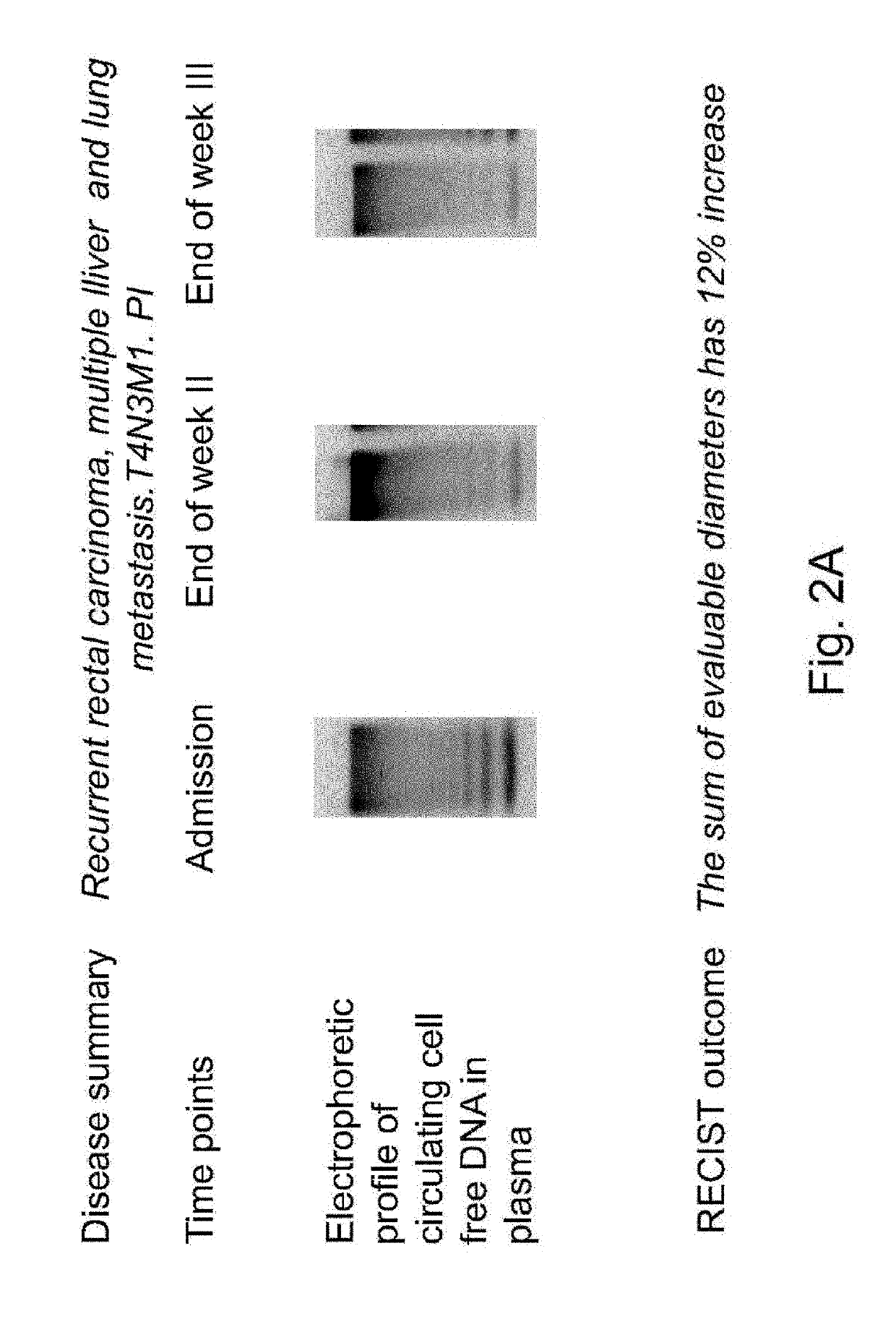
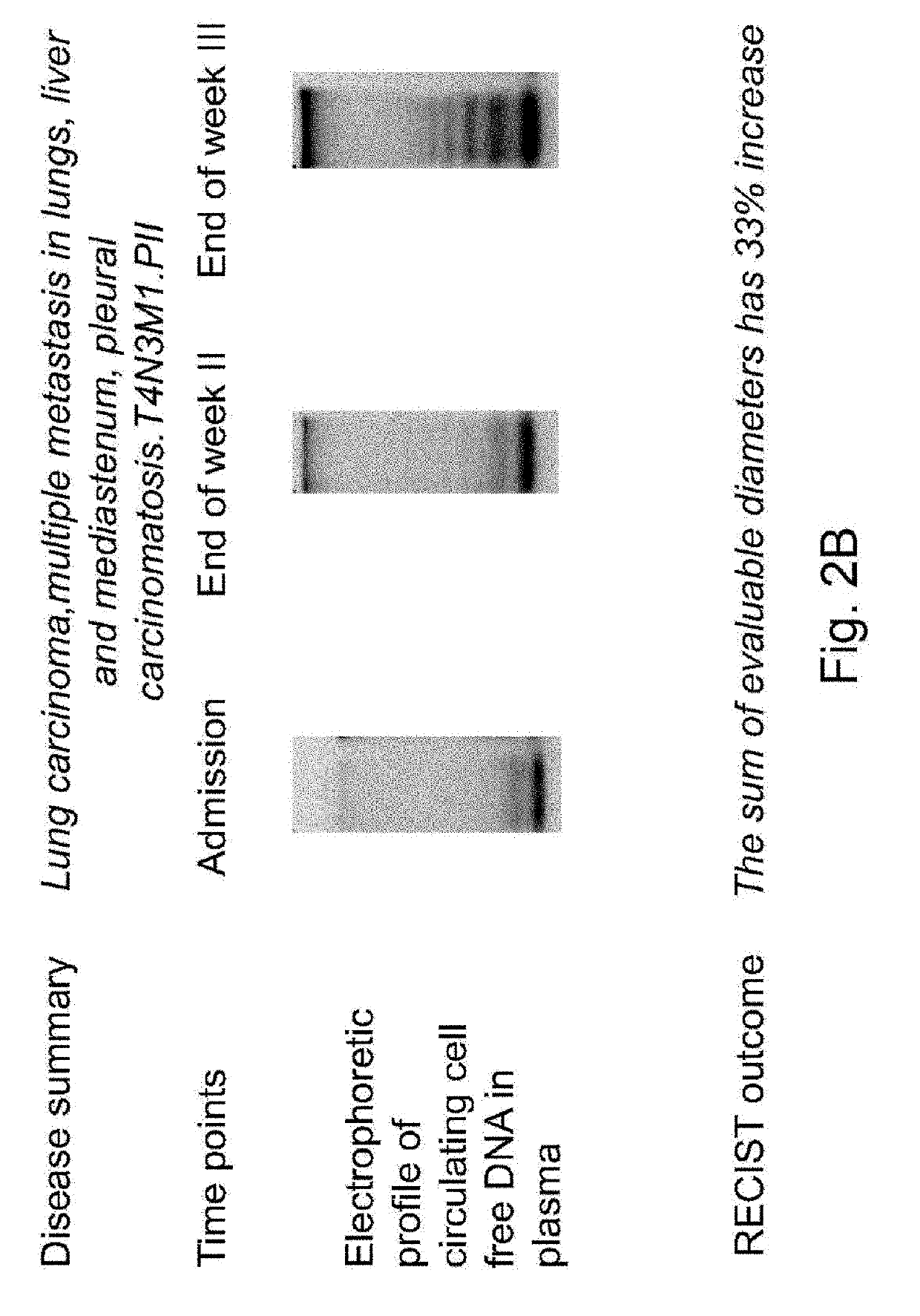
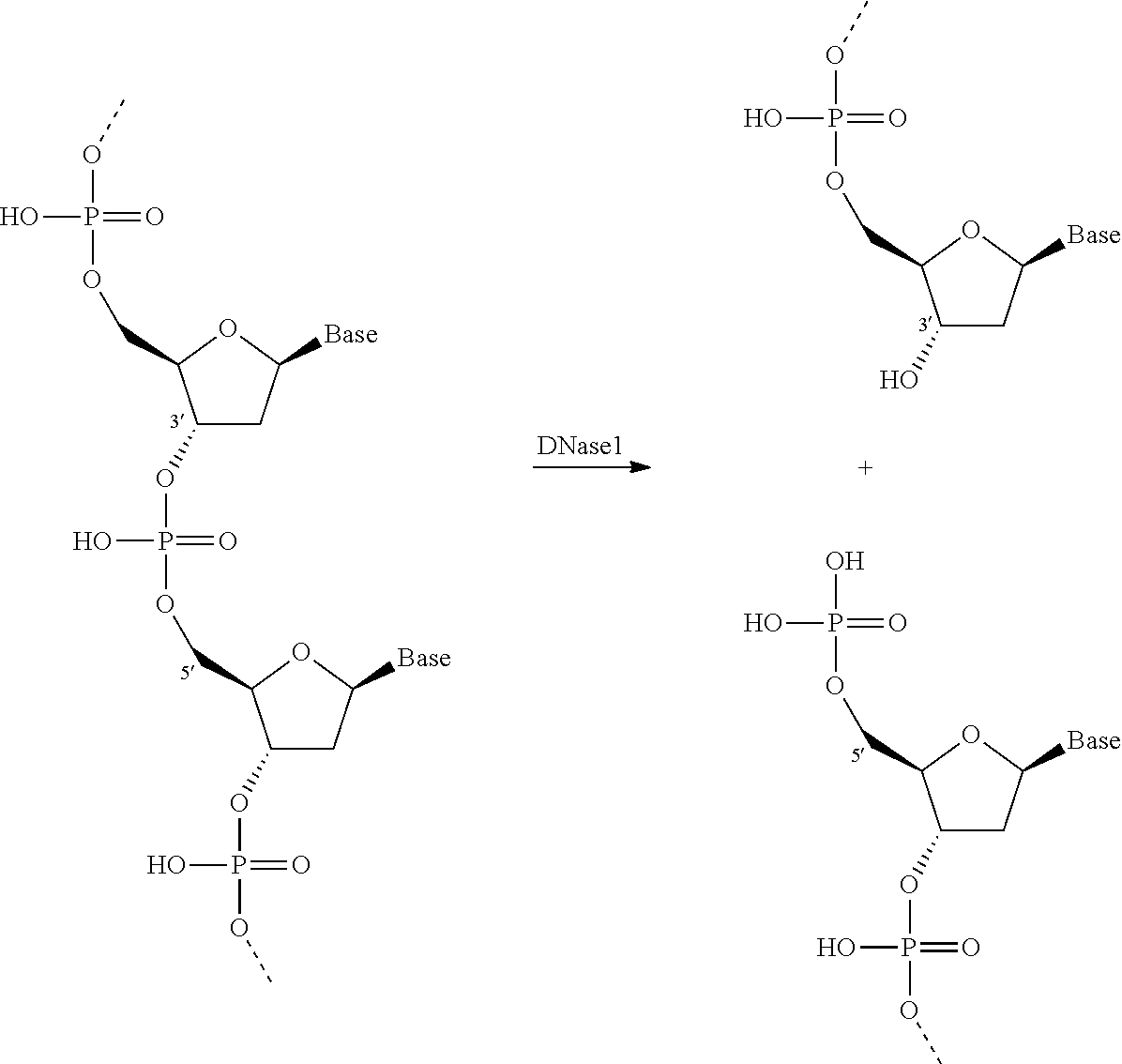
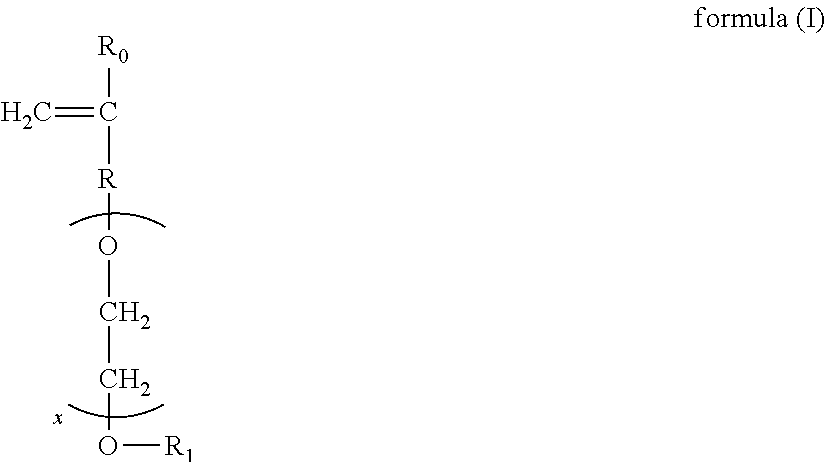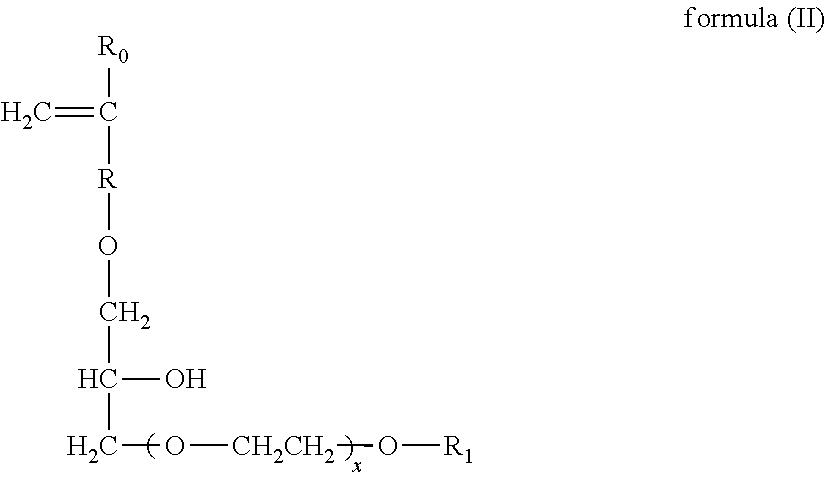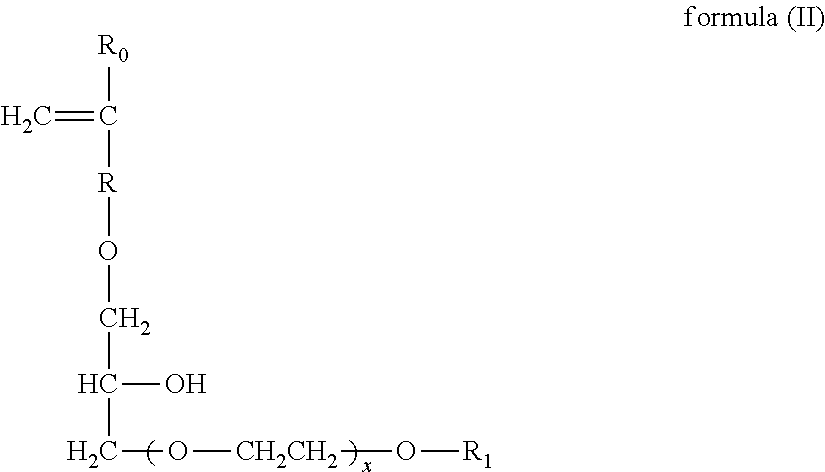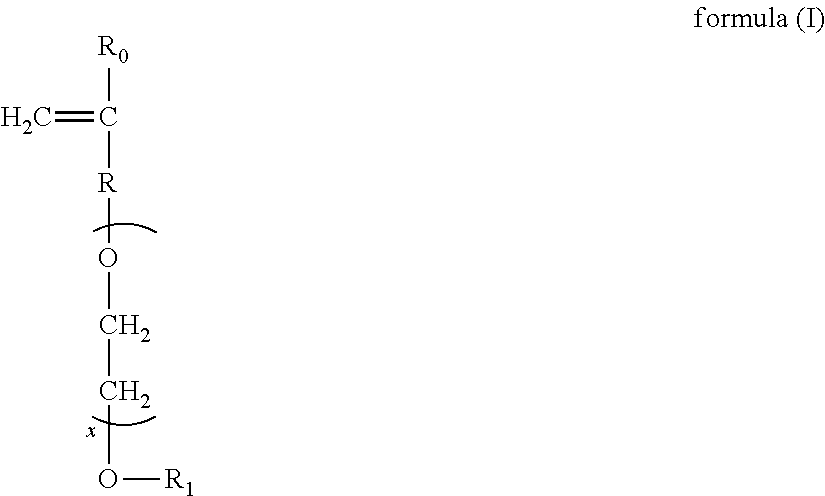Patents
Literature
62 results about "A deoxyribonuclease" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
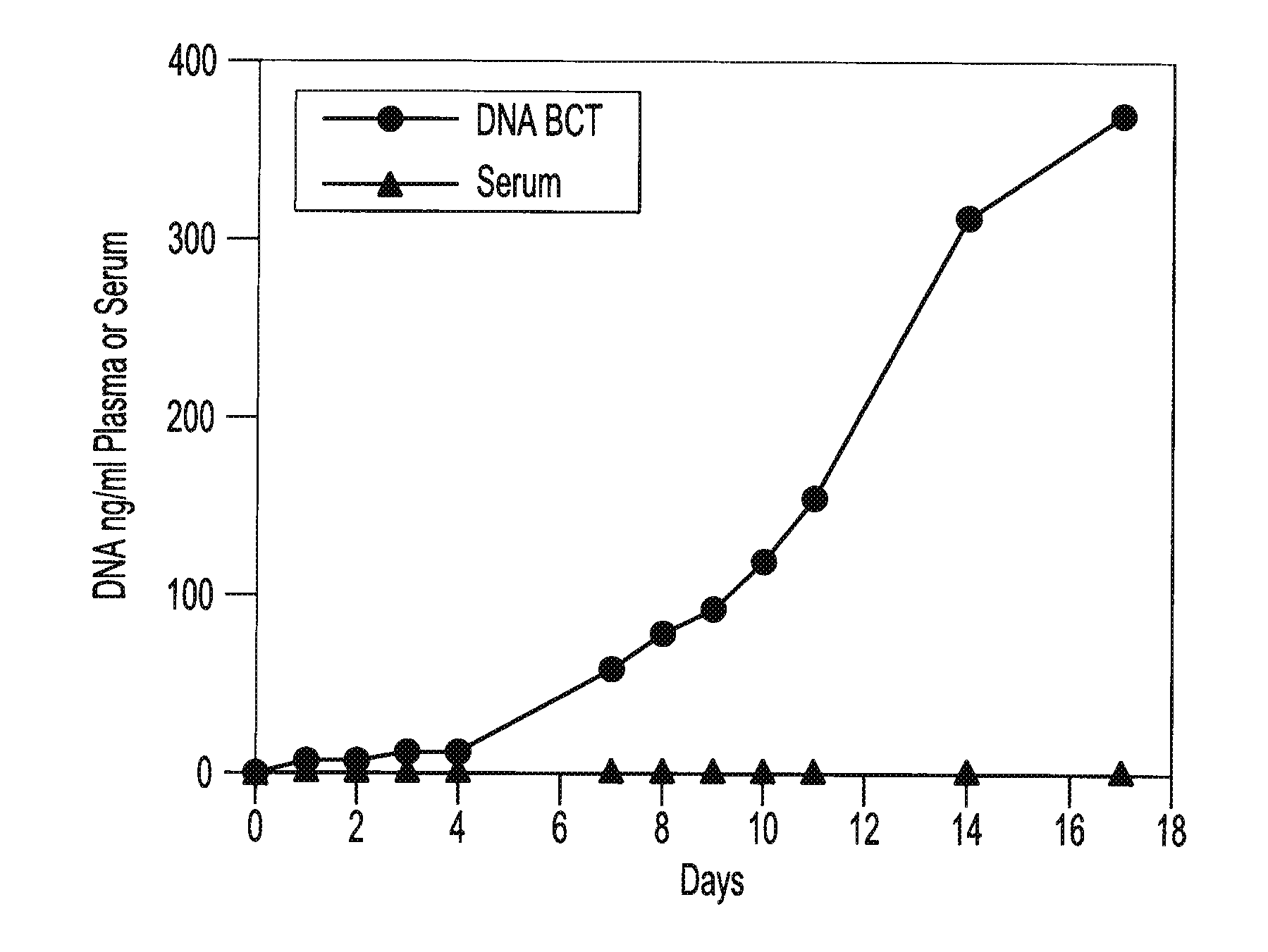
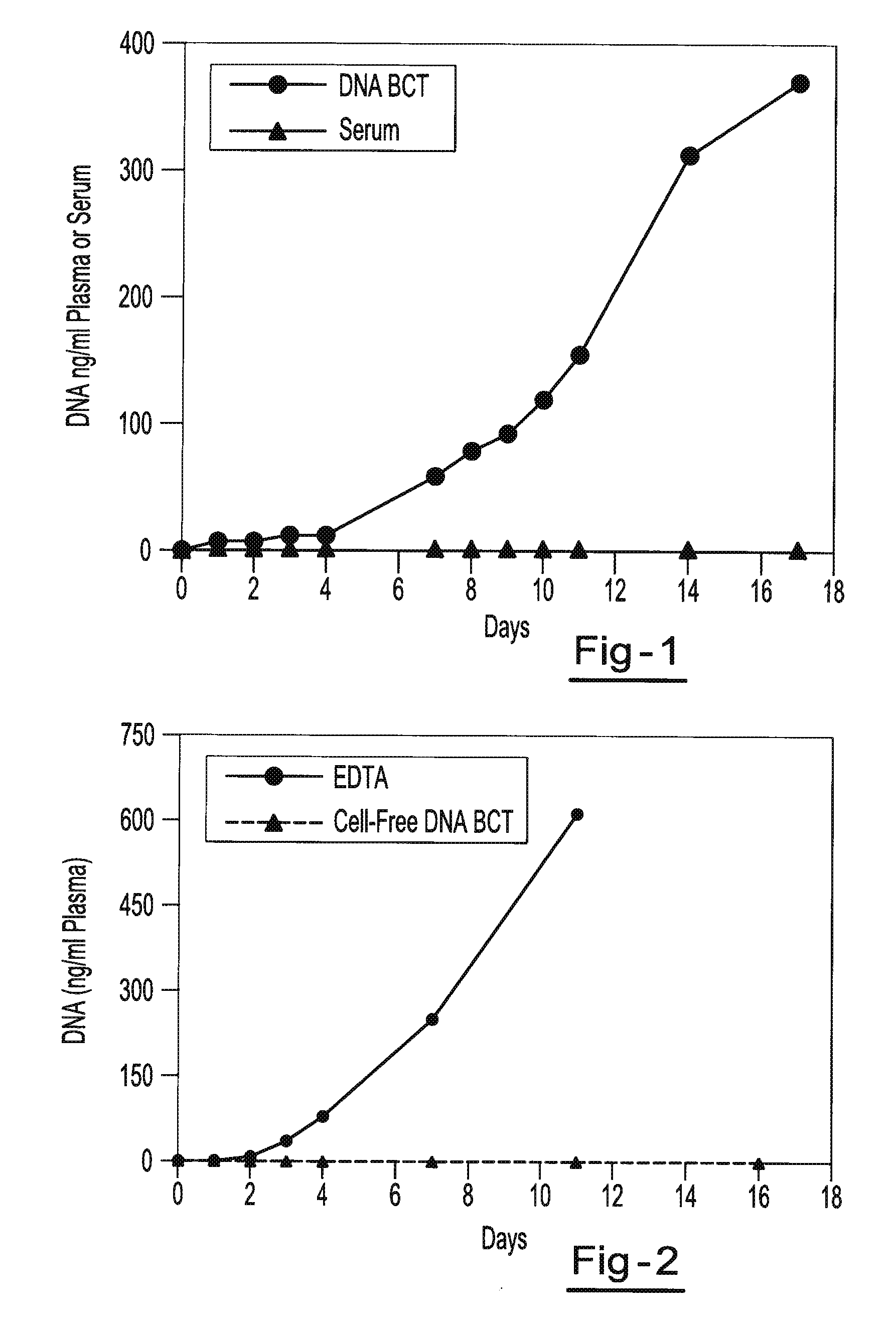
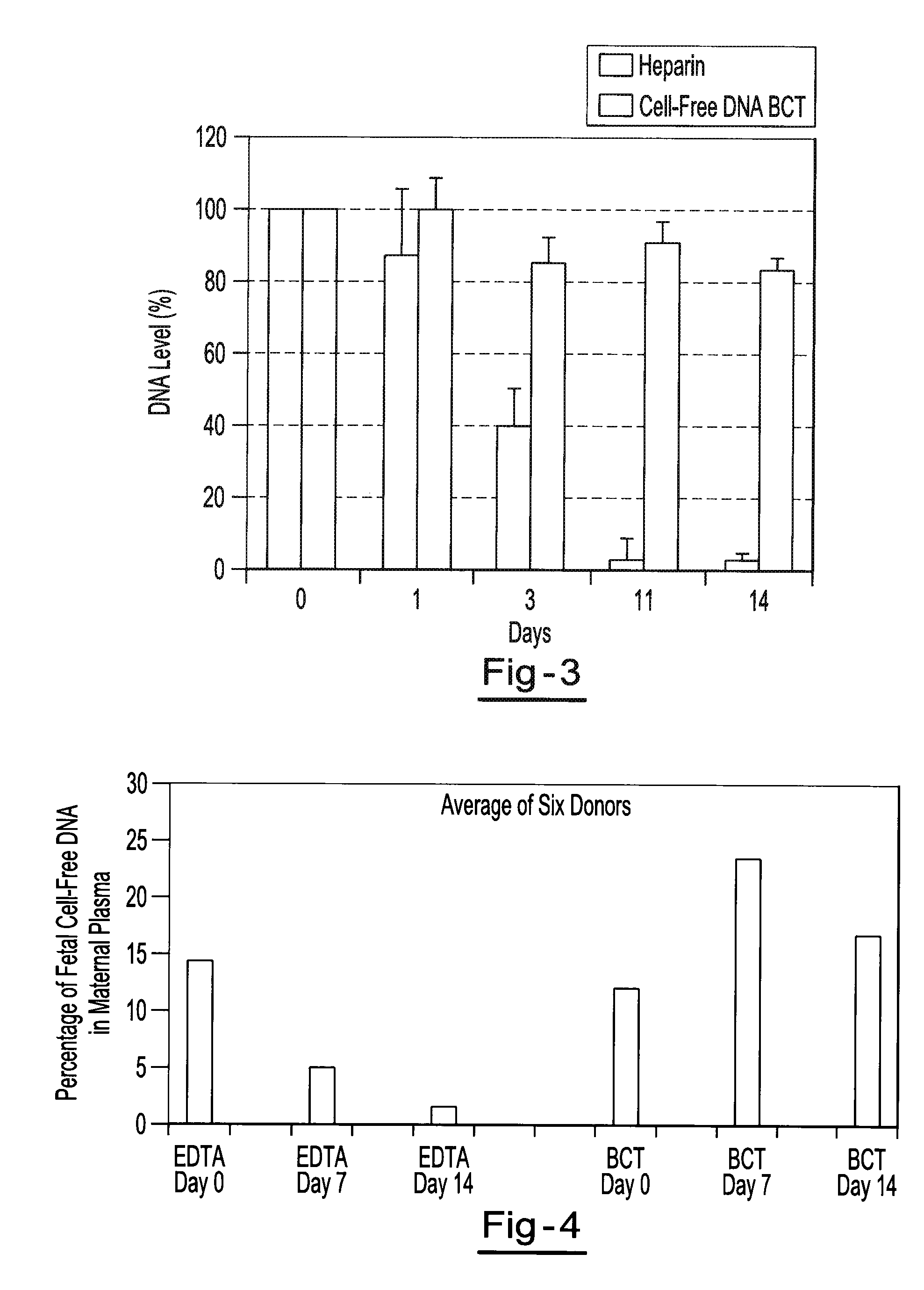
Preservation of fetal nucleic acids in maternal plasma
ActiveUS20100184069A1Maintain structural integrityInhibition releaseMicrobiological testing/measurementRibonucleaseLysis
A method for preserving and processing fetal nucleic acids located within maternal plasma is disclosed, wherein a sample of maternal blood containing fetal nucleic acids is treated to reduce both cell lysis of the maternal blood cells and deoxyribonuclease (DNase) and ribonuclease (RNase) activity within the fetal nucleic acids. The treatment of the sample aids in increasing the amount of fetal nucleic acids that can be identified and tested while maintaining the structure and integrity of the fetal nucleic acids.
Owner:STRECK LLC
Detergent composition
InactiveUS20160319228A1Non-ionic surface-active compoundsOrganic detergent compounding agentsNon ionicSURFACTANT BLEND
A cleaning composition comprising a nuclease enzyme, preferably a deoxyribonuclease and / or ribonuclease enzyme and a surfactant system comprising an anionic surfactant and a nonionic surfactant wherein the weight ratio of anionic to non-ionic surfactant is from 1.5:1 to 1:10.
Owner:THE PROCTER & GAMBLE COMPANY
Enzymatic DNA molecules
InactiveUS7141665B1Sugar derivativesMicrobiological testing/measurementNucleic acid sequencingEnzyme
The present invention discloses deoxyribonucleic acid enzymes—catalytic or enzymatic DNA molecules—capable of cleaving nucleic acid sequences or molecules, particularly RNA, in a site-specific manner, as well as compositions including same. Methods of making and using the disclosed enzymes and compositions are also disclosed.
Owner:THE SCRIPPS RES INST
Molecular and bioinformatics methods for direct sequencing
InactiveUS20160180018A1Fast and accurate methodMinimize degradationMicrobiological testing/measurementMicroorganism lysisBiotechnologyRNA - Ribonucleic acid
The present invention relates to methods for preparing an isolated biological sample containing at least one of DNA and RNA, such that the DNA and / or RNA is preserved in the sample at ambient temperatures for at least thirty days, the method comprising: contacting the isolated biological sample with a composition comprising a chaotropic agent, and subjecting the contacted sample to microbial cell lysis; and optionally, contacting the lysed biological sample with a slurry of size-selected silicon dioxide to form at least one of DNA-silicon dioxide complexes or RNA-silicon dioxide complexes in the sample; isolating at least one of DNA-silicon dioxide complexes or RNA-silicon dioxide complexes from the sample; and, separating at least one of DNA and RNA from the silicon dioxide and collecting at least one of the DNA and RNA.The present invention further relates to methods for preparing an isolated biological sample, the method comprising, separating the components in an isolated biological sample according to their size, wherein the components are at least one of DNA and RNA; purifying and isolating SSU rRNA from the biological sample using a composition comprising a ribonuclease inhibitor and a deoxyribonuclease to remove DNA from the sample, reverse transcribing the SSU rRNA into ds cDNA using random primers for SSU rRNA.The present invention also relates to computer implemented methods comprising, receiving an isolated sample prepared according to the methods of the invention, sequencing the sample, and providing the sequence with a sequence identifier (ID), the sequence comprising a plurality of groups of k-mers, each group of k-mers defining a node in a multi-level hierarchy which defines the relationship between the groups of k-mers; providing each group of k-mers with a respective group identifier (ID), determining the frequency of the k-mers in each group; generating a group signature array for each group of k-mers, each group signature array comprising the k-mers in each group that have the most increased frequency compared with the sibling k-mers; generating a signature map comprising each group signature array and at least one of the identifiers, the identifier of at least one parent group and the identifier of at least one child group; and outputting the signature map to be used to classify the sequence.
Owner:16S TECH INC
Detergent Composition
InactiveUS20170152462A1Improve washing effectReduce redepositionHydrolasesDetergent mixture composition preparationA deoxyribonucleaseChemistry
The present invention concerns a detergent and a pharmaceutical composition comprising a deoxyribonuclease (DNase), wherein the DNase is obtained from a fungal source. It further concerns a laundering method and the use of DNase. The present invention further relates to polypeptides having DNase activity, as well as methods of producing the polypeptides.
Owner:NOVOZYMES AS
Method of treating a fabric
InactiveUS20160319225A1Inorganic/elemental detergent compounding agentsSurface-active detergent compositionsBenzeneRibonuclease
A method of treating a textile in which a textile is contacted with an aqueous solution comprising a nuclease enzyme, preferably a deoxyribonuclease or ribonuclease enzyme and low levels of alkyl benzene sulphonate surfactant, and optionally rinsing and drying the textile.
Owner:THE PROCTER & GAMBLE COMPANY
Detergent Composition
ActiveUS20170107457A1Improve washing effectReduce redepositionHydrolasesDetergent mixture composition preparationNucleotideDNase activity
The present invention concerns a detergent and a pharmaceutical composition comprising a deoxyribonuclease (DNase), wherein the DNase is obtained from a fungal source. It further concerns a laundering method and the use of DNase. The present invention further relates to polypeptides having DNase activity, nucleotides encoding the polypeptide, as well as methods of producing the polypeptides.
Owner:NOVOZYMES AS
Method for extracting total ribonucleic acid from plants with polysaccharide and polyphenol by using silica membrane
The invention relates to a method for extracting total ribonucleic acid from plants with polysaccharide and polyphenol by using a silica membrane, and belongs to the field of nucleic acid purification. The method comprises the following steps of: adding a lysis solution for the plants with polysaccharide and polyphenol into a plant material with polysaccharide and polyphenol, which is ground by liquid nitrogen, performing centrifugal separation on the mixture, taking the supernate obtained through centrifugal separation out, transferring into a filtration column for filtration, adding absolute ethanol into filtrate, uniformly mixing, transferring into a silica membrane adsorbing column for adsorbing centrifugation, adding a protein removing solution and a deoxyribonuclease solution for protein removal centrifugation twice, adding a rinsing solution for desalting centrifugation, performing drying centrifugation, and adding deoxyribonuclease water for rinsing centrifugation to obtain a ribonucleic acid solution. The method has the characteristics of high efficiency, quickness and conciseness; and the purified ribonucleic acid (RNA) can be used for various downstream experiments suchas microarray analysis, in vitro translation, molecular cloning and the like.
Owner:TIANGEN BIOTECH BEIJING
Method of treating a fabric
ActiveUS20160319226A1Improve hydrophobicityIncrease depositionCationic surface-active compoundsNon-ionic surface-active compoundsRibonucleaseNuclease
Method for treating a hydrophobically-modified textile with an aqueous solution of a nuclease enzyme, preferably a deoxyribonuclease or ribonuclease enzyme, rinsing and drying the textile. Use of the finishing step for improved cleaning with an aqueous solution of a nuclease enzyme, preferably a deoxyribonuclease or ribonuclease enzyme
Owner:THE PROCTER & GAMBLE COMPANY
Method for rapidly extracting total ribonucleic acid from blood
The invention relates to a method for rapidly extracting total ribonucleic acid from blood, belonging to the field of nucleic acid purification. The method comprises the following steps of: firstly, adding a rapid erythrocyte lysing solution into whole blood; performing centrifugal separation on a mixture of the rapid erythrocyte lysing solution and the whole blood; taking precipitate formed by the centrifugal separation out; adding suspension into the precipitate; adding a lysing solution and a protease K solution in sequence; uniformly mixing and then adding absolute ethyl alcohol; uniformly mixing and transferring into a silicon membrane adsorption column to perform adsorptive centrifugation; adding a deproteinizing solution and a deoxyribonuclease solution; performing deproteinizing centrifugation twice; adding a rinsing solution to perform desalting centrifugation; and finally performing drying centrifugation and adding water without ribonuclease, and eluting and centrifuging so as to obtain a ribonucleic acid solution. The method provided by the invention has the characteristics of high efficiency, rapidness and simplicity; and purified RNA (Ribonucleic Acid) can be used for various downstream experiments such as chip analysis, in vitro translation, molecular cloning and the like.
Owner:TIANGEN BIOTECH BEIJING
Detergent composition
PendingCN107820515AInorganic/elemental detergent compounding agentsNon-ionic surface-active compoundsRibonucleaseActive agent
A cleaning composition comprising a nuclease enzyme, preferably a deoxyribonuclease and / or ribonuclease enzyme and a surfactant system comprising an anionic surfactant and a nonionic surfactant wherein the weight ratio of anionic to non-ionic surfactant is from 1.5:1 to 1:10.
Owner:PROCTER & GAMBLE CO
Detergent composition
ActiveUS10131863B2Soap detergents with organic compounding agentsOrganic detergent compounding agentsNucleotideDNase activity
Owner:NOVOZYMES AS
Multi-enzyme system for separating different tissues-derived primary culture cells of normal human and mammal, application and related kit thereof
InactiveCN101914504AEasy to freezePromote recoveryHydrolasesNervous system cellsBiotechnologyNeutral protease
The invention relates to a multi-enzyme system for separating different tissues-derived primary culture cells of a normal human and a mammal. The system comprises a plurality of 0.0125 to 0.125 weight percent of trypsin, 0.02 to 0.2 weight percent of collagenase I-IV, 0.0015 to 0.015 weight percent of hyaluronidase, 0.02 to 0.2 weight percent of neutral protease, 0.0015 to 0.015 weight percent of deoxyribonuclease I, 1 to 10 U / ml papain, 0.015 to 0.15 weight percent of pronase, 5 to 50 U / ml elastase and 0.01 to 0.1 weight percent of separase. The invention also relates to the application of the multi-enzyme system and a kit formed by the multi-enzyme system. The multi-enzyme system of the invention has the characteristics of reasonable design and capacity of separating the different tissues-derived primary culture cells of the normal human and the mammal highly specifically and highly sensitively; and the separated primary culture cells have the characteristics of high survival rate, high purity, easy cryopreservation and recovery, can be used for medicament experimental evaluation, medicament toxicity test and medicament therapeutic judgment, and are suitable for large-scale popularization and application.
Owner:刘东旭
Method of treating a fabric
ActiveCN107667166AInorganic/elemental detergent compounding agentsCationic surface-active compoundsRibonucleaseNuclease
Owner:PROCTER & GAMBLE CO
Mixed enzyme for skin tissue dissociation, preparation method of mixed enzyme, dissociation kit and dissociation method
ActiveCN112708610AHigh activityImprove experimental efficiencyCell dissociation methodsEpidermal cells/skin cellsDNA - Deoxyribonucleic acidDeoxyribonuclease I
The invention discloses a mixed enzyme for skin tissue dissociation. The mixed enzyme is composed of collagenase II, trypsin and deoxyribonuclease I, wherein the concentration of the collagenase II is 2-4 mg / mL; the concentration of the trypsin is 1-3 mg / mL; and the concentration of the deoxyribonuclease I is 0.01-0.05 mg / mL. Besides, the invention further discloses a dissociation method for the skin tissue. The skin tissue is pretreated with a specific buffer solution, and tissue dissociation is conducted through the specific mixed enzyme, so that a skin tissue single-cell suspension with large number of single cells, good activity, few fragments and low clustering rate can be efficiently, rapidly and stably obtained. The screened and optimized mixed enzyme and concentration have higher pertinence and higher efficiency; more costs and time are saved compared with those in a method of combining a few collagenases in a broad spectrum; and a good tool is provided for single-cell related research of a skin tissue sample which is always difficult to dissociate.
Owner:上海伯豪生物技术有限公司
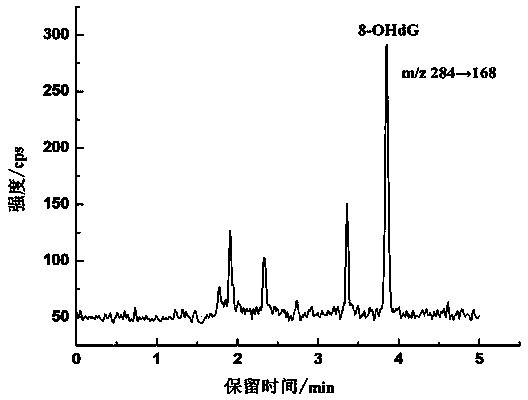
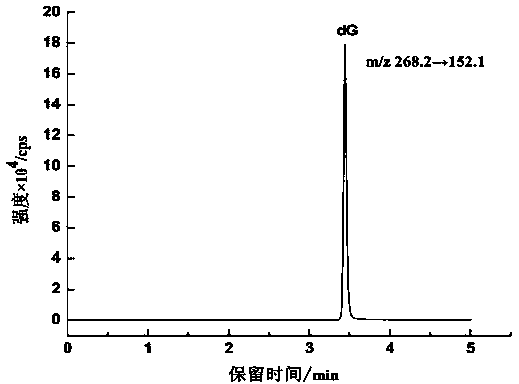
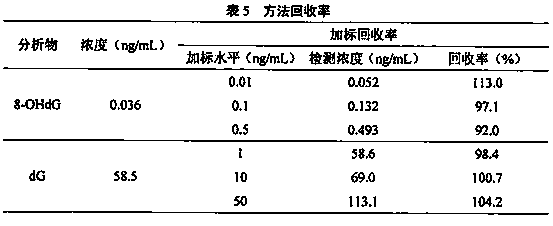
Method for detecting 8-hydroxy-2'-deoxyguanosine (8-OHdG) and 2'-deoxyguanosin (dG) in cell DNA
InactiveCN104181250AHigh sensitivityAvoid interferenceComponent separationDeferoxamine mesylateHydrolysate
The invention belongs to a cell DNA detection technology and in particular relates to a method for detecting 8-hydroxy-2'-deoxyguanosine (8-OHdG) and 2'-deoxyguanosin (dG) in cell DNA. The method can be used for quantitatively detecting the change of the content of 8-OHdG and dG in cell DNA, caused by exposure of harmful ingredients in cigarette smoke, so that the purpose of evaluating DNA oxidation damage caused by induction of the harmful ingredients can be achieved. According to the method, deoxyribonuclease I and alkaline phosphatase are used for hydrolyzing DNA in a cell, and deferoxamine mesylate is added into enzymatic hydrolysate, so that the oxidation of dG in the enzymolysis process is avoided; enzymatic hydrolysate is detected by HPLC-MS / MS; the cell DNA oxidation damage degree is evaluated according to the value of 8-OHdG / 10<6>dG. By virtue of the method, the interference of high-concentration metal ions and artificial dG oxidation is removed, and thus the result is relatively accurate; the method is low in detection limit, high in sensitivity, good in repeatability and high in recovery rate.
Owner:ZHENGZHOU TOBACCO RES INST OF CNTC
Method of treating a fabric
A method of treating a fabric, comprising contacting the fabric with an aqueous liquor comprising a fabric care component selected from the group consisting of cationic softening-compounds, silicone softening-compounds, paraffins, dispersible polyolefins, waxes and mixtures thereof; and contacting the fabric with an aqueous liquor comprising a nuclease enzyme, preferably a deoxyribonuclease or ribonuclease enzyme. The aqueous liquor may be provided by adding a cleaning or treatment composition to water. Preferably the composition comprises a surfactant, the wash liquor comprising from 0.05 to 4 g / l of a surfactant.
Owner:THE PROCTER & GAMBLE COMPANY
Method to improve safety and efficacy of Anti-cancer therapy
InactiveUS20170100463A1Avoid toxicityGood curative effectPeptide/protein ingredientsHydrolasesCytotoxicityCancer therapy
The invention relates to the use of a deoxyribonuclease (DNase) enzyme for prevention or amelioration of toxicity associated with various cytostatic and / or cytotoxic chemotherapeutic compounds and radiation therapy.
Owner:CLS THERAPEUTICS
Synthesis method and application of fluorescence DNA-polyphenylene acetylene hydrogel
InactiveCN103053517AThe synthesis method is simpleMild conditionsBiocideDisinfectantsFluorescenceSinglet oxygen
The invention discloses a synthesis method for synthesizing a fluorescence DNA-polyphenylene acetylene hydrogel, and the method adopts DNA as a hydrogen netlike frame to synthesize a novel DNA hybrid hydrogel together with polyphenylene acetylene by electrostatic interaction. The synthesis method is simple and mild in condition; the hydrogel can effectively coat the polyphenylene acetylene in the netlike frame of the DNA so as to prevent the polyphenylene acetylene from having contact with bacteria; the hydrogel can be applied to sterilization regulation and control; when the hydrogel is added with deoxyribonuclease, the netlike frame of the DNA is damaged to release polyphenylene acetylene, the dissociative polyphenylene acetylene can approach to the bacteria or enter the bacteria to generate singlet oxygen or active oxygen substance by the irradiation of white light, so that the bacteria are killed; and the deoxyribonuclease can be utilized to realize controllable sterilization.
Owner:SHAANXI NORMAL UNIV
Detergent composition
InactiveUS20180105772A1Improve the immunityDifferent propertyHydrolasesSurface-active detergent compositionsCombinatorial chemistryA deoxyribonuclease
The present invention concerns a detergent comprising a deoxyribonuclease (DNase). The present invention further relates to methods and uses of the detergent comprising a deoxyribonuclease (DNase) for laundering.
Owner:NOVOZYMES AS
Rapid construction method of genome DNA sequencing library, and matched kit
PendingCN109853047AGet rid of the ultrasound stepLow costLibrary creationEnzyme digestionMagnetic bead
The invention discloses a rapid construction method of a genome DNA sequencing library, and a matched kit. The method comprises the following steps: (1) selecting DNase I deoxyribonuclease I as a DNAfragmenting enzyme to fragment a sample DNA by enzyme digestion to obtain fragmented DNA; (2) directly adding a dA tail to the fragmented DNA to obtain DNA with the dA tail; (3) connecting the DNA with the dA tail with a joint to obtain a joint connected product; (4) carrying out magnetic-bead purification on the joint connected product; (5) carrying out polymerase chain reaction (PCR) amplification on the joint connected product to obtain an amplification product; (6) carrying out magnetic-bead purification on the amplification product to obtain a genome DNA sequencing library product; and (7) carrying out fragment distribution inspection and library evaluation on the library product by adopting an agarose gel electrophoresis detection method. According to the invention, the DNase I is selected as the DNA fragmenting enzyme, so that the steps of tail end repair and phosphorylation are reduced, the library construction time is shorter, and cost is lower.
Owner:YEASEN BIOTECHNOLOGY (SHANGHAI) CO LTD
Method to improve safety and efficacy of anti-cancer therapy
ActiveUS10617743B2Avoid toxicityGood curative effectOrganic active ingredientsHydrolasesChemical compoundEfficacy
The invention relates to the use of a deoxyribonuclease (DNase) enzyme for prevention or amelioration of toxicity associated with various cytostatic and / or cytotoxic chemotherapeutic compounds and radiation therapy.
Owner:CLS THERAPEUTICS
Mesenchymal stem cell separated from placenta blood vessel with digestive enzyme composition
ActiveCN109628388AGood differentiation effectHigh purityCell dissociation methodsSkeletal/connective tissue cellsBlood Vessel TissueHyaluronidase
The invention relates to a mesenchymal stem cell separated from the placenta blood vessel with a digestive enzyme composition, in particular to the digestive enzyme composite used for a method for separating the placenta mesenchymal stem cell from the placenta blood vessel. A buffering solution containing tissue digestive enzymes is provided with added digestive enzymes which include pancreatic enzymes, deoxyribonuclease I, collagenase II, collagenase IV and hyaluronidase. In addition, the invention further relates to the method for separating the mesenchymal stem cell from the placenta bloodvessel, and the method includes the steps that the placenta is sterilized; the placenta vessel is separated from the placenta; cutting, cleaning and bloodiness filtering removal are conducted, so thatplacenta blood vessel tissue is obtained; mixed enzyme liquor is added for digestion; digestion is stopped, and interstitial fluid is filtered and collected; cell sediments obtained through centrifuging are original placenta mesenchymal stem cells, re-suspending is conducted, sampling is conducted, and the number nucleated cells and the survival rate of the nucleated cells are calculated; the obtained cells are subjected to refrigeration preservation or continuous passage and / or identified, detected and subjected to refrigeration preservation and database creation. Through the method, the efficiency of separating the mesenchymal stem cell from the placenta blood vessel can be improved effectively.
Owner:BOYALIFE
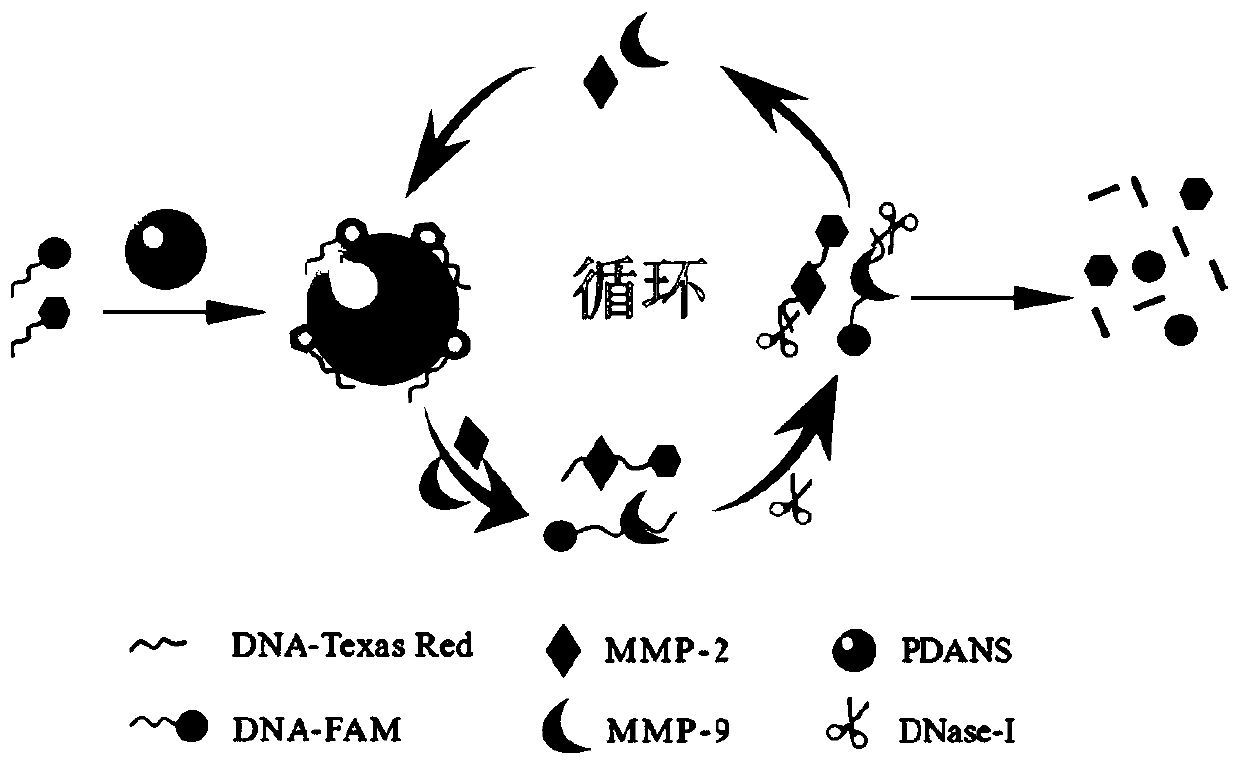
System for simultaneously detecting matrix metalloproteinase-9 and matrix metalloproteinase-2, preparation method thereof and application thereof
PendingCN110824164AAchieve improvementHigh sensitivityMicrobiological testing/measurementBiological material analysisAptamerDNA - Deoxyribonucleic acid
The invention discloses a system for simultaneously detecting matrix metalloproteinase-9 and matrix metalloproteinase-2, a preparation method thereof and application thereof. The system comprises a polydopamine nano microsphere-fluorescein modified nucleic acid aptamer nano sensor and deoxyribonuclease, and the sensor is formed by adding MMP-9-aptamer-FAM and MMP-2-aptamer-Texas Red into polydopamine nano microspheres. According to the invention, polydopamine nano microspheres with an excellent quenching effect are prepared, a PDANS-modified fluorescein nucleic acid aptamer nano sensor is constructed through non-public valence bond combination, DNase-I is added, aptamer is hydrolysed, an object to be detected is dissociated, cyclic reaction is carried out, the fluorescence signal amplification is realized, and the sensitivity of a system is improved. The system is simple in preparation, the reaction condition is mild, the cost is low, and a new tool and a new method are provided for simultaneous and rapid detection of the matrix metalloproteinase-9 and the matrix metalloproteinase-2.
Owner:CHINA PHARM UNIV
Treatment of diseases by liver expression of an enzyme which has a deoxyribonuclease (DNASE) activity
The invention relates to the liver-specific delivery and / or expression of an enzyme which has a deoxyribonuclease (DNase) activity for enhanced clearance of cell free DNA (cfDNA) accumulated in hepatic porto-sinusoidal circulation and the use of such liver-specific delivery and / or expression for treatment of various diseases and conditions, including cancer and neurodegeneration.
Owner:CLS治疗有限公司
Method of detecting methylated DNA in sample
InactiveUS20120219949A1Improve detection accuracyNovel methodMicrobiological testing/measurementSingle strand dnaDeoxyribonuclease I
In order to simply obtain a methylated DNA sample, a method including bringing a sample which may include methylated DNA into contact with a protein capable of binding to methylated DNA to bind the methylated DNA in the sample to the protein, bringing the sample obtained into contact with at least one deoxyribonuclease to degrade DNA in the sample; and detecting methylated DNA which is not degraded by the deoxyribonuclease by the binding of the sample obtained to the protein is used. In the method, at least one type of deoxyribonucleases is a deoxyribonuclease different from a methylation-sensitive restriction enzyme capable of degradating a single stranded DNA.
Owner:SYSMEX CORP
TREATMENT OF DISEASES BY LIVER EXPRESSION OF AN ENZYME WHICH HAS A DEOXYRIBONUCLEASE (DNase) ACTIVITY
PendingUS20190241908A1Inhibit transferEfficient and preferential targetingNervous disorderHydrolasesCell freeBlood vessel
The invention relates to the liver-specific delivery and / or expression of an enzyme which has a deoxyribonuclease (DNase) activity for enhanced clearance of cell free DNA (cfDNA) accumulated in hepatic porto-sinusoidal circulation and the use of such liver-specific delivery and / or expression for treatment of various diseases and conditions, including cancer and neurodegeneration.
Owner:CLS THERAPEUTICS
Method of treating a fabric
ActiveUS10513671B2Cationic surface-active compoundsOrganic detergent compounding agentsRibonucleaseNuclease
Method for treating a hydrophobically-modified textile with an aqueous solution of a nuclease enzyme, preferably a deoxyribonuclease or ribonuclease enzyme, rinsing and drying the textile. Use of the finishing step for improved cleaning with an aqueous solution of a nuclease enzyme, preferably a deoxyribonuclease or ribonuclease enzyme.
Owner:PROCTER & GAMBLE CO
Method of detecting methylated DNA in sample
In order to simply obtain a methylated DNA sample, a method including bringing a sample which may include methylated DNA into contact with a protein capable of binding to methylated DNA to bind the methylated DNA in the sample to the protein, bringing the sample obtained into contact with at least one deoxyribonuclease to degrade DNA in the sample; and detecting methylated DNA which is not degraded by the deoxyribonuclease by the binding of the sample obtained to the protein is used. In the method, at least one type of deoxyribonucleases is a deoxyribonuclease different from a methylation-sensitive restriction enzyme capable of degradating a single stranded DNA.
Owner:SYSMEX CORP
Method for rapidly extracting total ribonucleic acid from blood
The invention relates to a method for rapidly extracting total ribonucleic acid from blood, belonging to the field of nucleic acid purification. The method comprises the following steps of: firstly, adding a rapid erythrocyte lysing solution into whole blood; performing centrifugal separation on a mixture of the rapid erythrocyte lysing solution and the whole blood; taking precipitate formed by the centrifugal separation out; adding suspension into the precipitate; adding a lysing solution and a protease K solution in sequence; uniformly mixing and then adding absolute ethyl alcohol; uniformly mixing and transferring into a silicon membrane adsorption column to perform adsorptive centrifugation; adding a deproteinizing solution and a deoxyribonuclease solution; performing deproteinizing centrifugation twice; adding a rinsing solution to perform desalting centrifugation; and finally performing drying centrifugation and adding water without ribonuclease, and eluting and centrifuging so as to obtain a ribonucleic acid solution. The method provided by the invention has the characteristics of high efficiency, rapidness and simplicity; and purified RNA (Ribonucleic Acid) can be used forvarious downstream experiments such as chip analysis, in vitro translation, molecular cloning and the like.
Owner:TIANGEN BIOTECH BEIJING
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com