Patents
Literature
207results about How to "Cutting portion" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
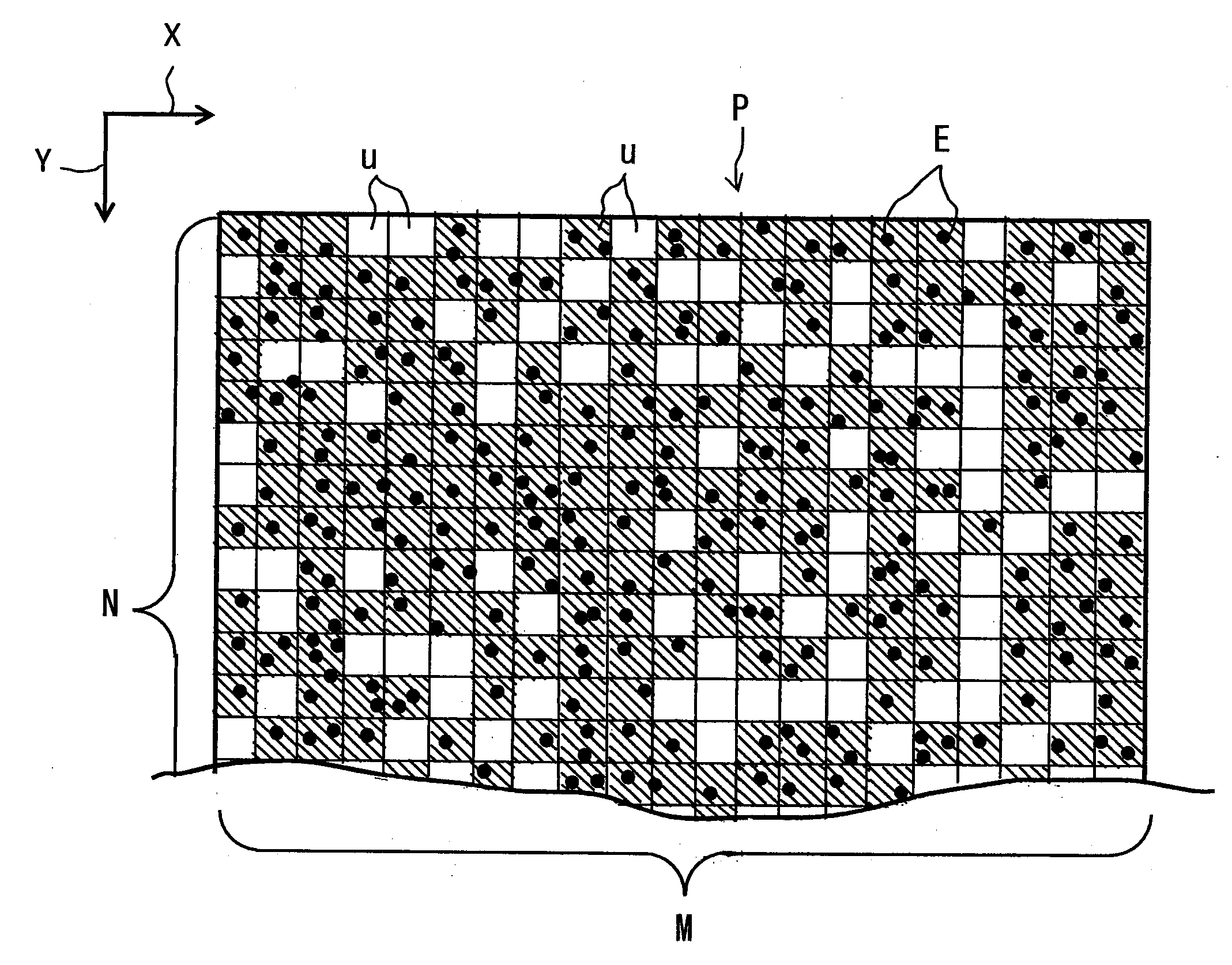
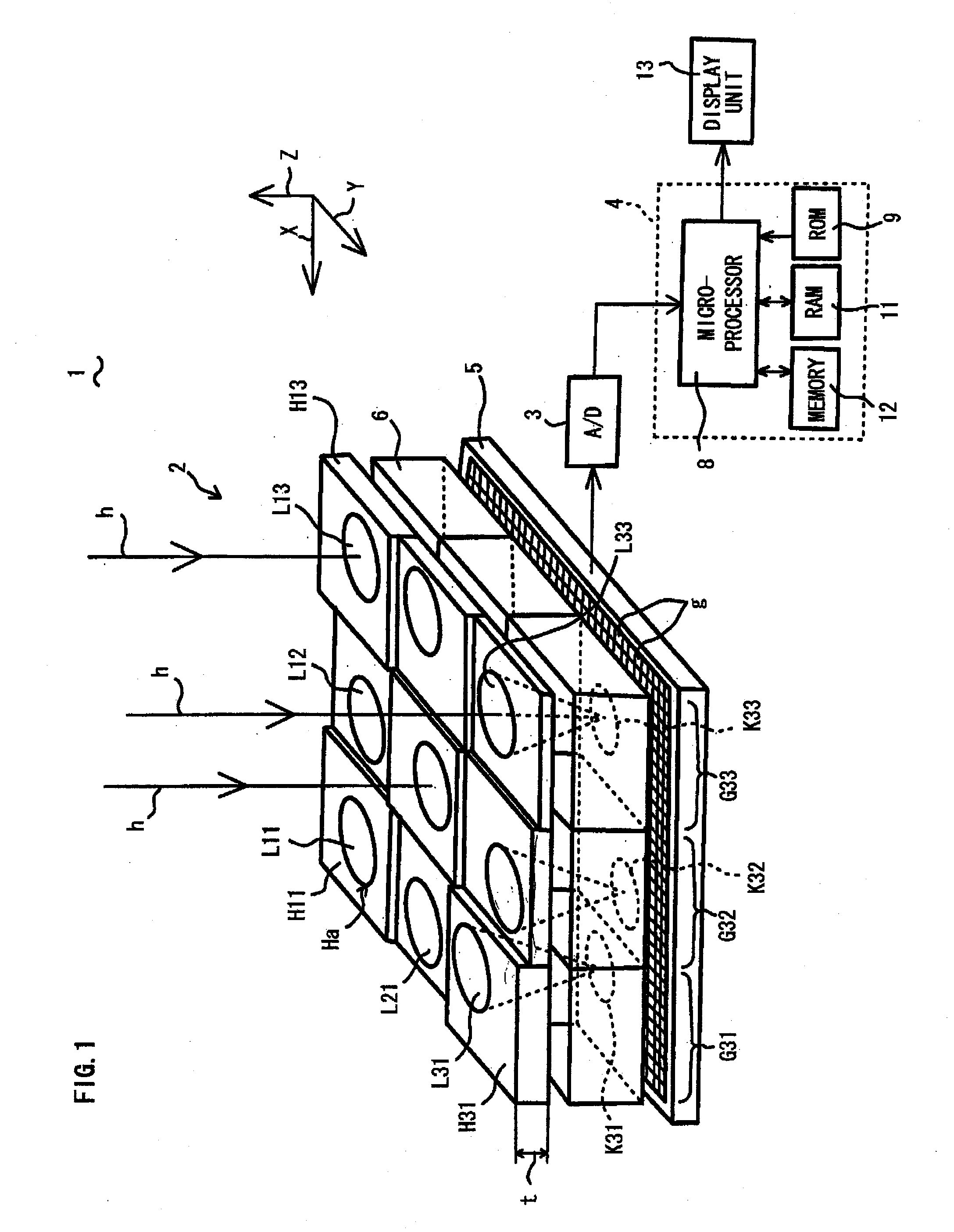
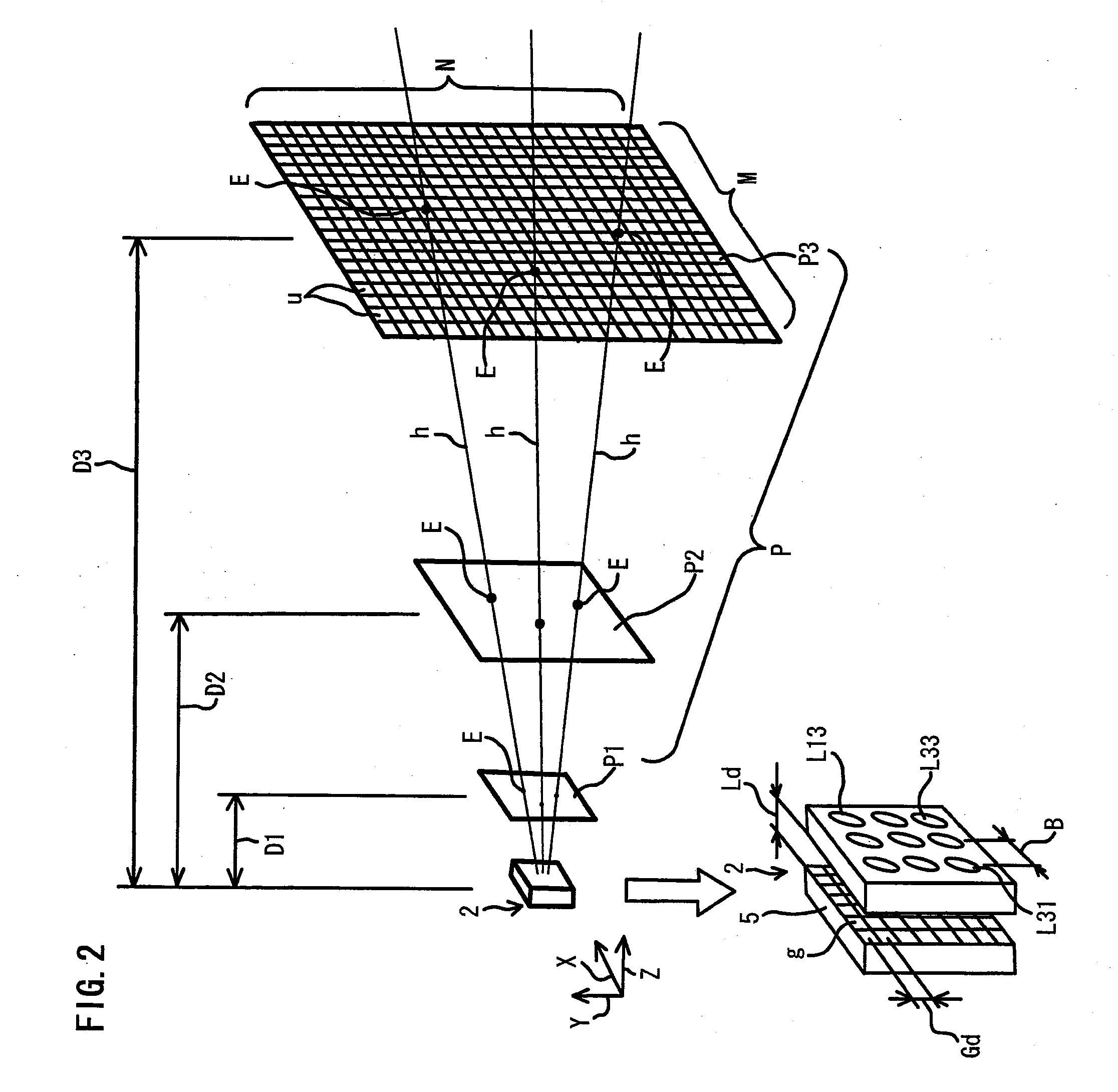
Optical Condition Design Method for a Compound-Eye Imaging Device
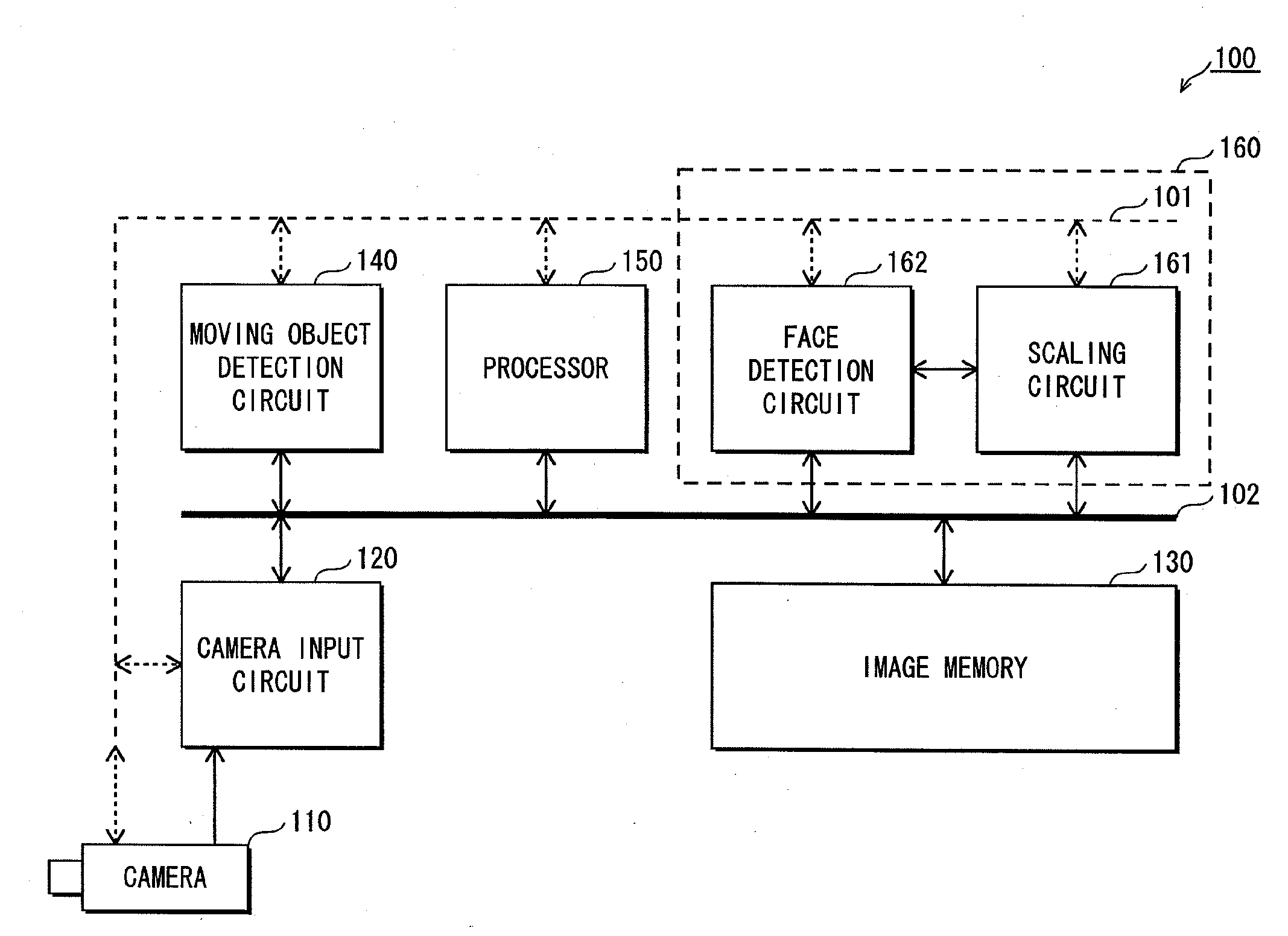
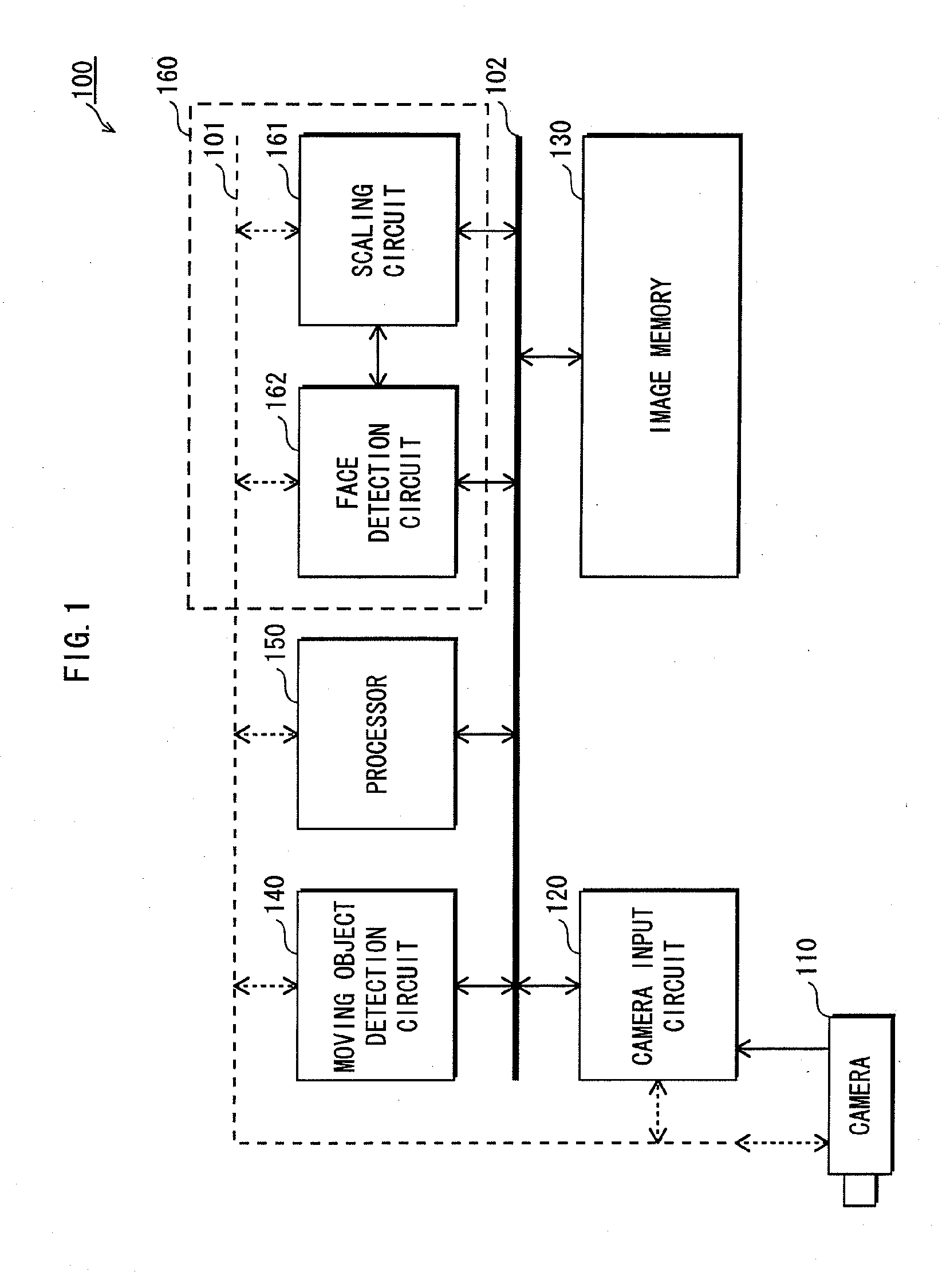
InactiveUS20100053600A1Cutting portionHigh definitionTelevision system detailsSolid-state devicesObservation pointHigh definition
An imaginary object plane is set in front of an imaging device body (plane setting step). A part of optical conditions of optical lenses are changed as variables, and positions of points (pixel observation points) on the imaginary object plane where lights coming from pixels of a solid-state imaging element and back-projected through the optical lenses are calculated (pixel observation point calculating step). The dispersion in position of the calculated pixel observation points is evaluated (evaluating step). Finally, a set of values of the variables giving maximum evaluated dispersion of the calculated pixel observation points is determined as optimum optical condition of the optical lenses (condition determining step). This reduces the number of pixels which image the same portions of the target object, making it possible to reduce portions of the same image information in multiple unit images, and to stably obtain a reconstructed image having a high definition.
Owner:FUNAI ELECTRIC CO LTD +1
Rapid solar-thermal conversion of biomass to syngas
ActiveUS20080086946A1Improve reaction kineticsWide rangeElectrical coke oven heatingSolar heating energySyngasReactor design
Methods for carrying out high temperature reactions such as biomass pyrolysis or gasification using solar energy. The biomass particles are rapidly heated in a solar thermal entrainment reactor. The residence time of the particles in the reactor can be 5 seconds or less. The biomass particles may be directly or indirectly heated depending on the reactor design. Metal oxide particles can be fed into the reactor concurrently with the biomass particles, allowing carbothermic reduction of the metal oxide particles by biomass pyrolysis products. The reduced metal oxide particles can be reacted with steam to produce hydrogen in a subsequent process step.
Owner:UNIV OF COLORADO THE REGENTS OF
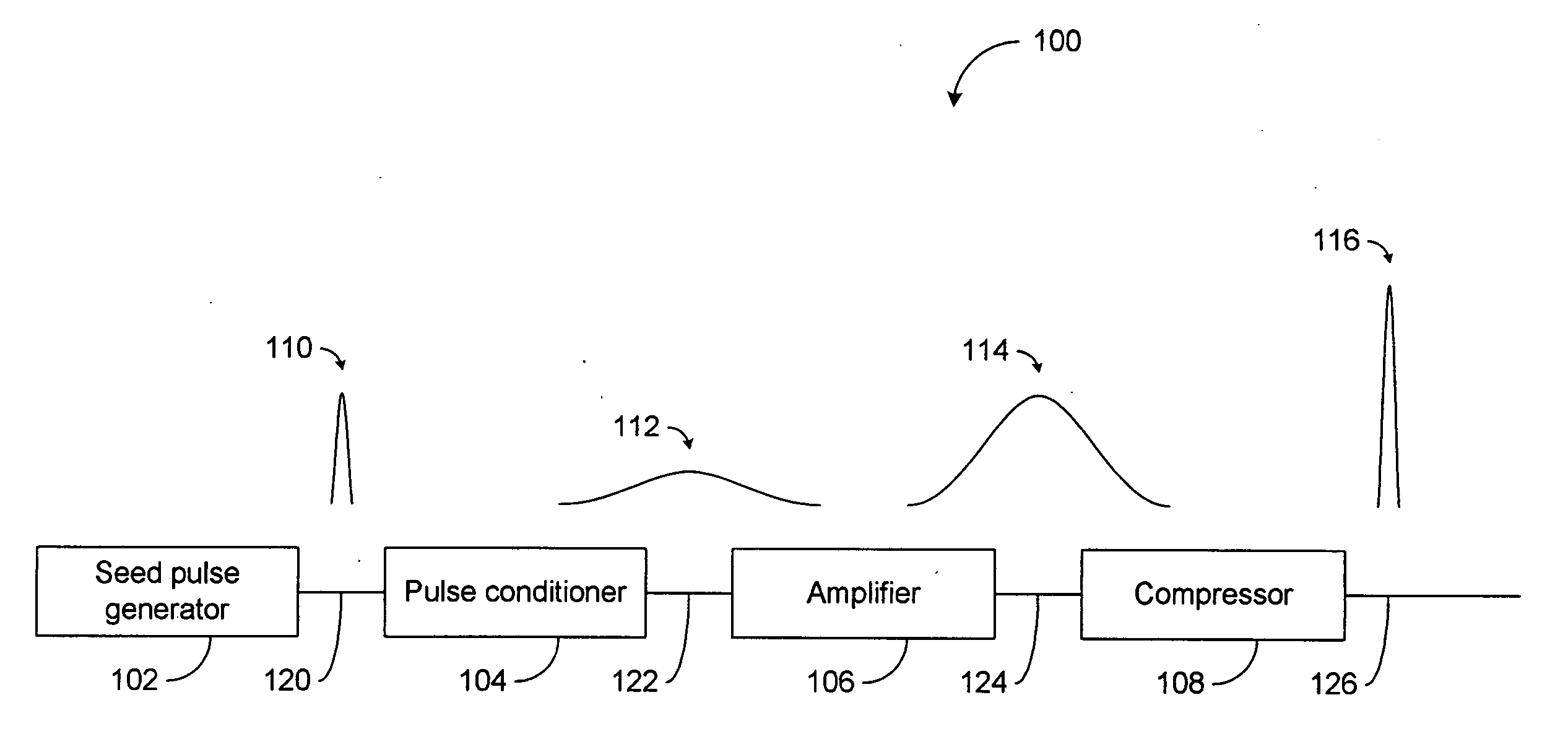
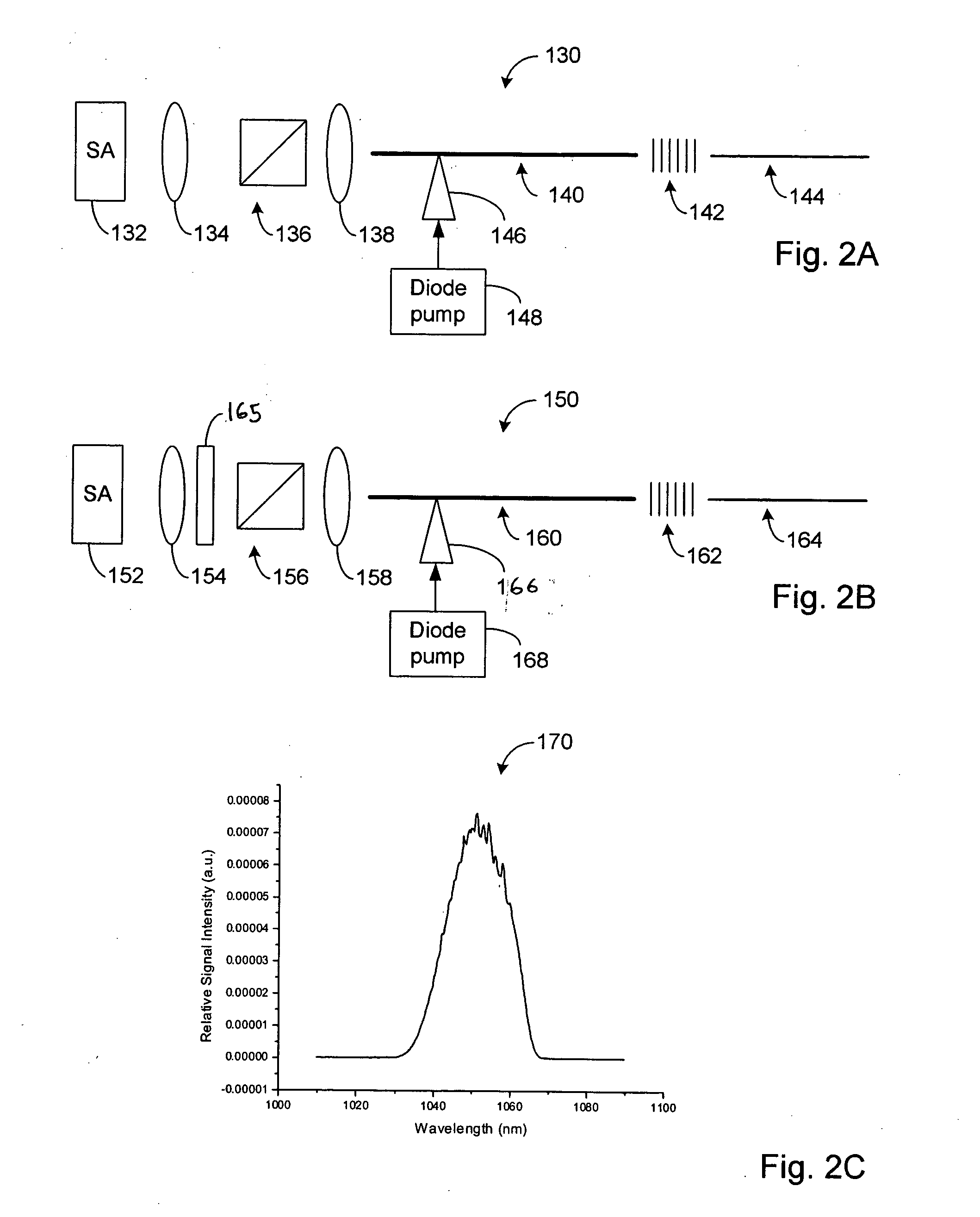
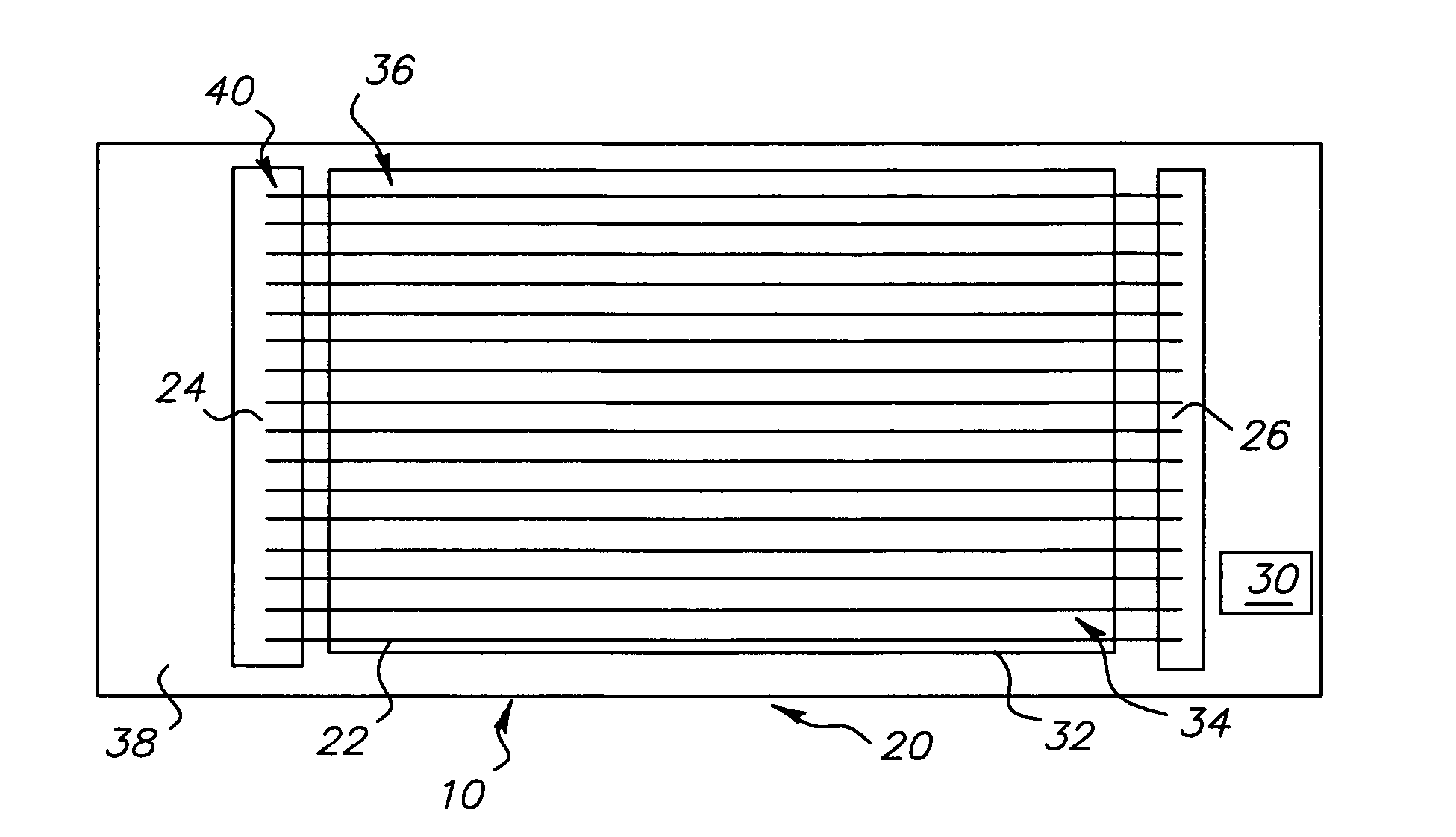
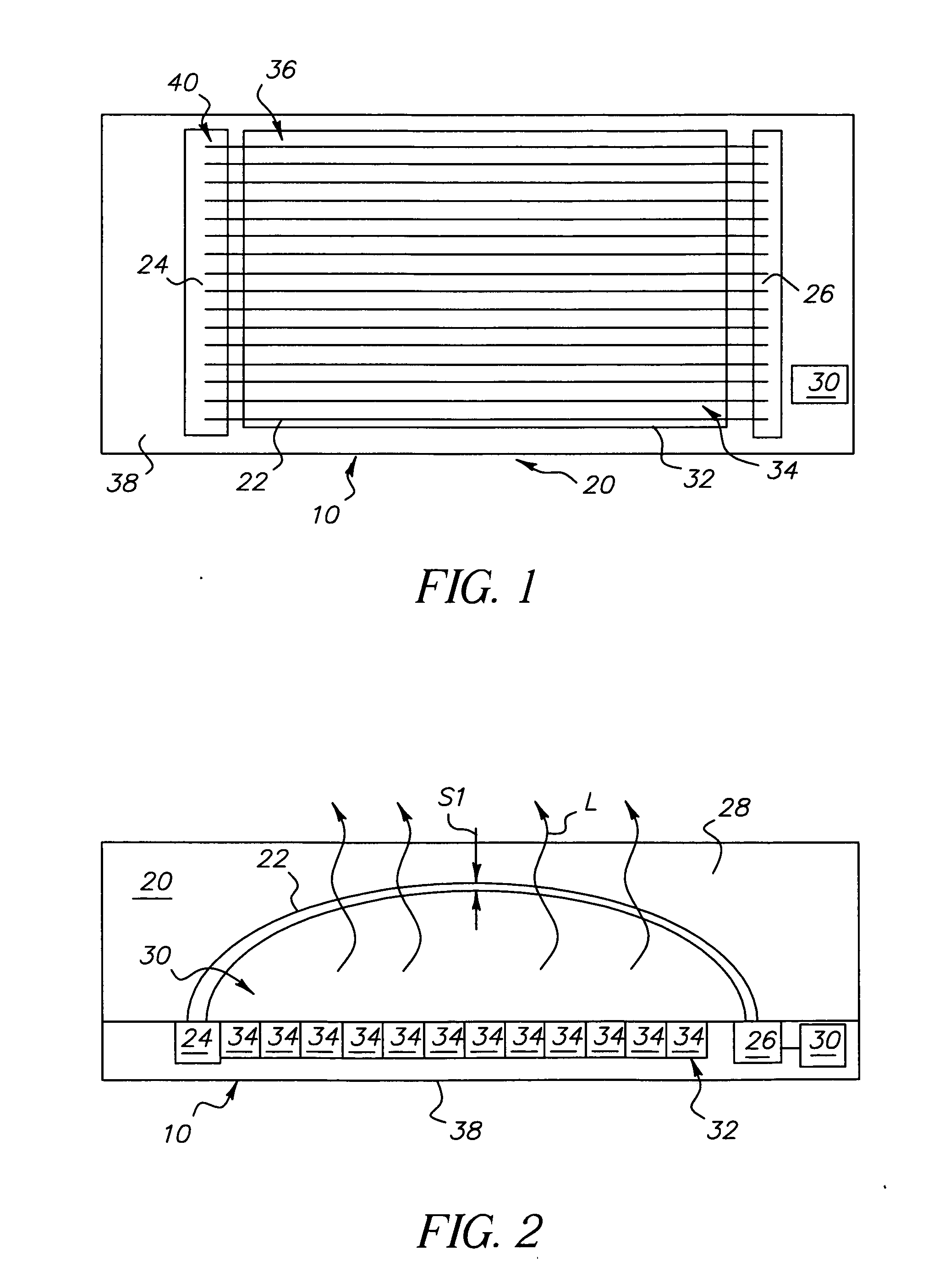
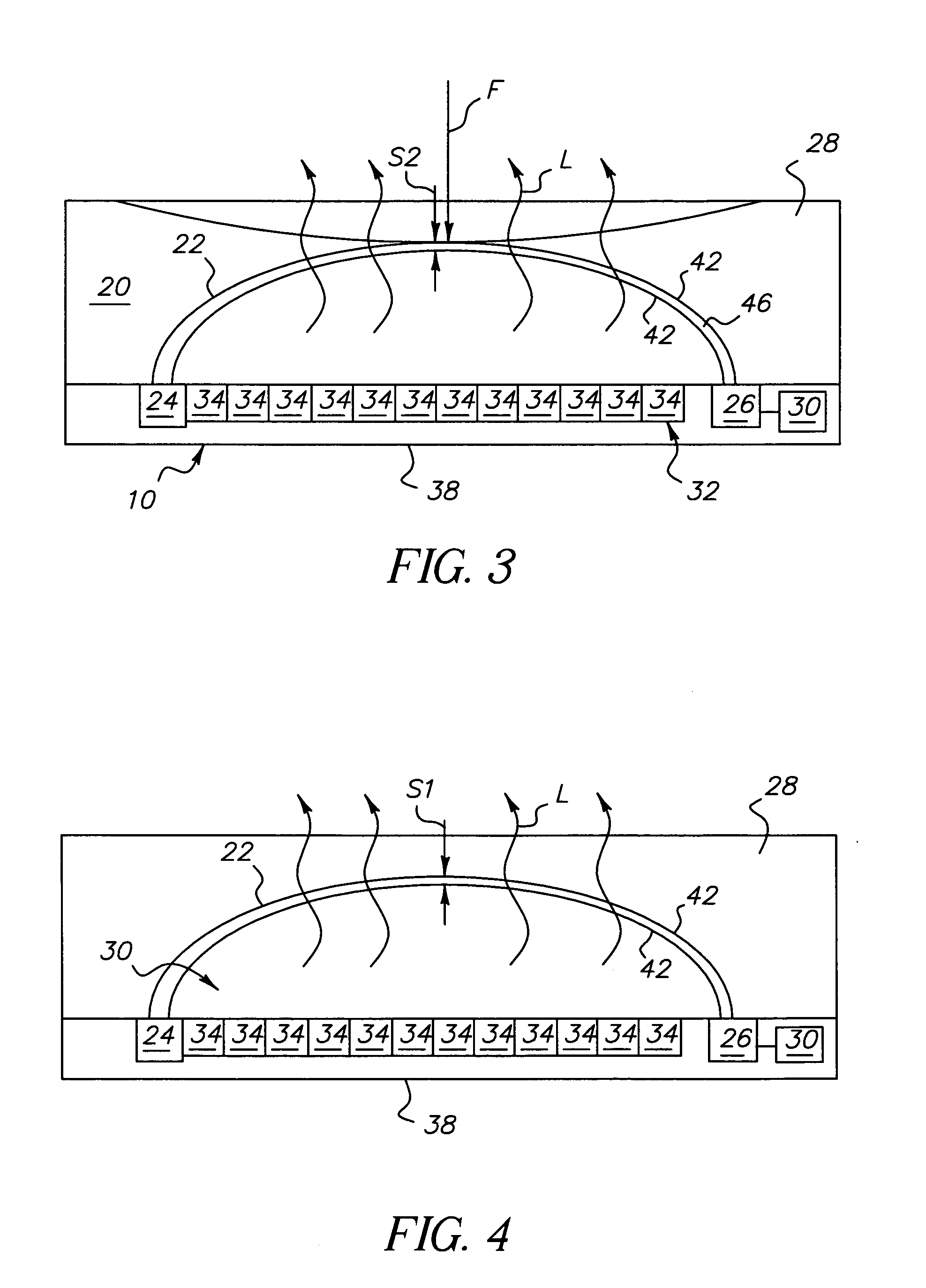
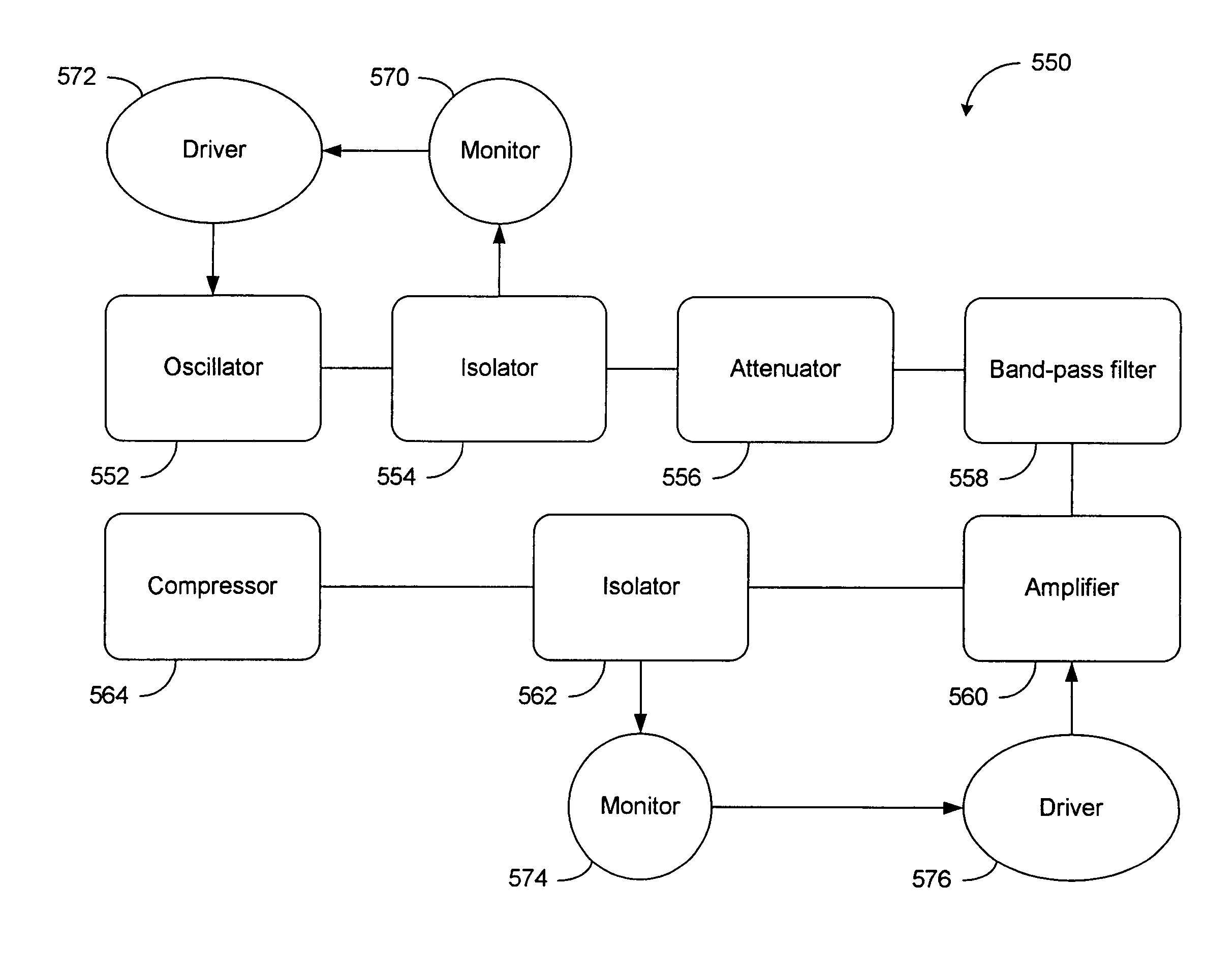
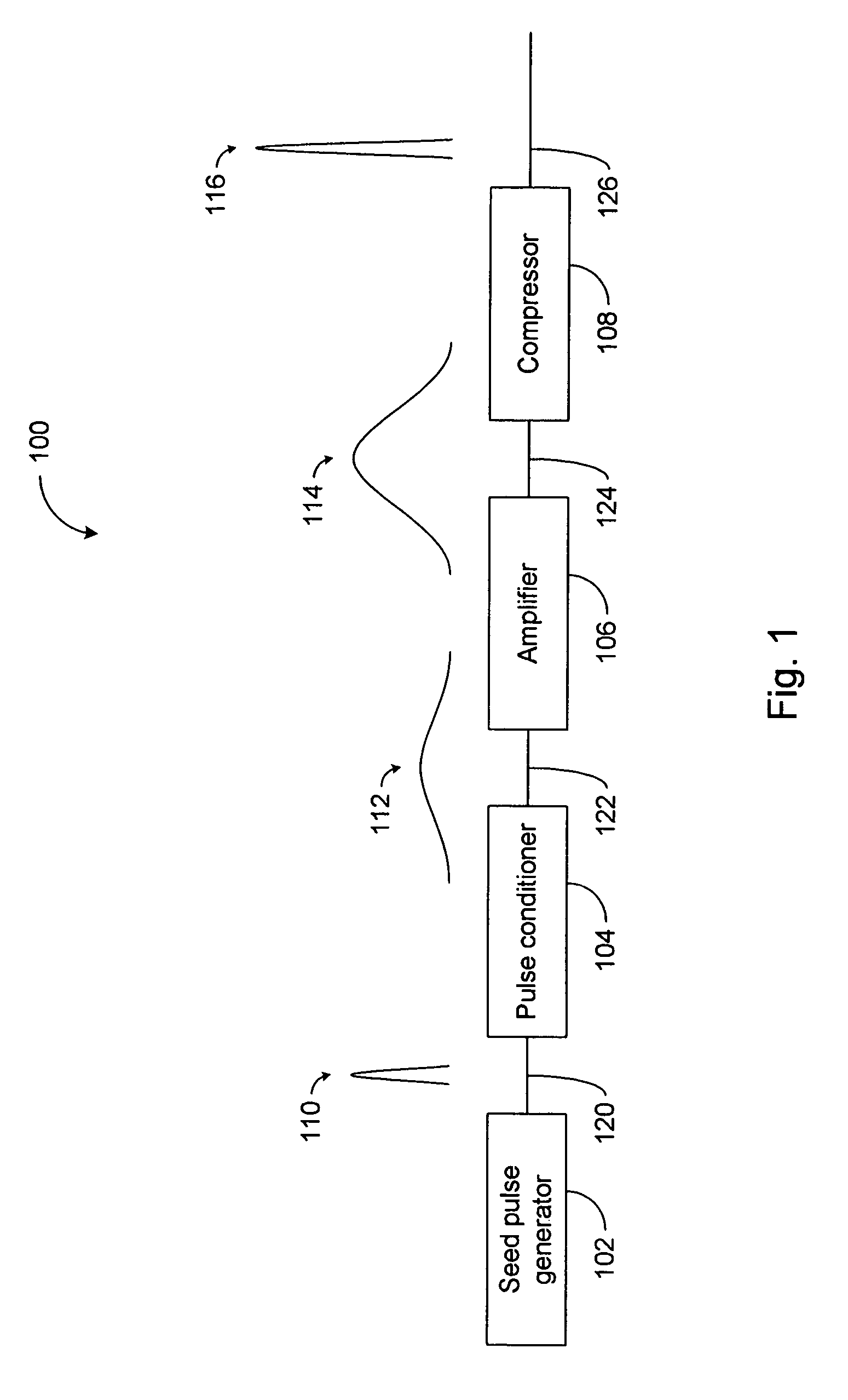
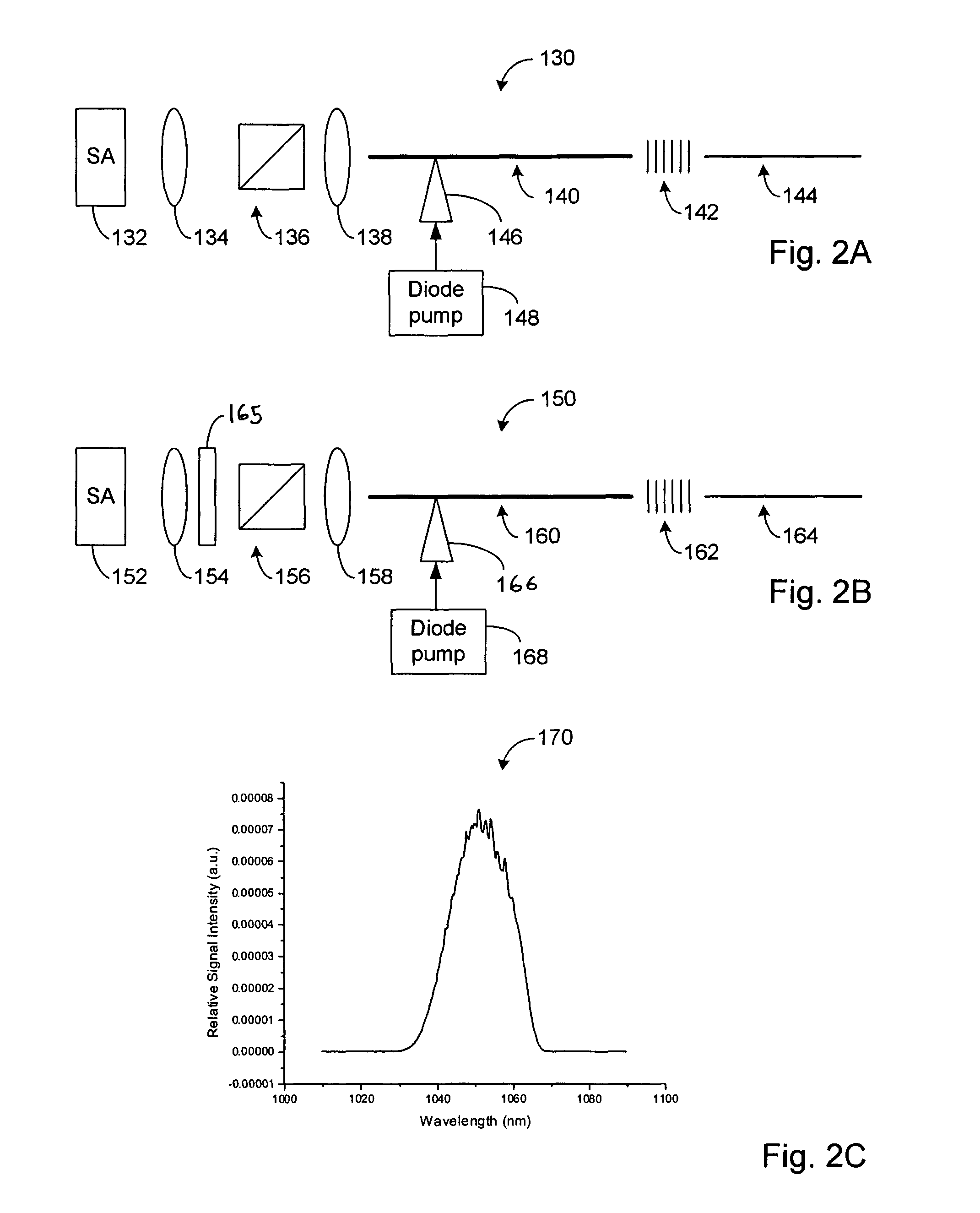
High power short pulse fiber laser
InactiveUS20050226278A1Reduce widthWeaken energyLaser using scattering effectsLaser arrangementsAudio power amplifierPigtail
A pulsed laser comprises an oscillator and amplifier. An attenuator and / or pre-compressor may be disposed between the oscillator and amplifier to improve performance and possibly the quality of pulses output from the laser. Such pre-compression may be implemented with spectral filters and / or dispersive elements between the oscillator and amplifier. The pulsed laser may have a modular design comprising modular devices that may have Telcordia-graded quality and reliability. Fiber pigtails extending from the device modules can be spliced together to form laser system. In one embodiment, a laser system operating at approximately 1050 nm comprises an oscillator having a spectral bandwidth of approximately 19 nm. This oscillator signal can be manipulated to generate a pulse having a width below approximately 90 fs.
Owner:IMRA AMERICA
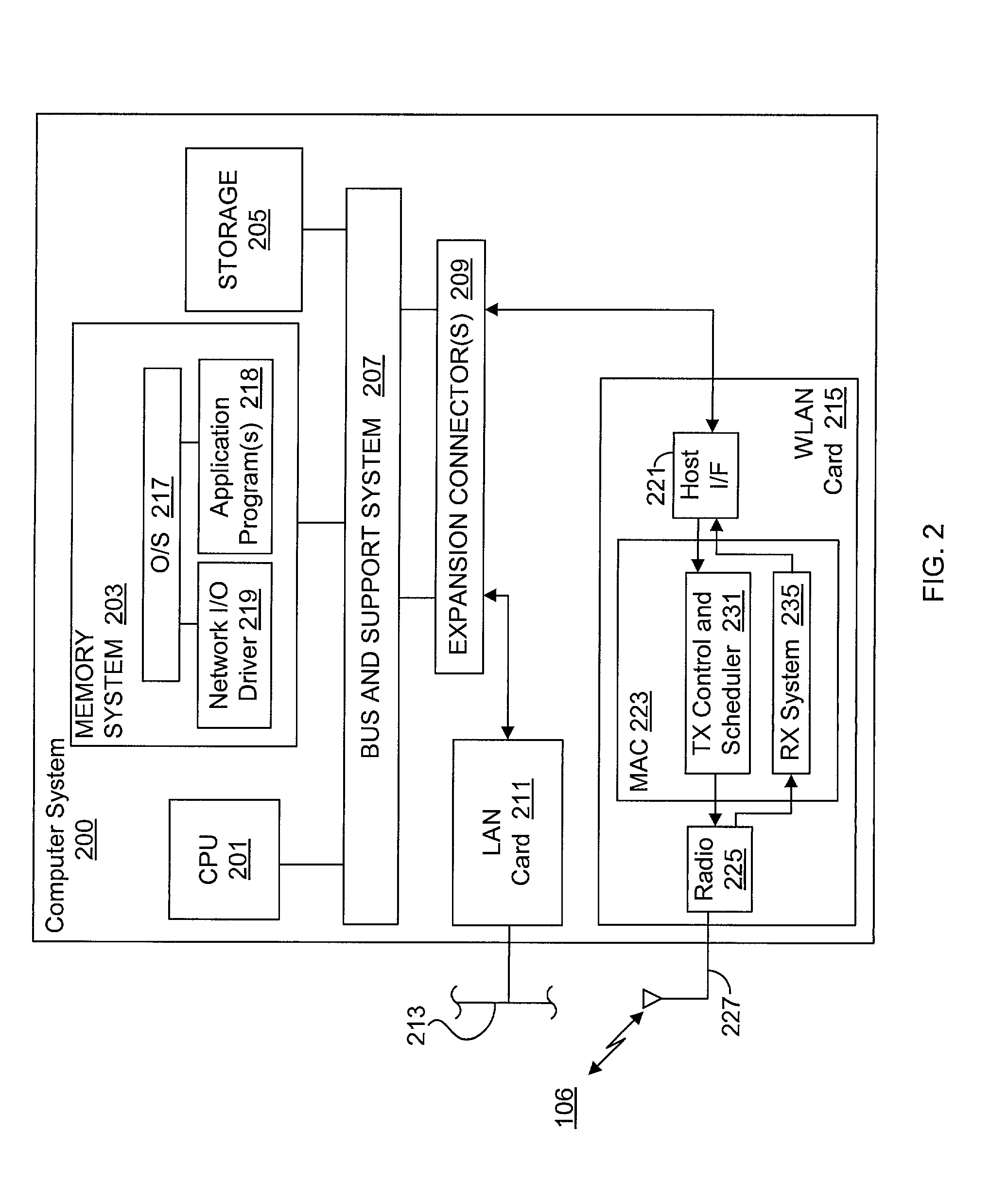
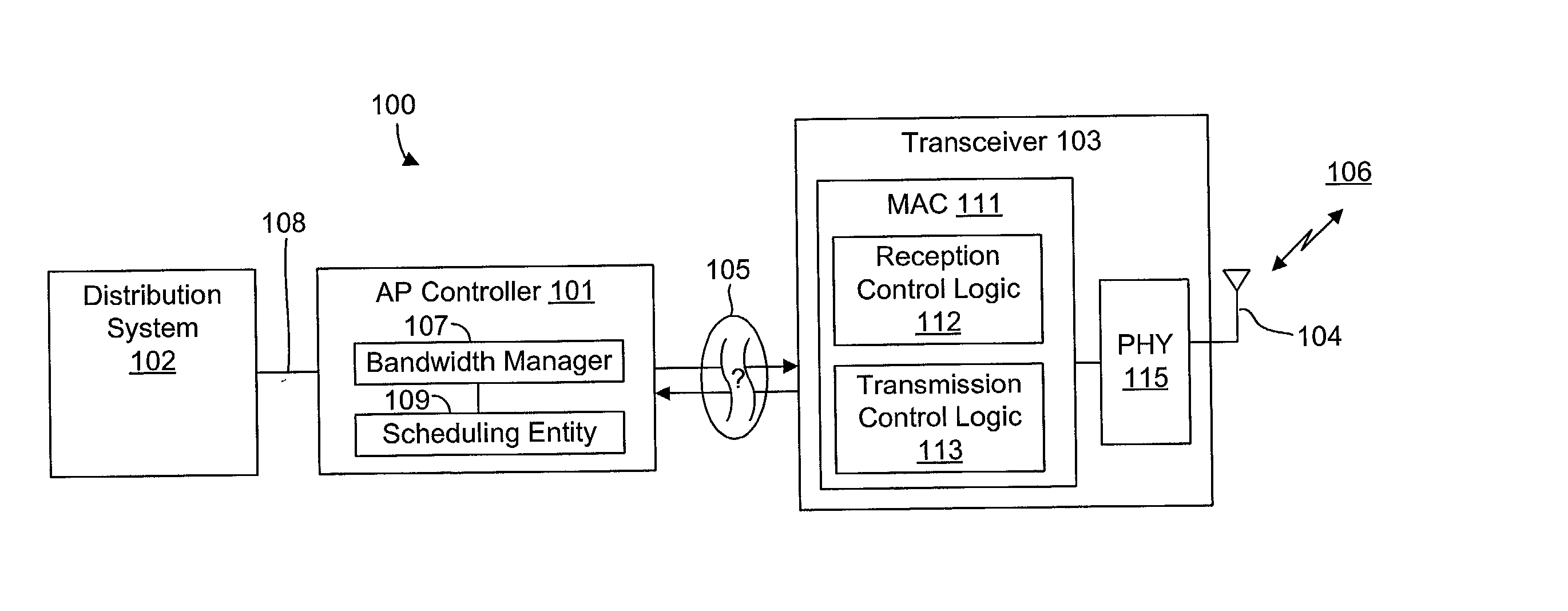
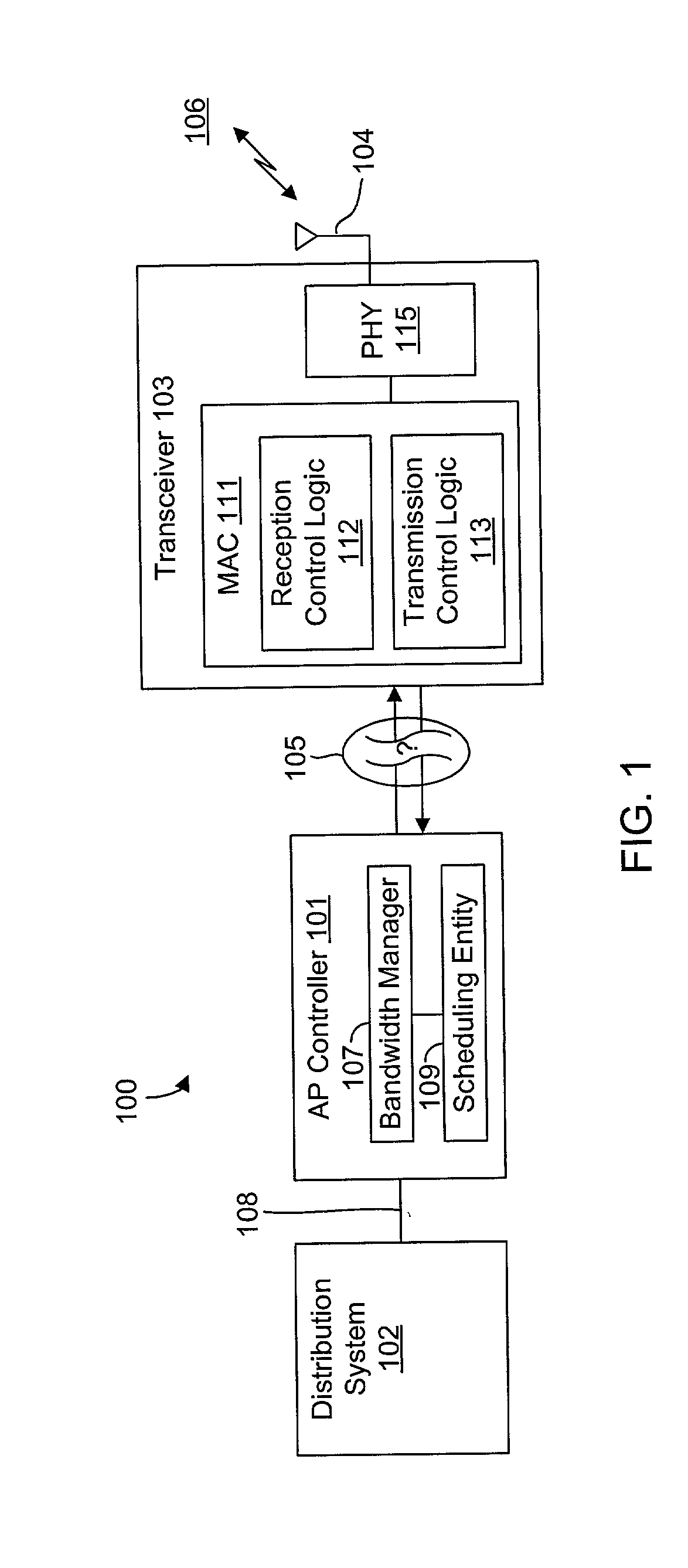
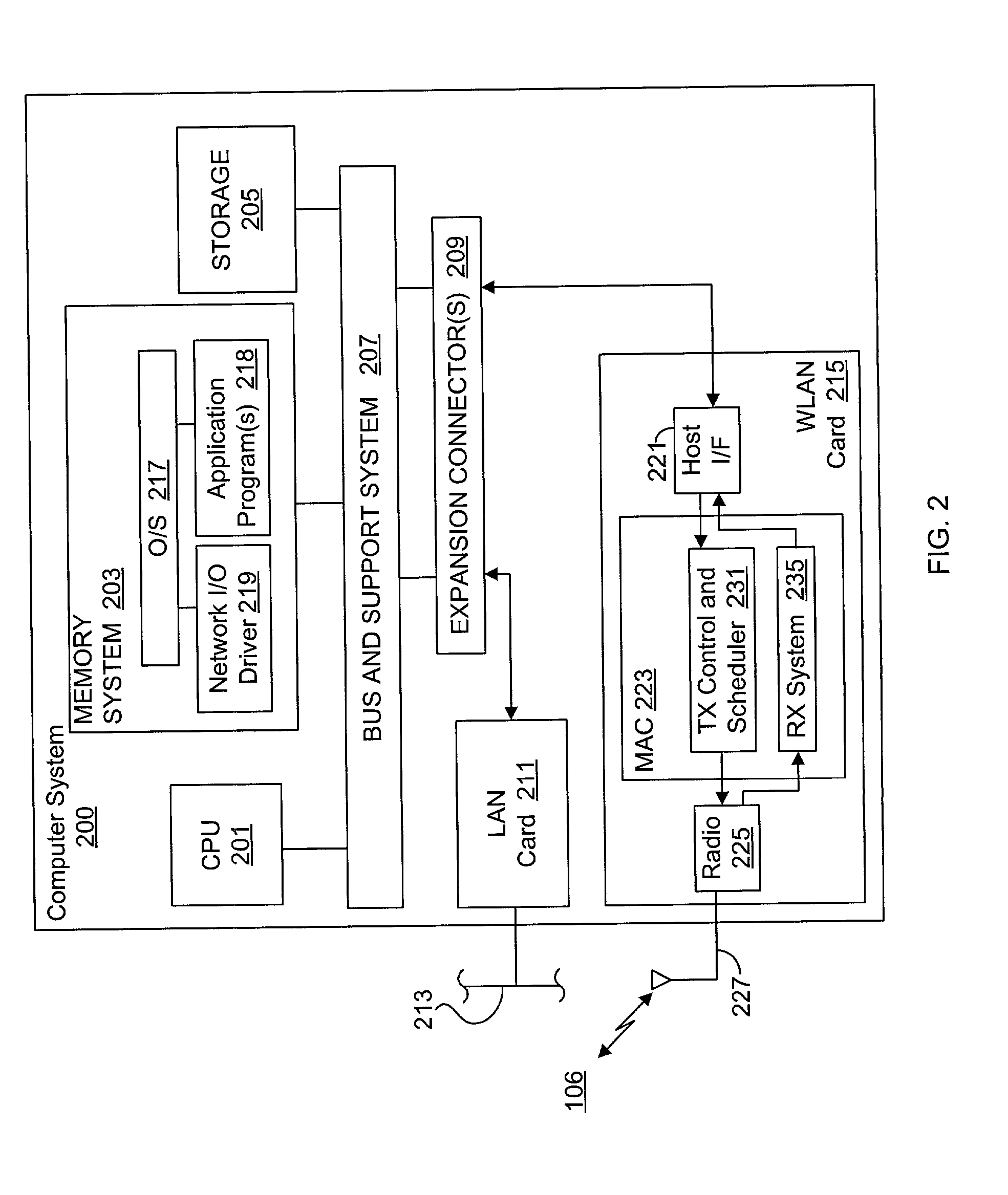
System and method for synchronizing data trasnmission across a variable delay interface
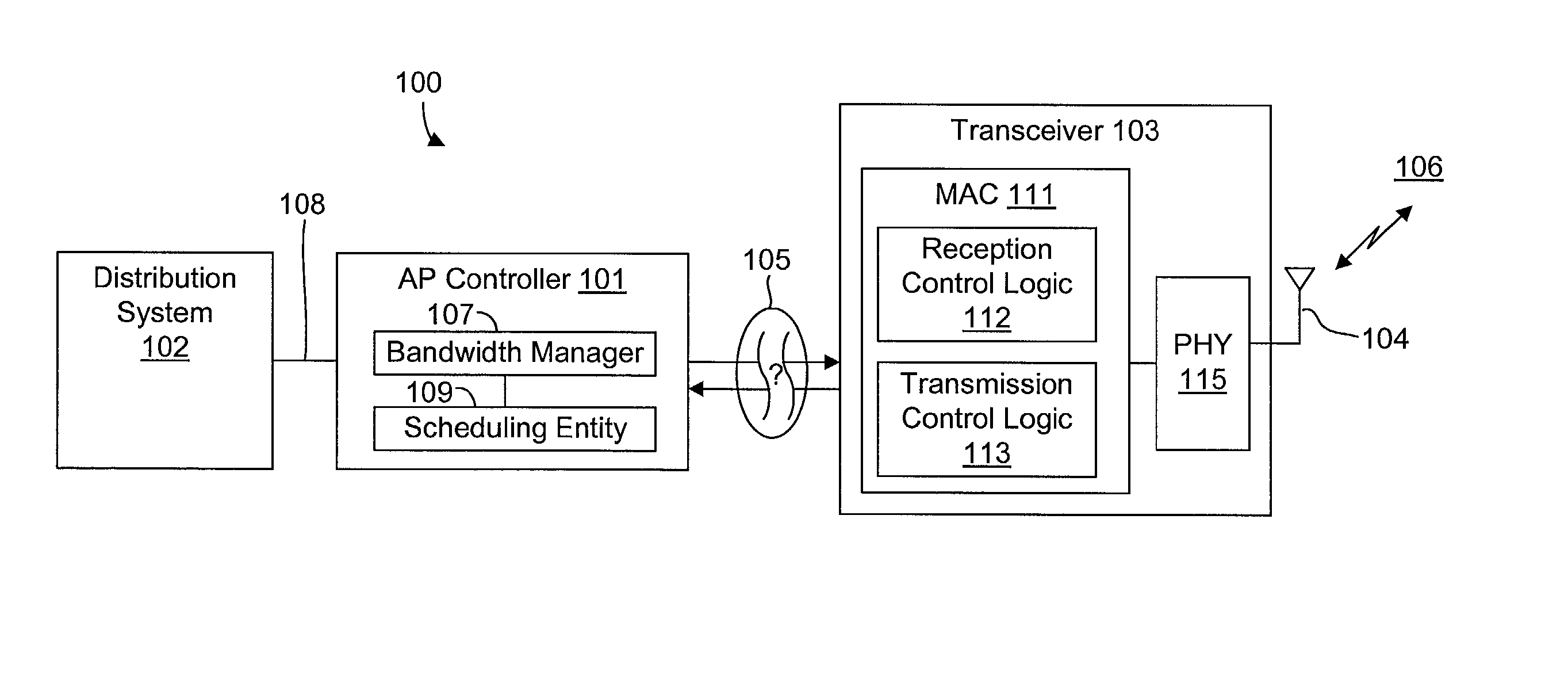
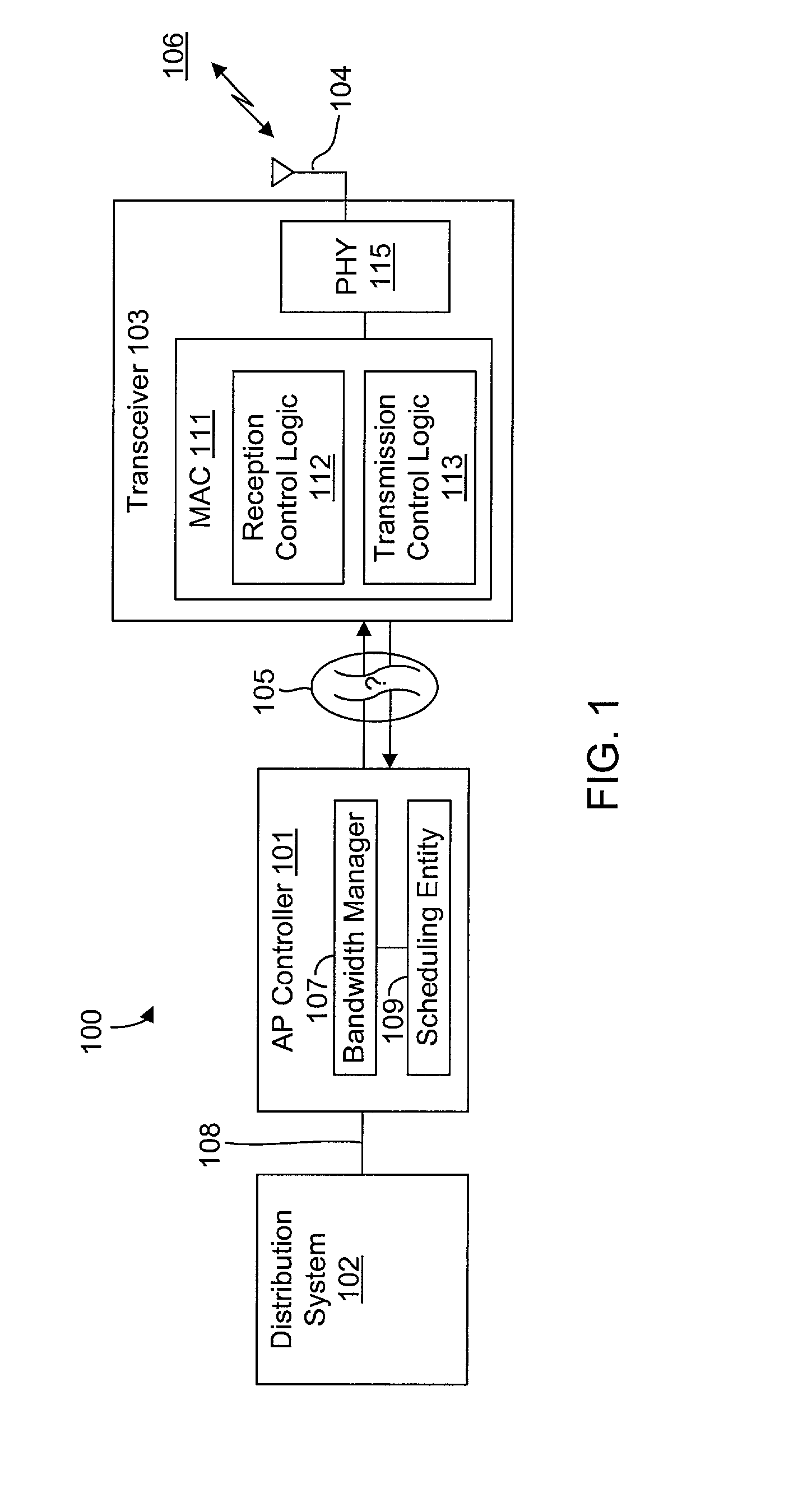
InactiveUS20020089927A1Significant overheadLimitation in wireless bandwidthError prevention/detection by using return channelFrequency-division multiplex detailsSufficient timeComputerized system
A method of synchronizing data transmission between a host computer system and a transmitter across an interface with variable delay or latency. The host computer system marks transition frames between successive transmission intervals and transfers the outgoing frames across the variable interface to the transmitter. The transmitter enqueues outgoing frames into one or more FIFO transmission queue(s) and processes the enqueued frames as appropriate for the communication protocol in use. Marked frames are detected as they reach the head of the appropriate transmit queue. In particular, while bypassing is not active, the transmitter transmits unmarked frames until the end of the current interval, or until there is insufficient time in the interval to transmit another frame or until a marked frame is detected. While bypassing is not active, the transmitter terminates transmission from the transmit queue when a marked frame is detected during each interval. While bypassing is active, the transmitter discards unmarked frames without transmission until a marked frame is detected. During each interval, the transmitter activates bypassing if a marked frame has not been detected and deactivates bypassing if a marked frame is detected while bypassing is active. The transmitter enables queue mark operation if a marked frame is detected while queue mark operation is not enabled. The transmitter increments a bypass counter each time an interval ends without detecting a marked frame, and disables queue mark operation if the bypass counter reaches a predefined limit.
Owner:CONEXANT
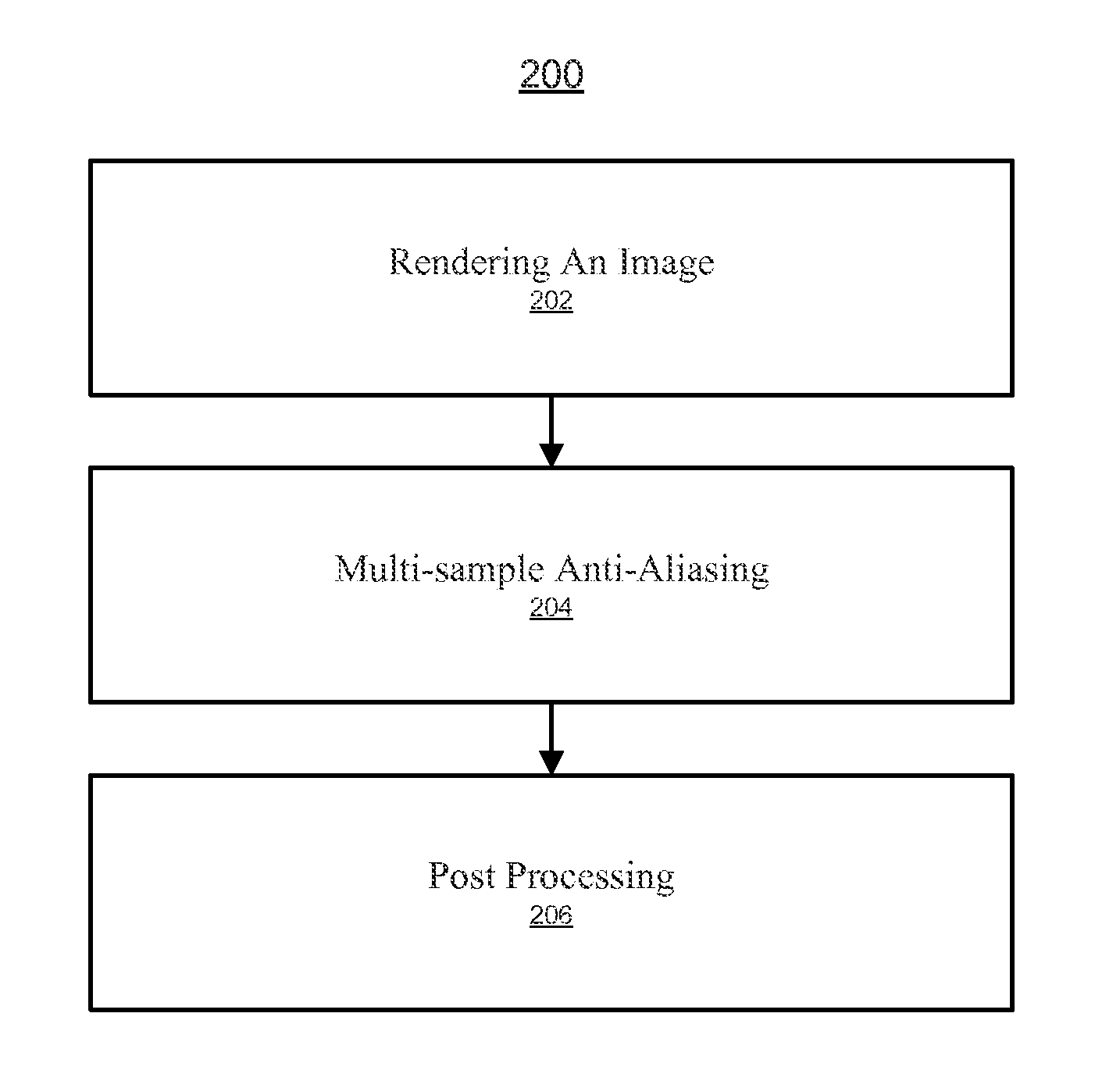
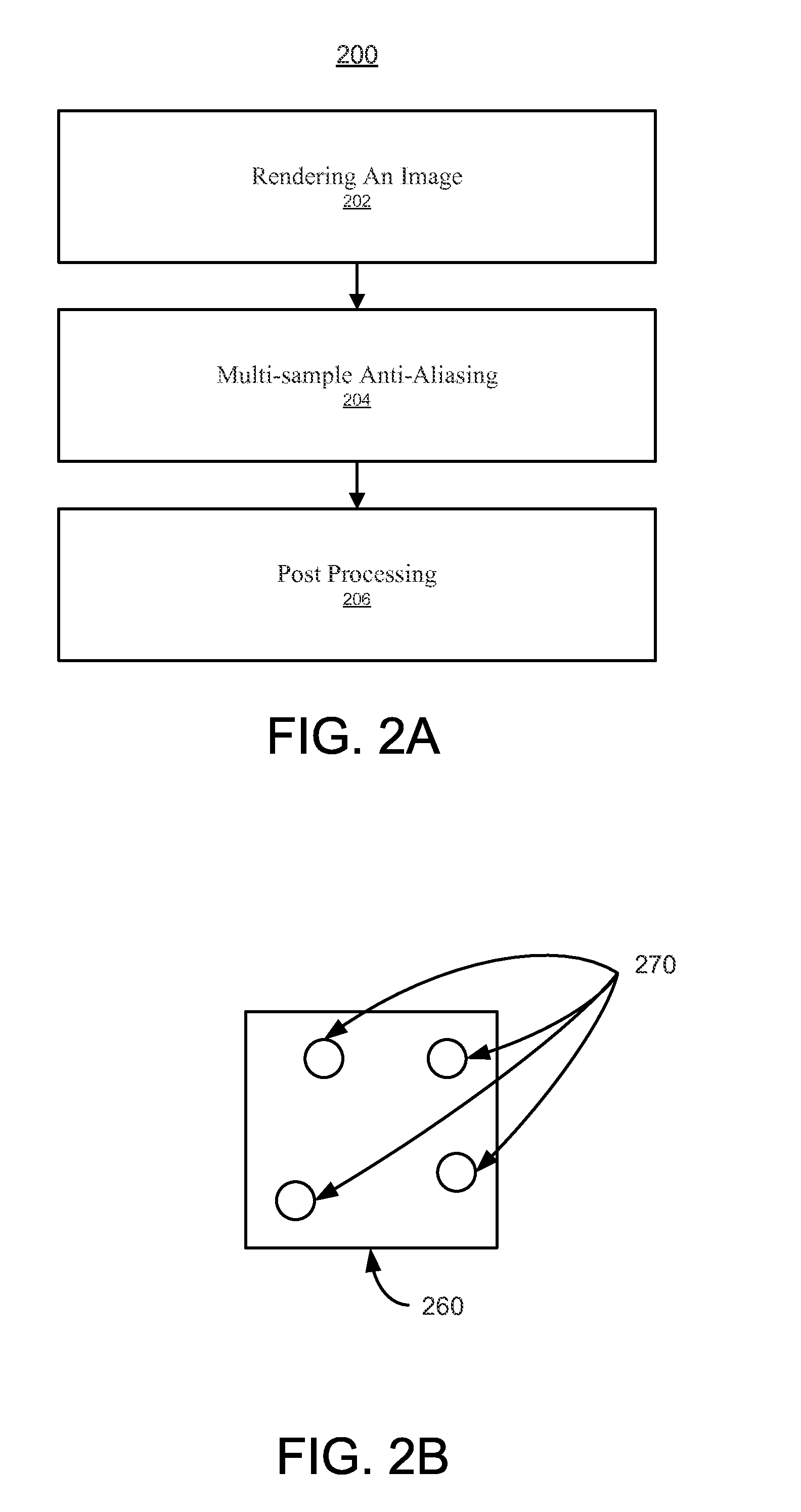
System and method for enhanced multi-sample Anti-aliasing
ActiveUS20140354675A1Improve visual qualityEnhanced and accurate visual resultImage enhancementImage analysisPattern recognitionAnti-aliasing
A system and method for enhanced multi-sample anti-aliasing. The method includes determining a sampling pattern corresponding to a pixel and adjusting the sampling pattern based on a visual effect (e.g., post-processing visual effect). The method further includes accessing a first plurality of samples based on the sampling pattern. The first plurality of samples may comprise a second plurality of samples within the pixel and a third plurality of pixels outside of the pixel. The method further includes performing anti-aliasing filtering of the pixel based on the first plurality of samples and the sampling pattern.
Owner:NVIDIA CORP
Clot engagement and removal systems
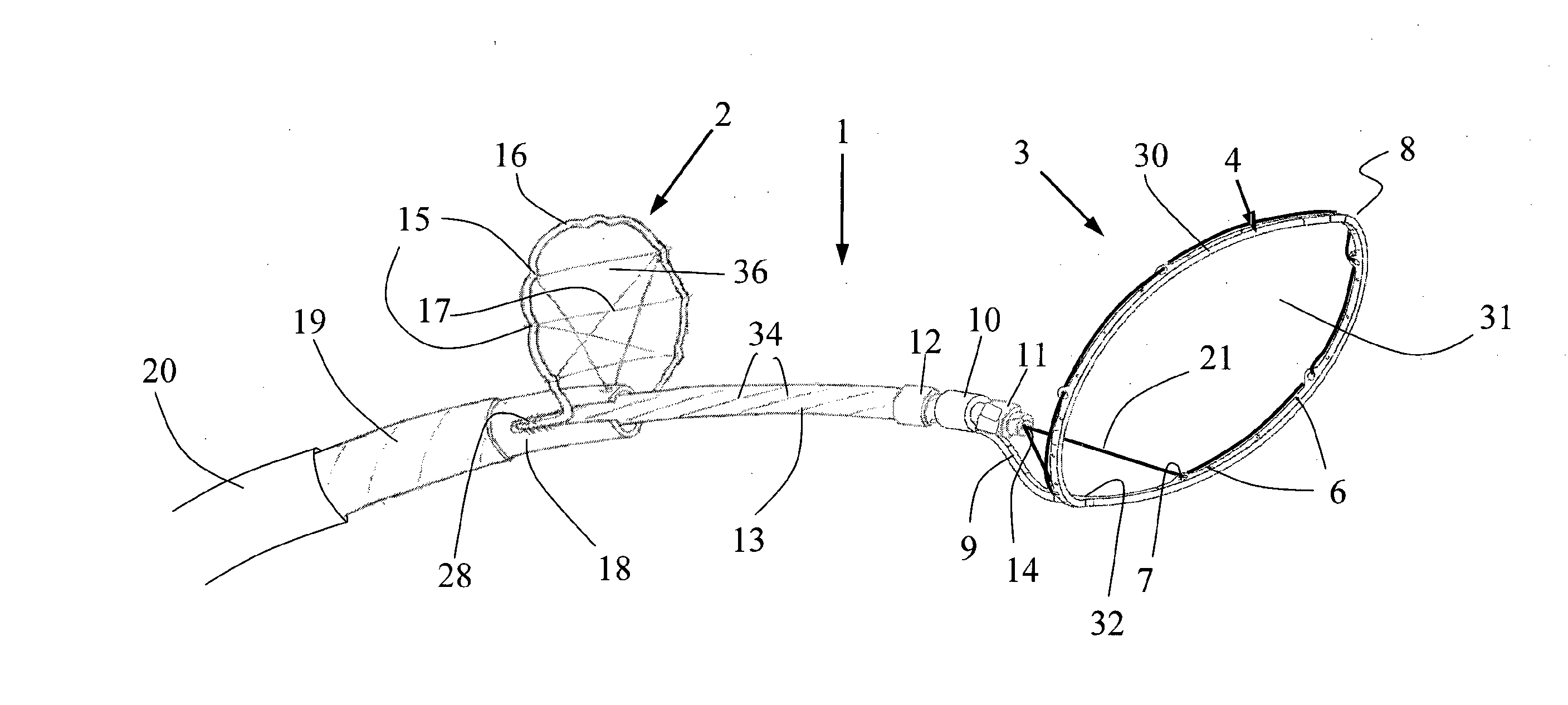
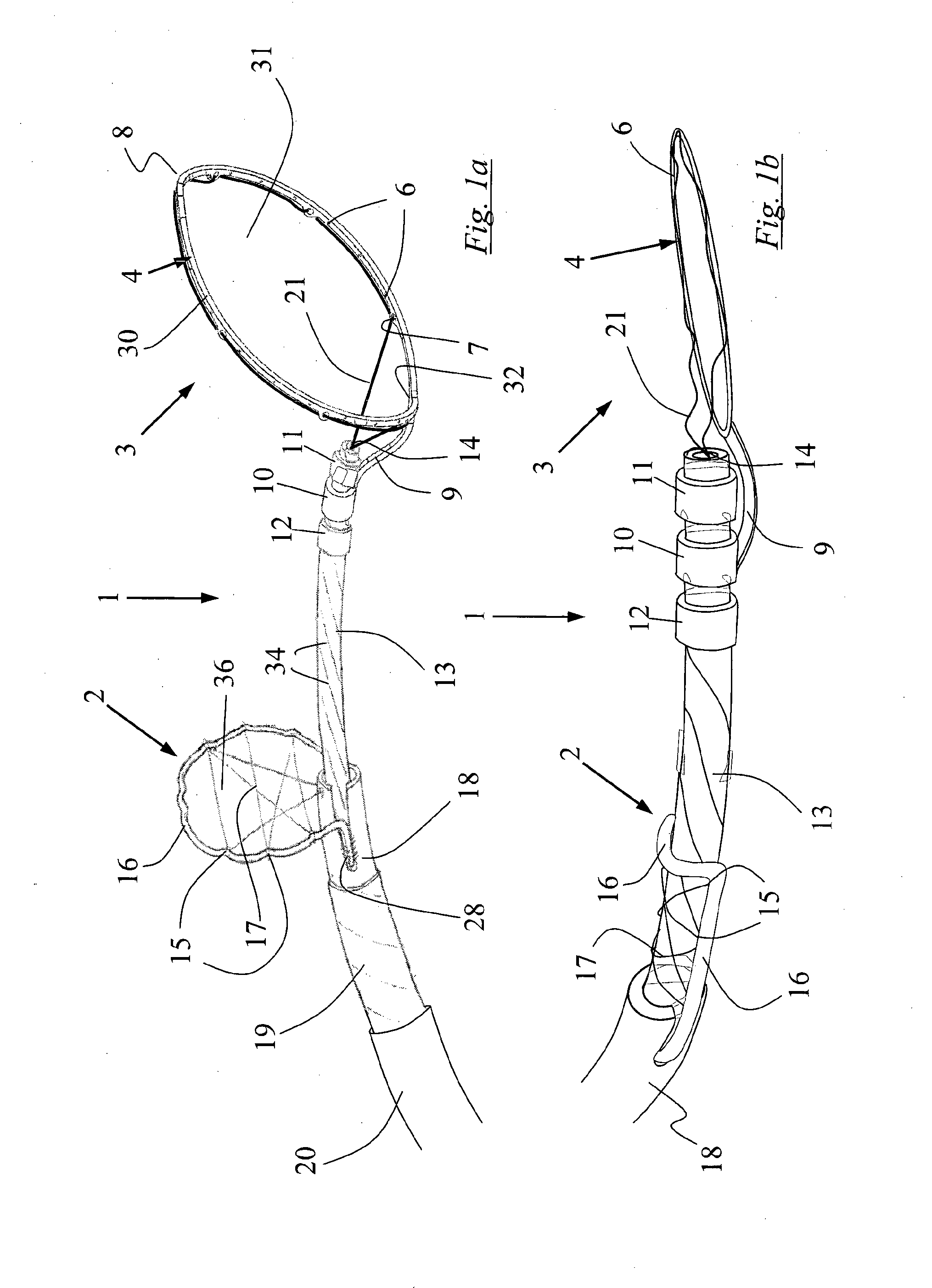
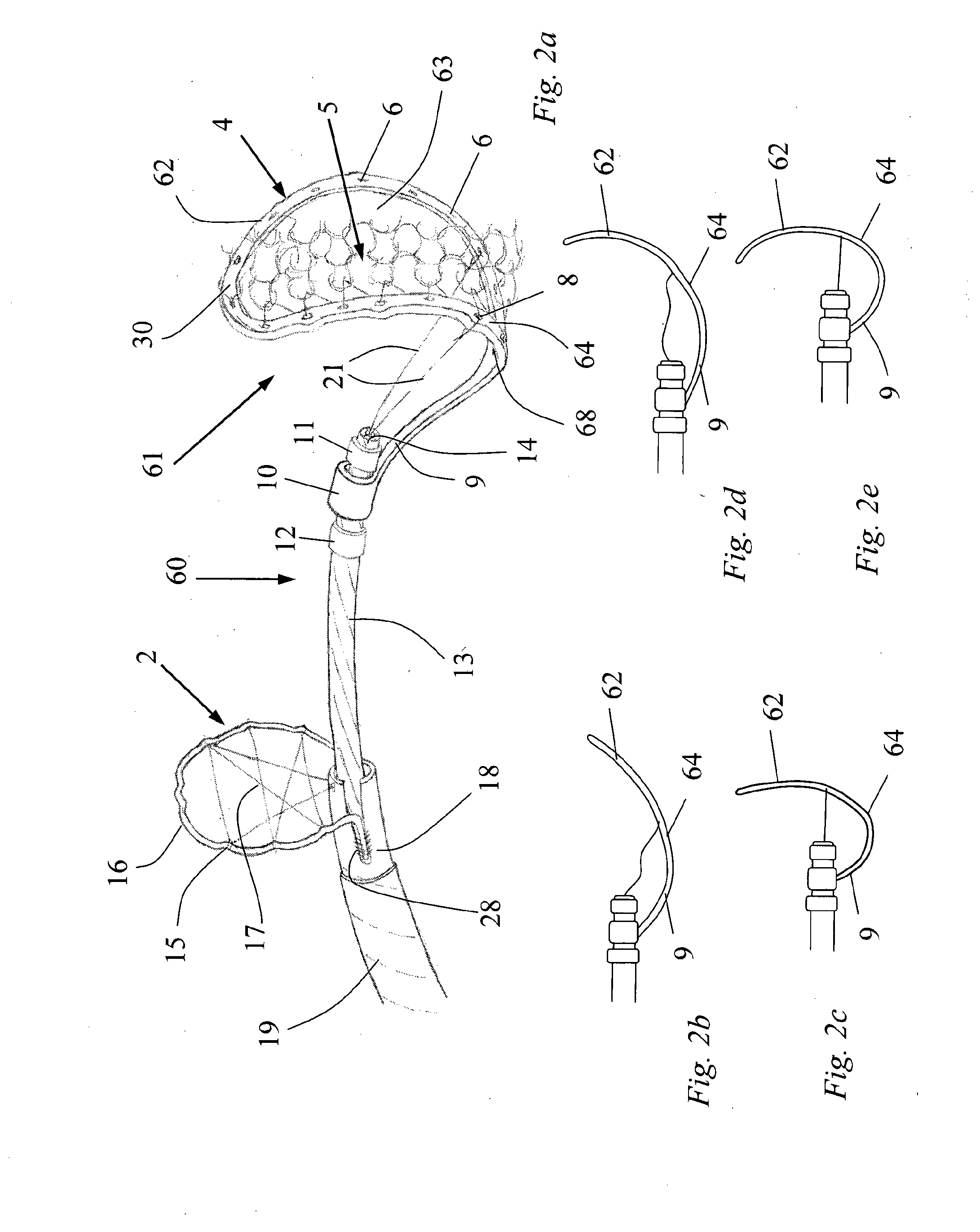
InactiveUS20130184739A1Cutting portionProtection from forceDilatorsExcision instrumentsArterial vesselBlood vessel
A device for the removal of clot obstructing the flow of blood through an arterial vessel comprises an elongate member, a clot engaging element and a capture basket. The elongate member extends in use from a point adjacent a target treatment site interior of the patient to a point exterior of the patient. The capture basket comprises a frame and a net, and has an expanded and a collapsed configuration. The clot engaging element comprises a plurality of struts having an expanded and a collapsed configuration. The plurality of struts form a first section and a second section, the first section tapering outward and distally from the elongate member and connected to the second section. The second section comprises a plurality of cells defined by a plurality of struts and arranged around at least a portion of the circumference of an axis substantially parallel to that of the elongate member. The clot engaging element and the capture basket are restrained in the collapsed configuration for delivery to the target site. The clot engaging element is located adjacent the distal end of the elongate member and proximal of the capture basket.
Owner:NEURAVI
System and method for providing a selectable retry strategy for frame-based communications
InactiveUS20020089959A1Eliminate timeBroaden applicationError prevention/detection by using return channelNetwork traffic/resource managementTransceiverCommunications system
A frame-based communications system with selectable retry strategy including a controller that programs a retry strategy field of a frame descriptor of a frame, where the retry value indicates a selected one of a normal retry count, an alternative retry count, and at least one "no retry" option. The system includes a transceiver that attempts retransmission of the frame up to as many times indicated by a selected retry count or until expiration of a frame transmit duration, or that does not attempt retransmission if the retry value indicates no retry. The no retry options may include treating the first attempt as successful or returning an unsuccessful attempt as a failure. The frame itself may be programmed with a request to not acknowledge a successfully received frame to recapture the time normally allocated to acknowledgement frames. The transceiver may include acknowledgement logic that is configured to suppress an acknowledgement frame.
Owner:CONEXANT
System and method of repetitive transmission of frames for frame-based communications
InactiveUS20020089994A1Function increaseLower latencyError prevention/detection by using return channelNetwork traffic/resource managementCommunications systemFrame based
A communications system including a scheduling entity and a transceiver coupled across a variable timing interface. The scheduling entity forwards frames for transmission and identifies selected frames as persistent. The transceiver includes a queue, a frame manager and a transmission scheduler. The frame manager receives and enqueues forwarded frames and the transmission scheduler dequeues and transmits frames from the queue and forwards persistent frames back to the frame manager. The transmission scheduler includes persistence logic that detects a persistent mark and asserts a persistent signal that is detected by the transmission scheduler. The scheduling entity identifies a persistent frame by setting a bit in a transmit control field of the frame descriptor. The scheduling entity sends a clear persistence command to the transceiver to clear a persistent mark of an identified frame. The transceiver may be configured for wireless communications.
Owner:CONEXANT
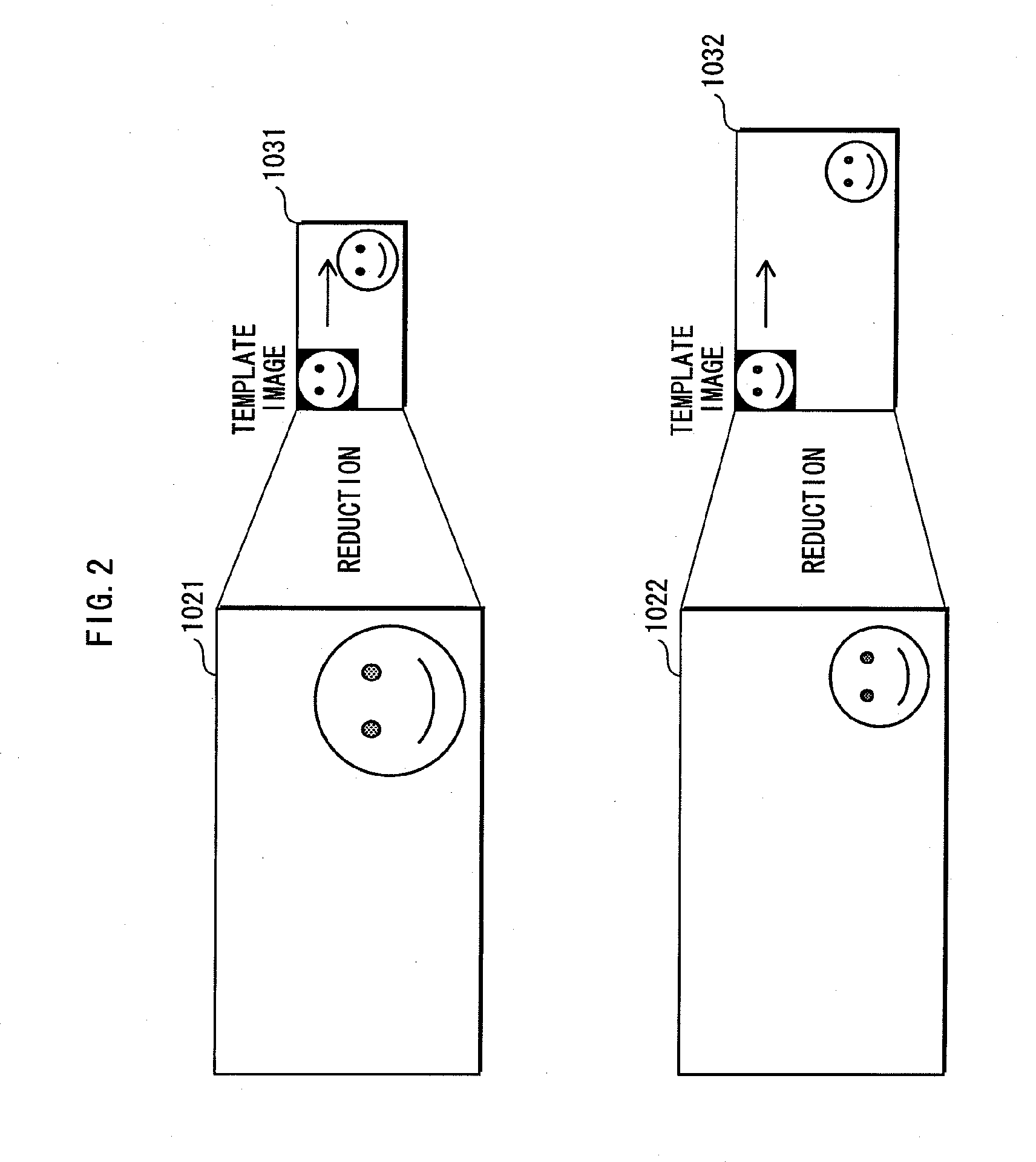
Detector, detection method, and integrated circuit for detection
InactiveUS20100232712A1Raise the possibilityEfficient detectionCharacter and pattern recognitionComputer visionIntegrated circuit
A detector detects a specified image in an input image. The detector includes an area determination unit for determining, in the input image, a detection target area in which the specified image potentially exists, a setting unit for setting positions of a plurality of matching target ranges substantially in the detection target area, each of the matching target ranges being a predetermined size, so that the matching target ranges cover the detection target area, and each matching target range overlaps a neighboring matching target range by a predetermined overlap width, and a matching unit for detecting the specified image by matching a portion of the input image encompassed by each matching target range set by the setting unit and a template image for detecting the specified image.
Owner:PANASONIC INTELLECTUAL PROPERTY CORP OF AMERICA
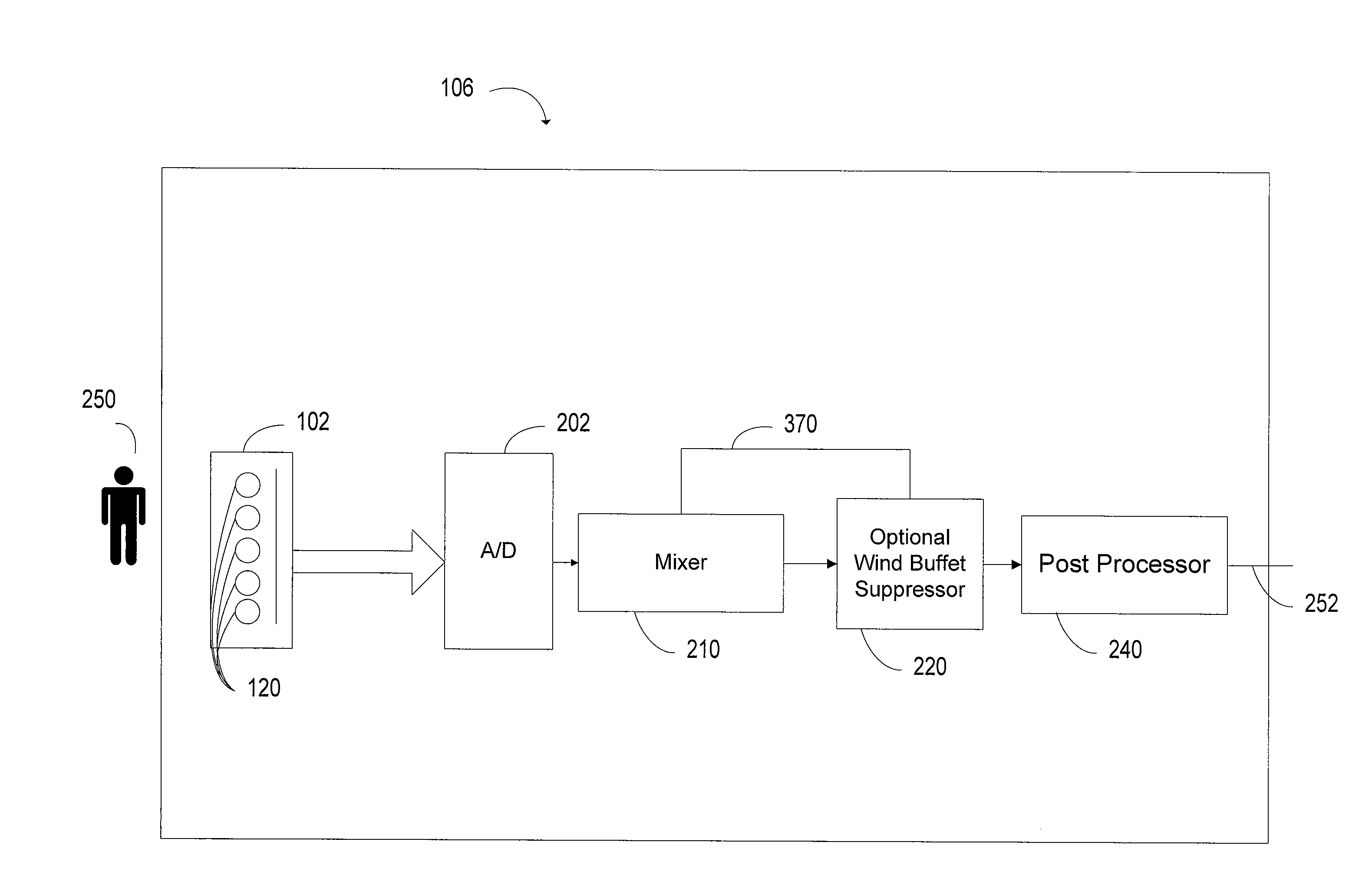
Mixer with adaptive post-filtering
ActiveUS20090116661A1Cutting portionSpeech analysisMicrophones signal combinationCoherence levelSelf adaptive
A noise reduction system includes multiple transducers that generate time domain signals. A transforming device transforms the time domain signals into frequency domain signals. A signal mixing device mixes the frequency domain signals according to a mixing ratio. Frequency domain signals are rotated in phase to generate phase rotated signals. A post-processing device attenuates portions of the output based on coherence levels of the signals.
Owner:BLACKBERRY LTD
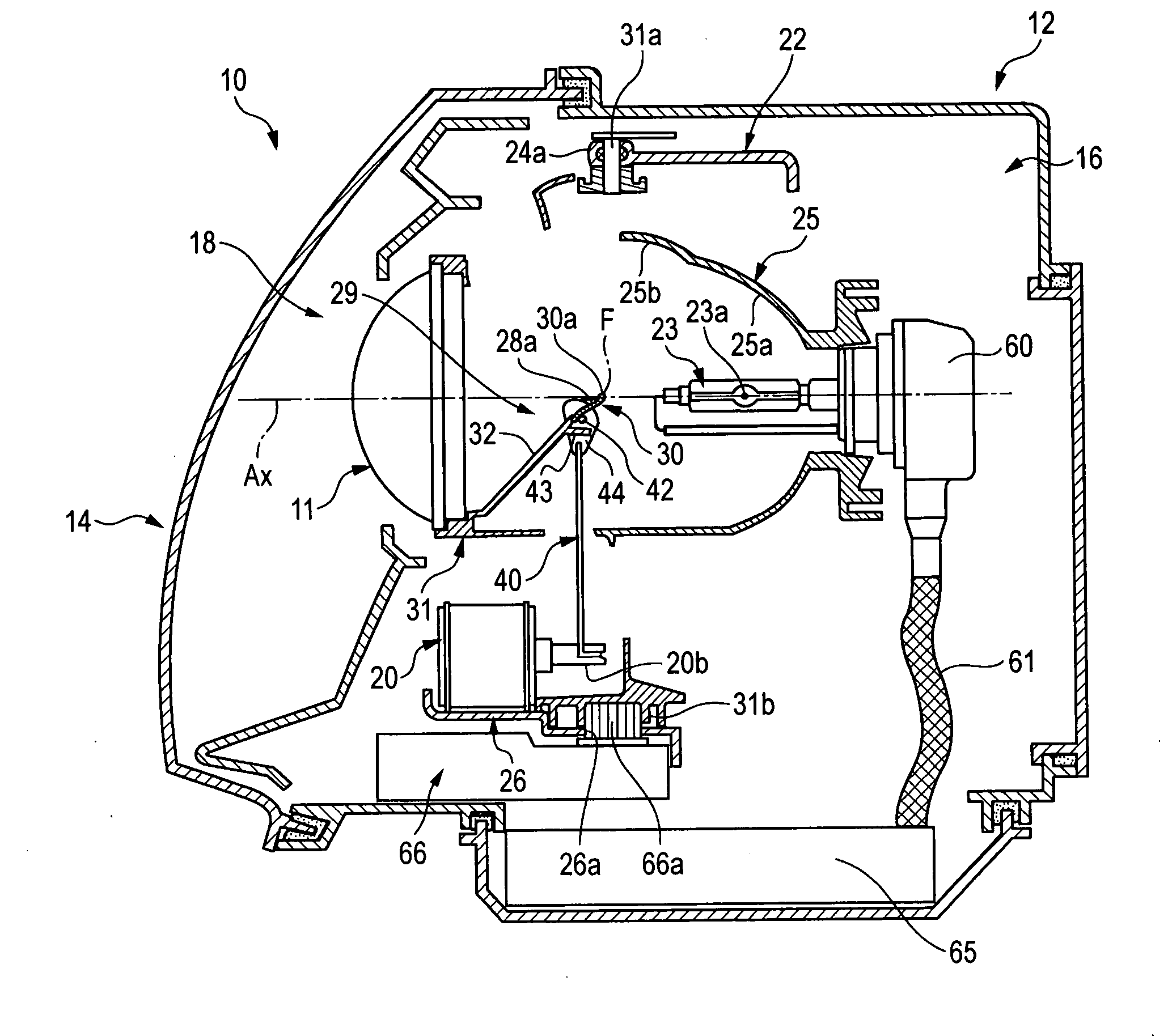
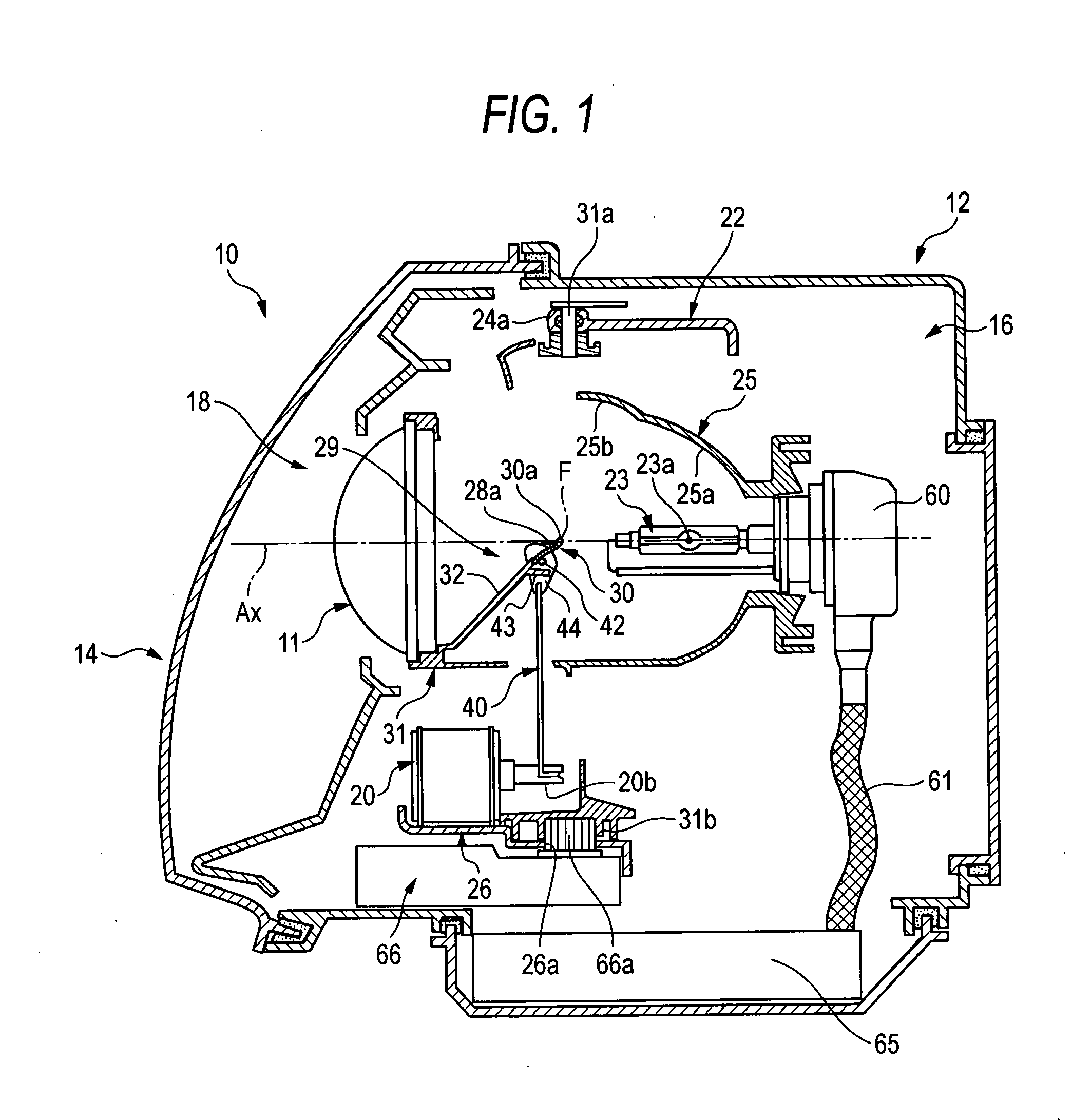
Vehicle headlamp
InactiveUS20070247865A1Easy to adjustCutting portionVehicle headlampsPoint-like light sourceIlluminanceOptoelectronics
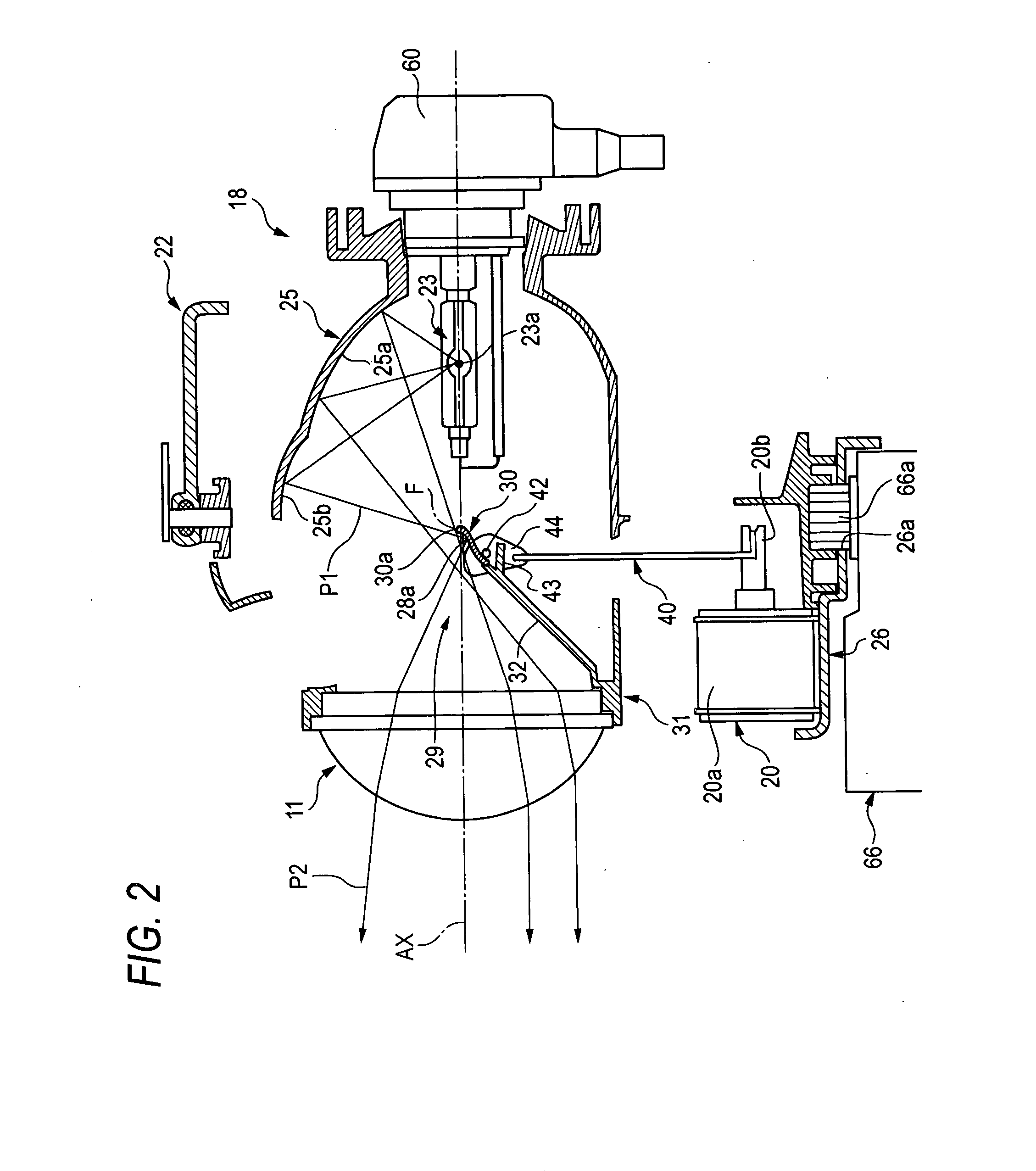
A lamp unit 18 is provided with a reflecting face 25b for an overhead sign and a light receiving face 28 for the overhead sign. The reflecting face 25b for an overhead sign reflects light from a light source 23a and is provided on an upper side of the light source 23a and rearward from a rear side focal point F of a projector lens 11. The light receiving face 28 for the overhead sign is provided on a front side of a movable shade 30 arranged between the projector lens 11 and the light source 23a. The light receiving face 28 forms overhead sign irradiating light P2 by reflecting light from the reflecting face 25b for the overhead sign to the projector lens 11. Illuminance reducer for reducing a portion of irradiating light is provided at a position of a vicinity of an upper end of the movable shade 30 of the light receiving face 28a for the overhead sign.
Owner:KOITO MFG CO LTD
Fuel cell having interdigitated flow channels and water transport plates
InactiveUS20010004501A1Increasing migrationHigh water removal rateCell electrodesWater management in fuel cellsEngineeringWater transport
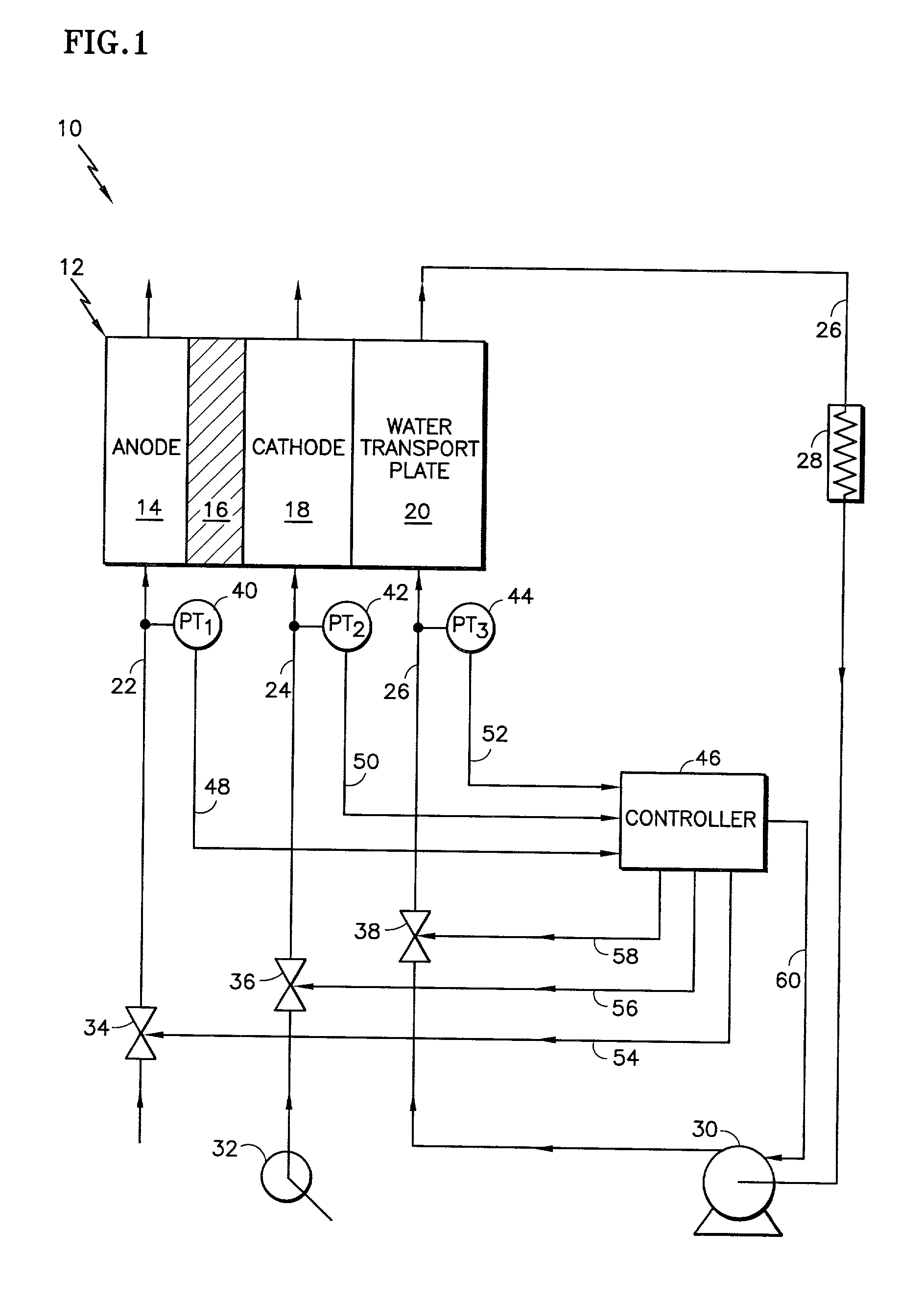
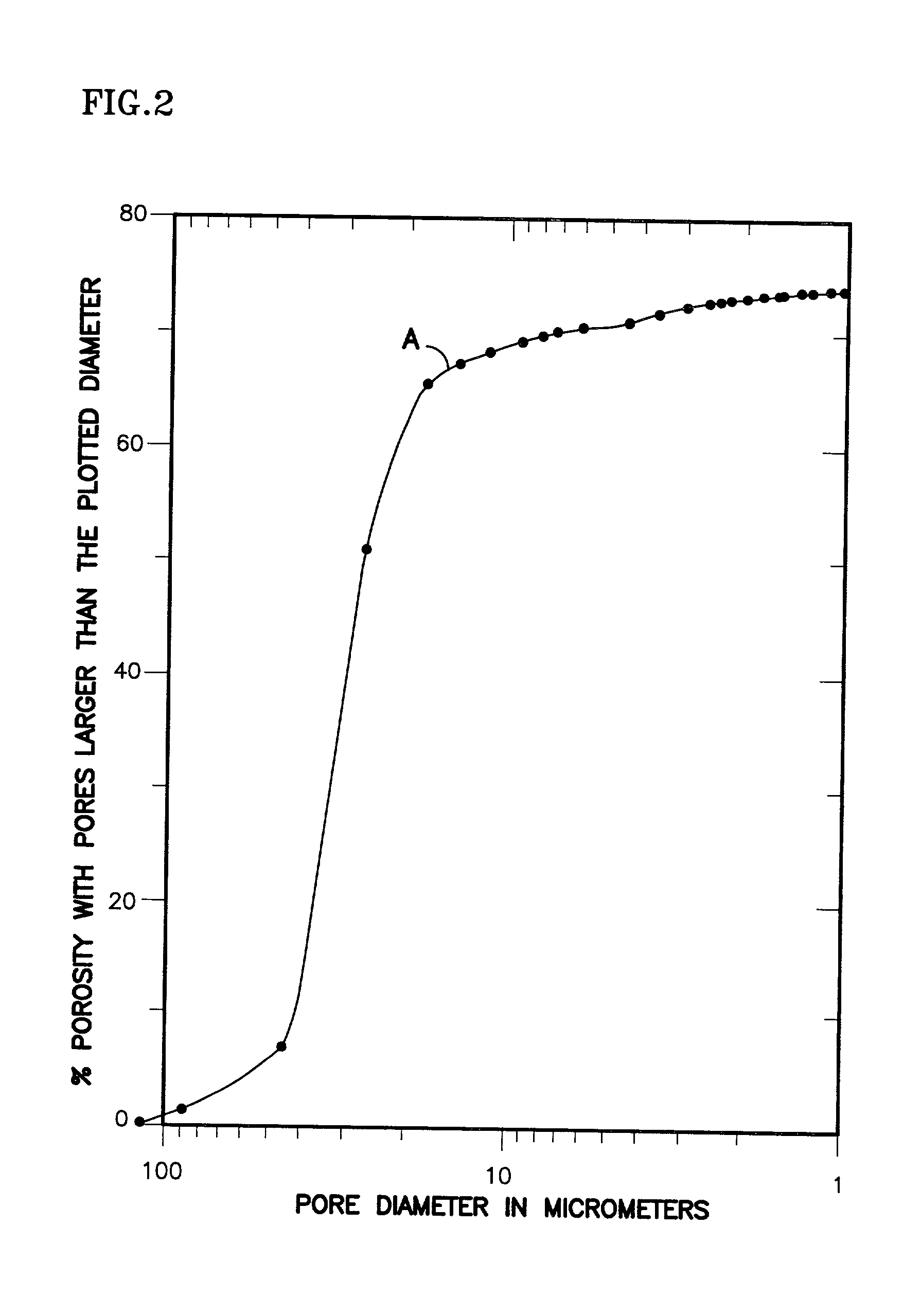
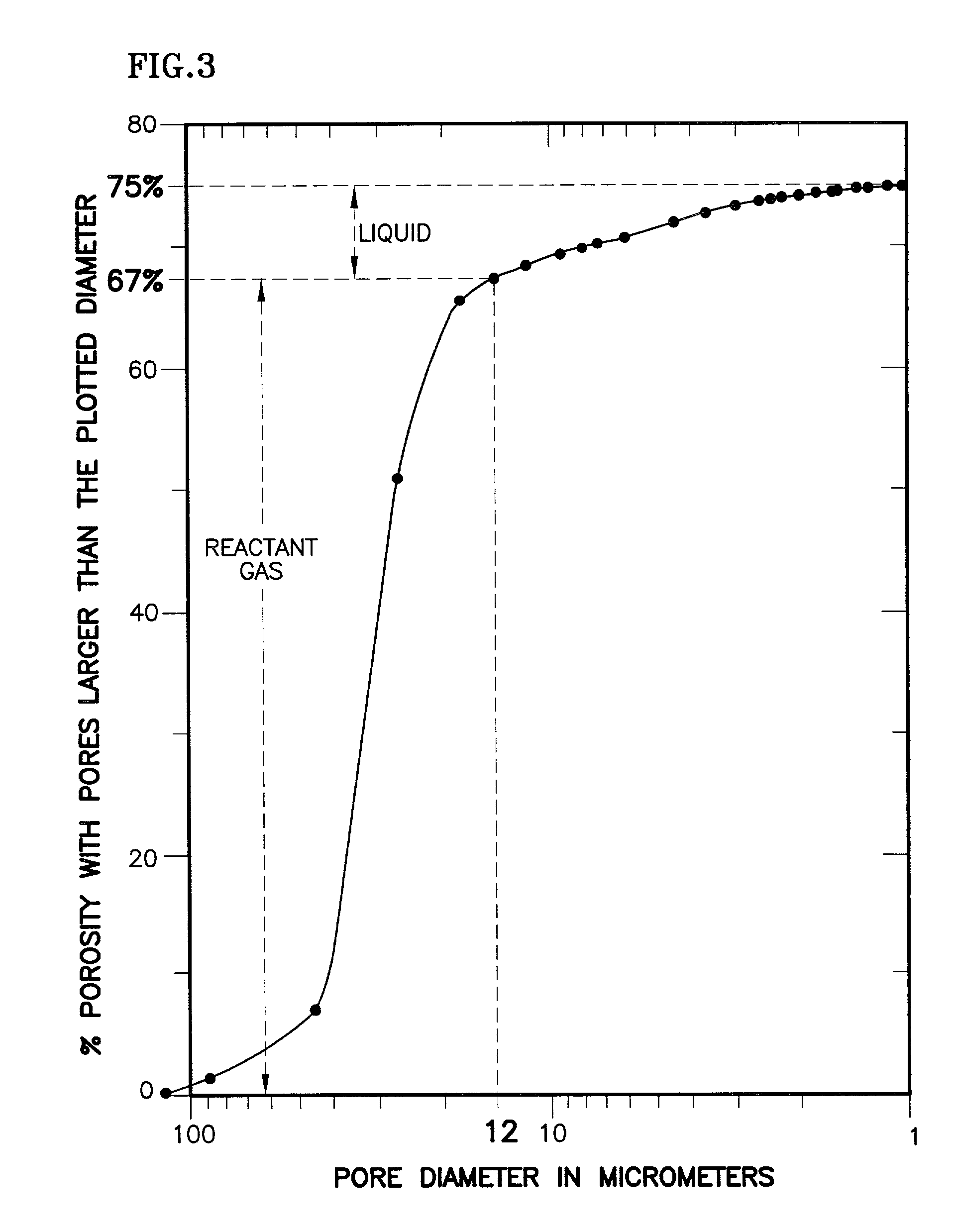
The present invention is a fuel cell power plant that includes a fuel cell having a membrane electrode assembly (MEA), which is disposed between an anode support plate and a cathode support plate, and porous water transport plates adjacent the anode and cathode support plates. The porous water transport plates have interdigitated flow channels for the reactant gas streams to pass therethrough and conventional flow channels for a coolant stream to pass therethrough. The fuel cell power plant also has means for creating a pressure differential between the reactant gas streams and the coolant stream such that the pressure of the reactant gas streams is greater than the coolant stream. Incorporating the interdigitated flow channels into the porous water transport plates and operating the fuel cell at a pressure differential allows the coolant water to saturate the water transport plates thereby forcing the reactant gases into the anode and cathode support plate. This, in turn, increases the mass transfer of such gases into the support plates, thereby increasing the electrical performance of the fuel cell. Current densities of about 1.6 amps per square meter are achieved with air stochiometries of not over 2.50.
Owner:AUDI AG
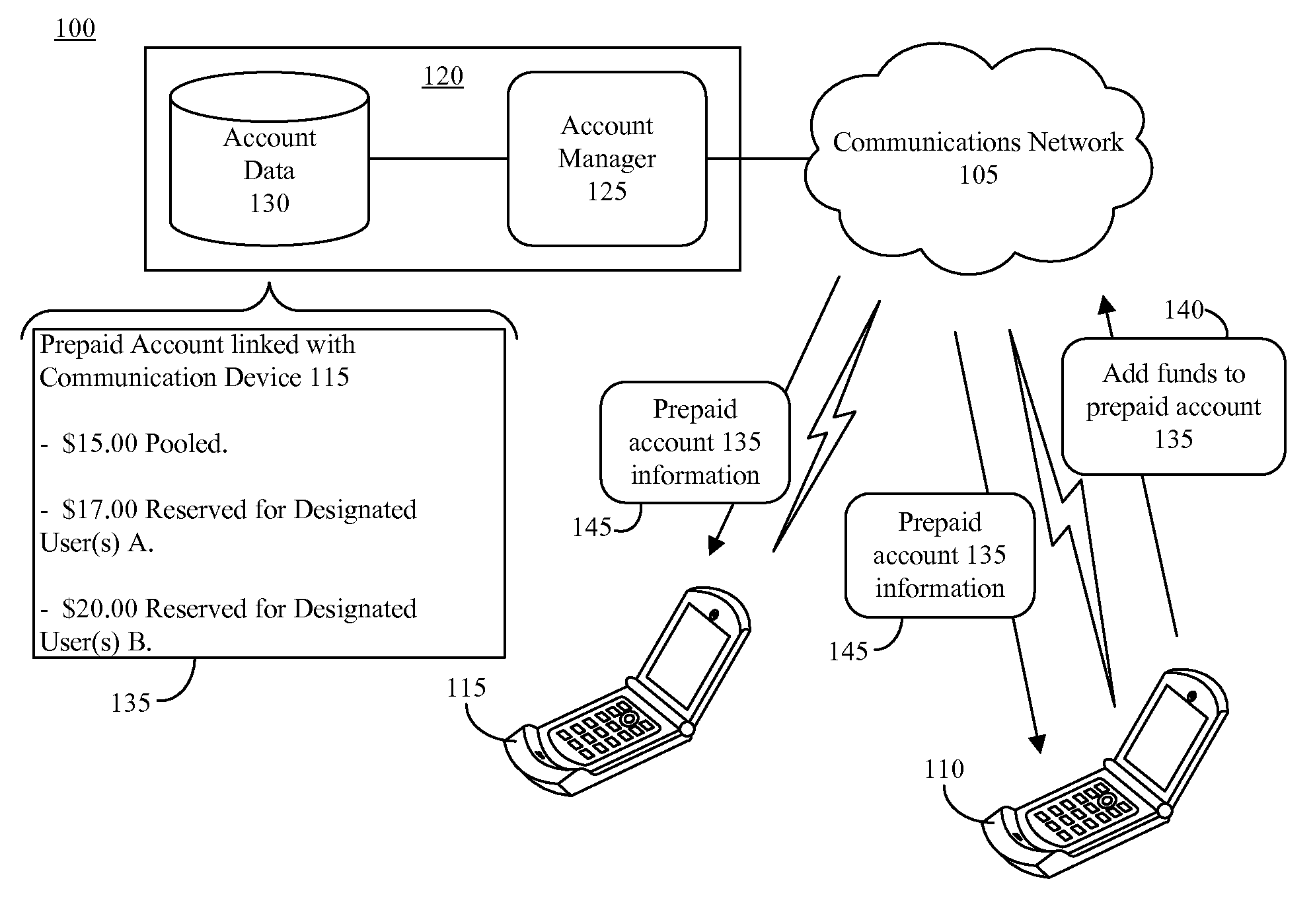
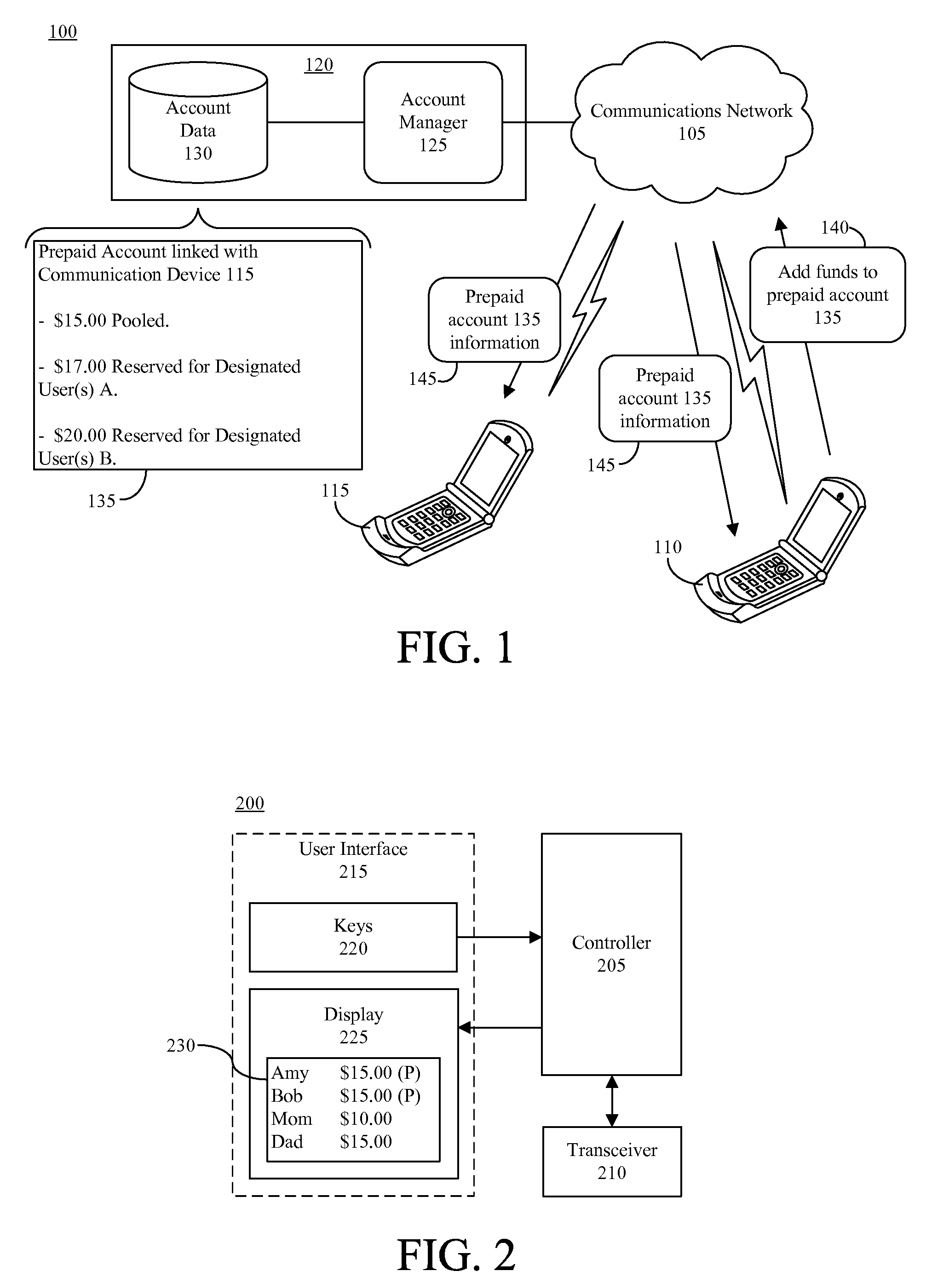
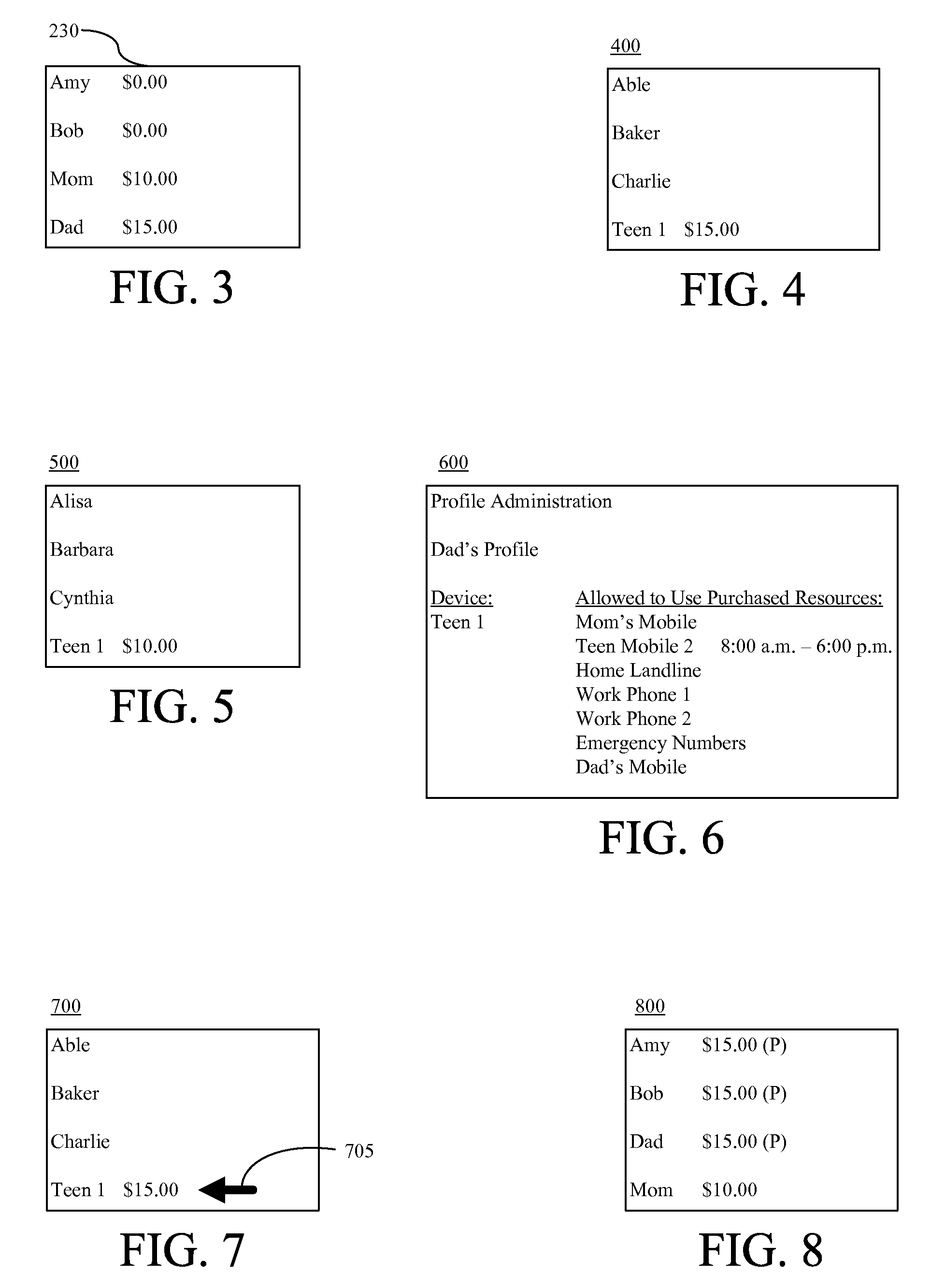
Prepaying usage time for another communication device
InactiveUS20080130849A1Improve balanceCutting portionAccounting/billing servicesPrepayment with prepaid account/card rechargingCommunication deviceMultimedia
A method of prepaying for usage time for a communication device can include increasing an available balance of a prepaid account linked to a communication device of a first user by a designated amount responsive to a request from a second user (910) and allocating a portion of the available balance of the prepaid account that is attributable to the designated amount for communicating with a designated user (915). The method further can include paying charges resulting from communications between the communication device of the first user and the designated user from the allocated portion of the available balance of the prepaid account (925).
Owner:GOOGLE TECH HLDG LLC
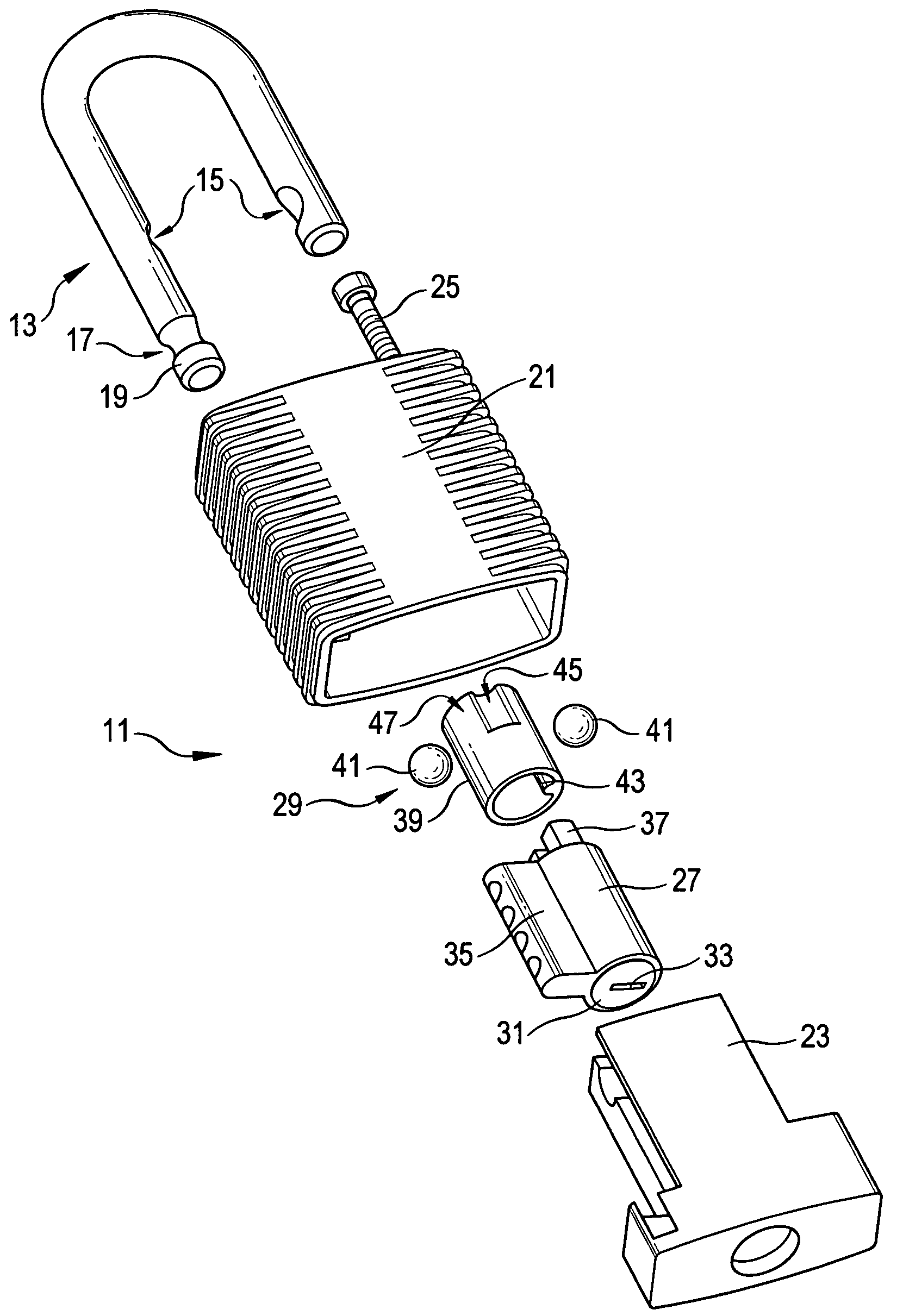
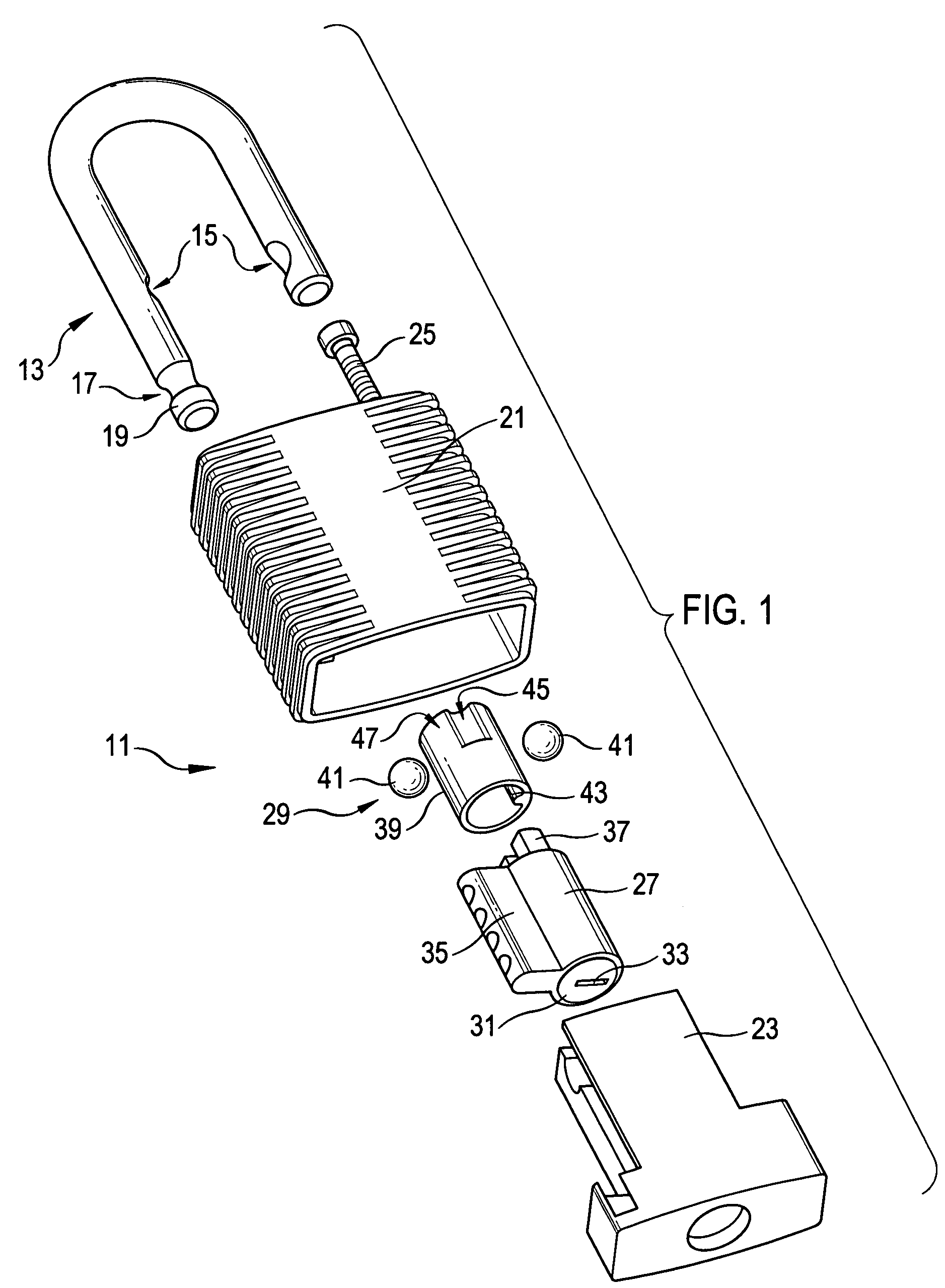
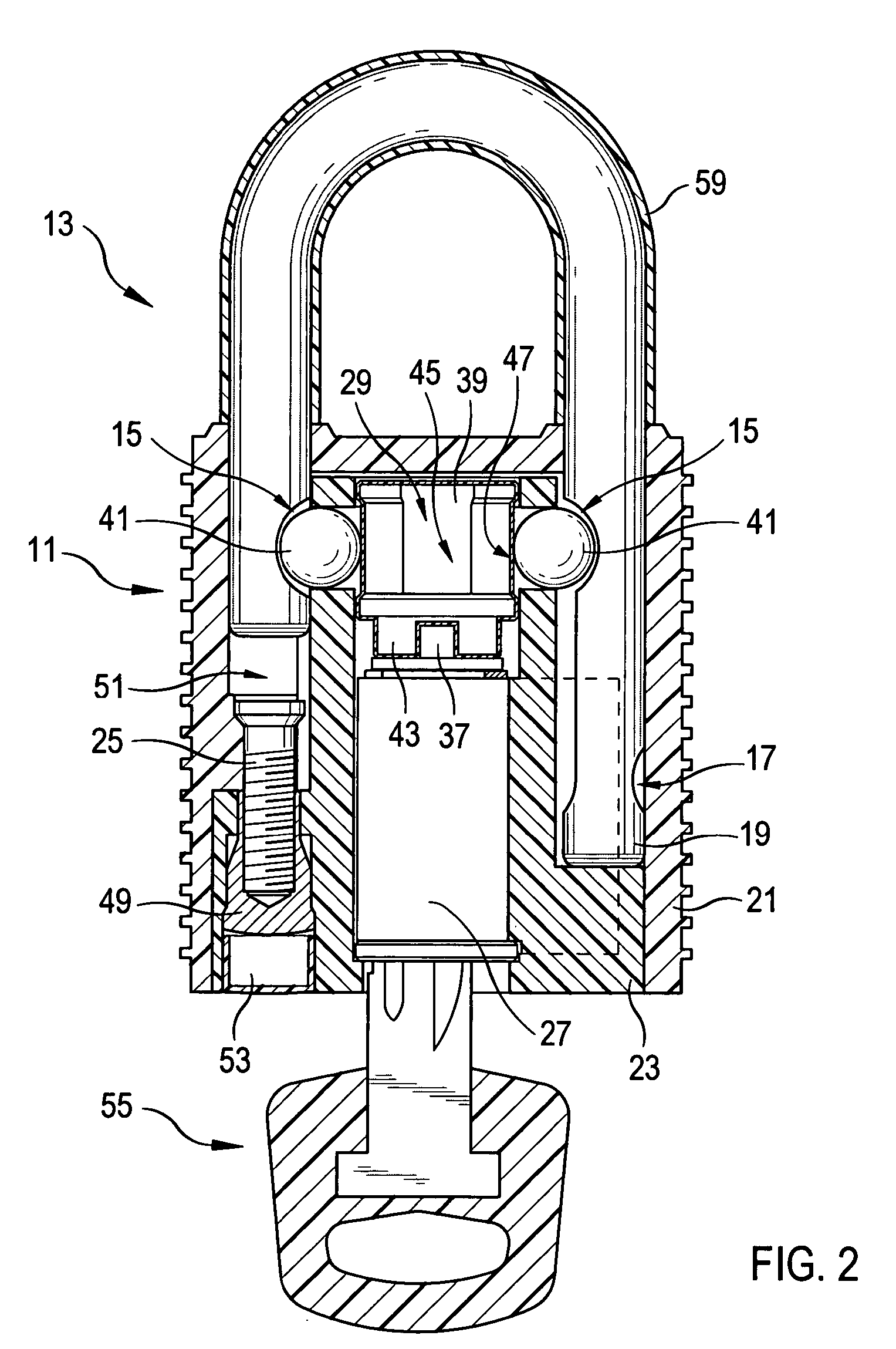
Padlock
ActiveUS7278283B2Improve mechanical stabilityImprove guidancePadlocksCarpet fastenersEngineeringElectrical and Electronics engineering
Owner:ABUS AUGUST BREMICKER SOEHNE AG
Bulk acoustic wave filter with a roughened substrate bottom surface and method of fabricating same
InactiveUS6943647B2Effect deterioratesEasy to integratePiezoelectric/electrostrictive device manufacture/assemblyPiezoelectric/electrostriction/magnetostriction machinesAcoustic waveBulk acoustic wave
A filter device comprises a substrate having a top surface and a bottom surface, and at least one acoustic wave situated on the top surface of the substrate, wherein the bottom surface of the substrate is roughened to reduce the reflection of an acoustic wave back to the acoustic wave filter. The effect achieved by the roughening of the bottom surface of the substrate that an acoustic wave which is generated by the acoustic wave filter and reaches the bottom surface of the substrate, is basically scattered so the acoustic wave that is actually reflected back to the acoustic wave device is reduced which, in turn, improves the performance characteristics of the acoustic wave filter.
Owner:NOKIA CORP +1
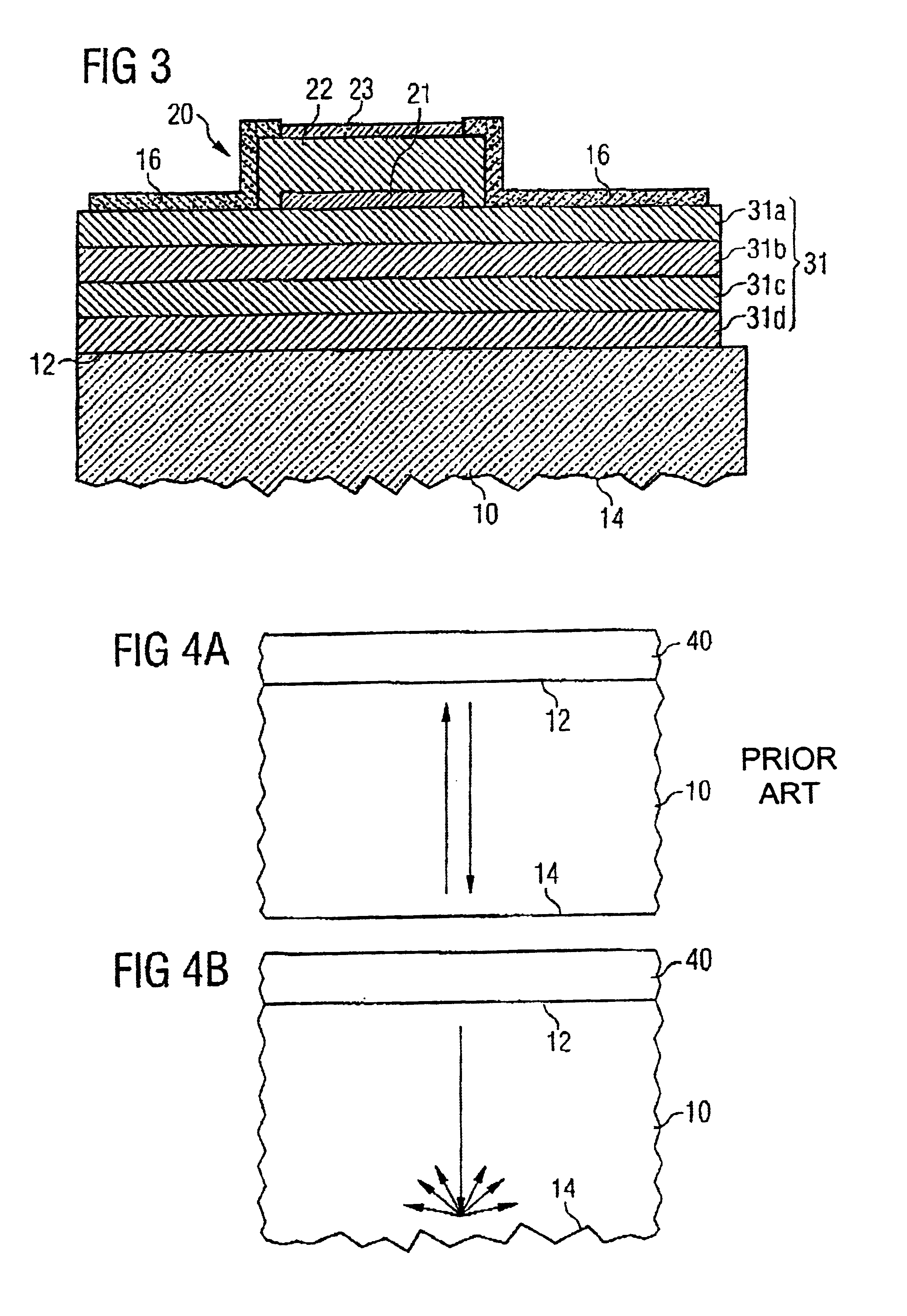
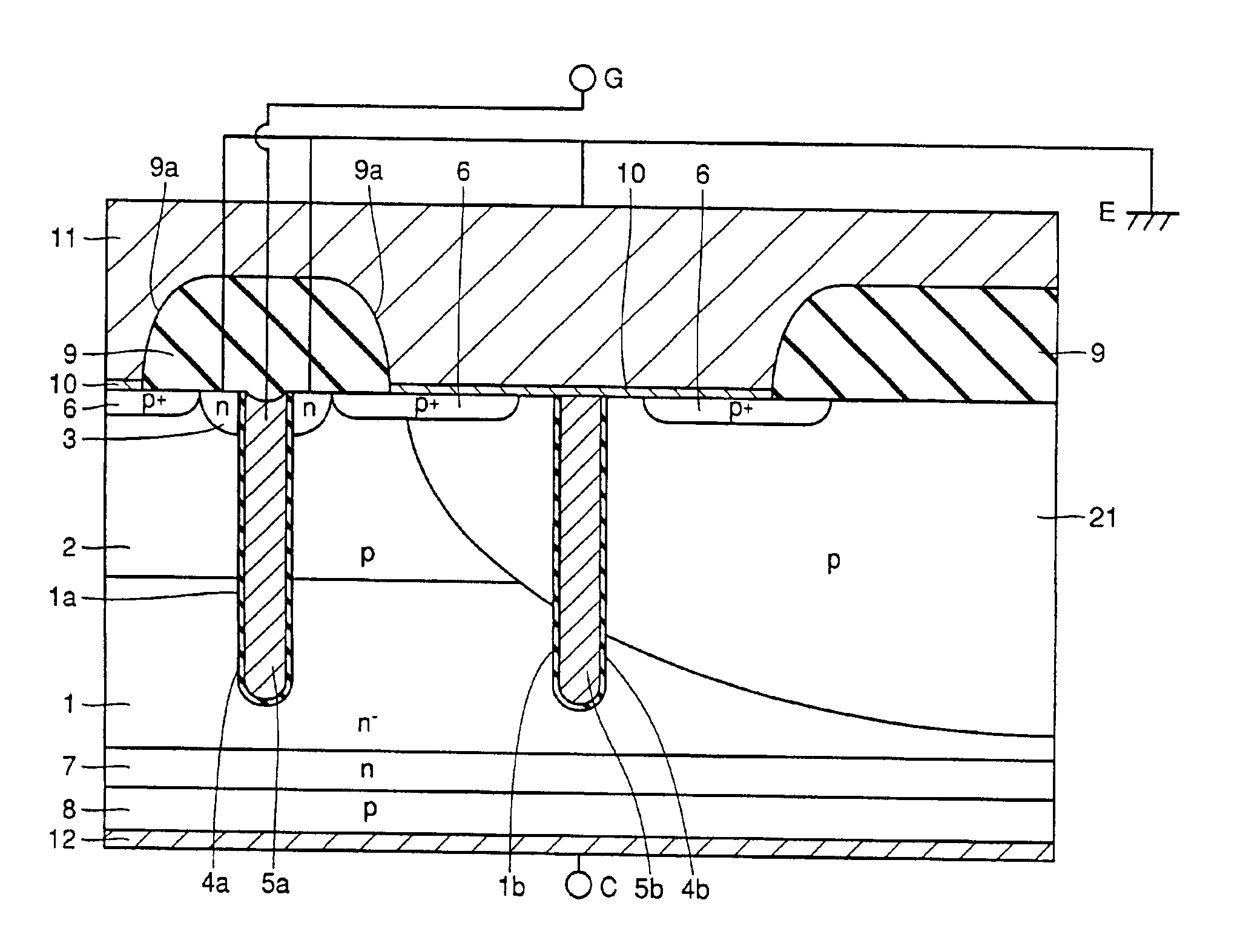
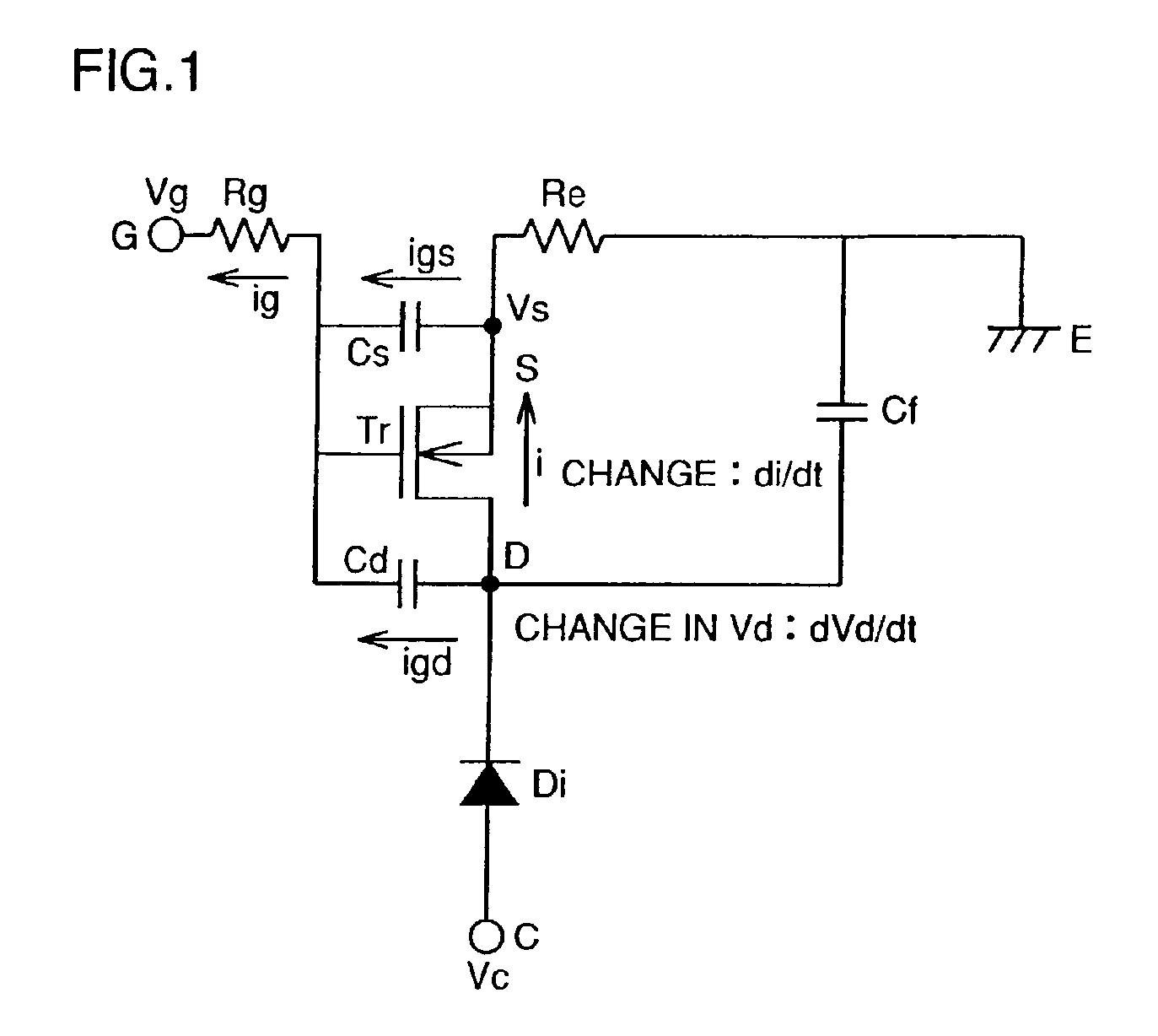
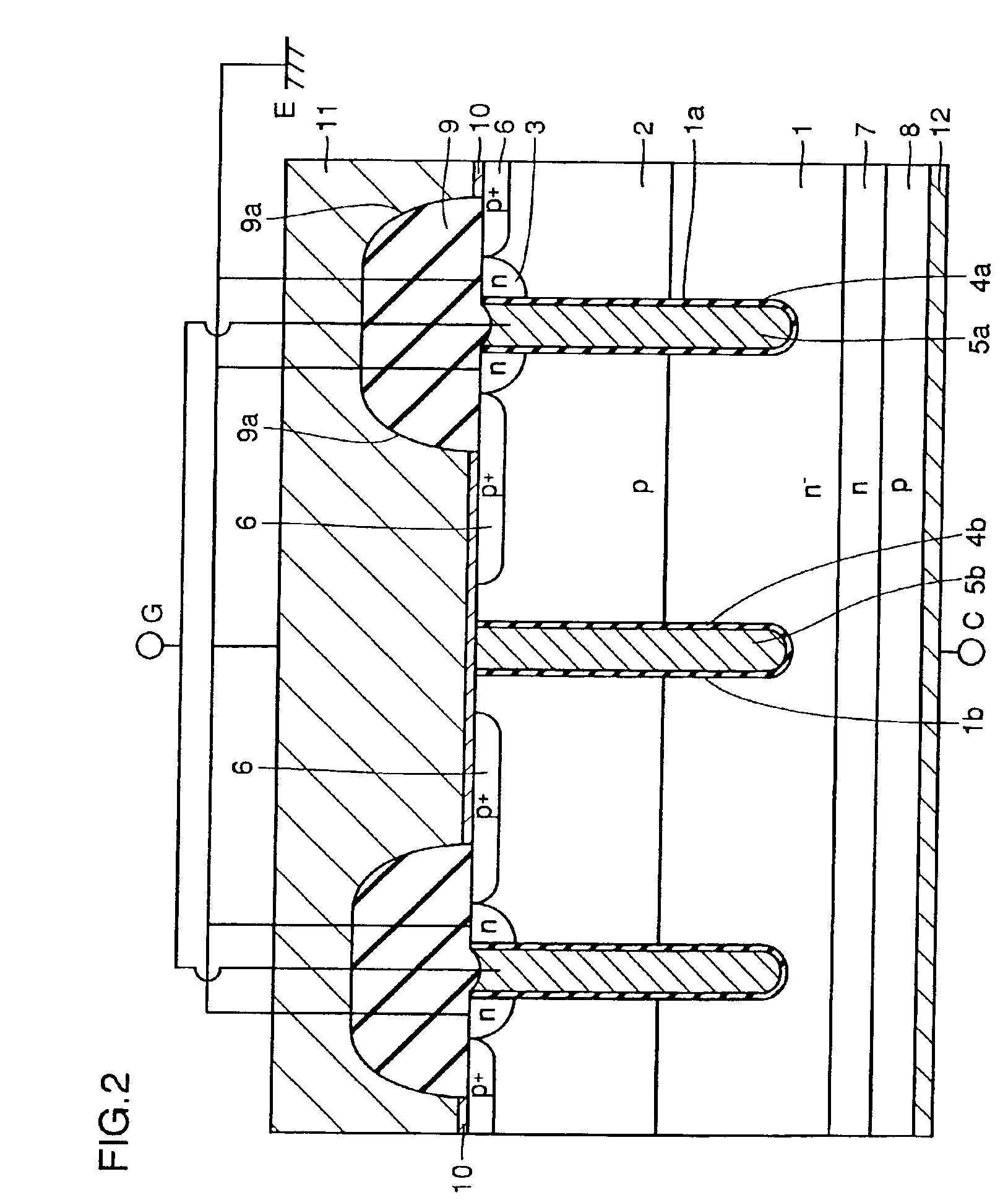
High voltage withstanding semiconductor device
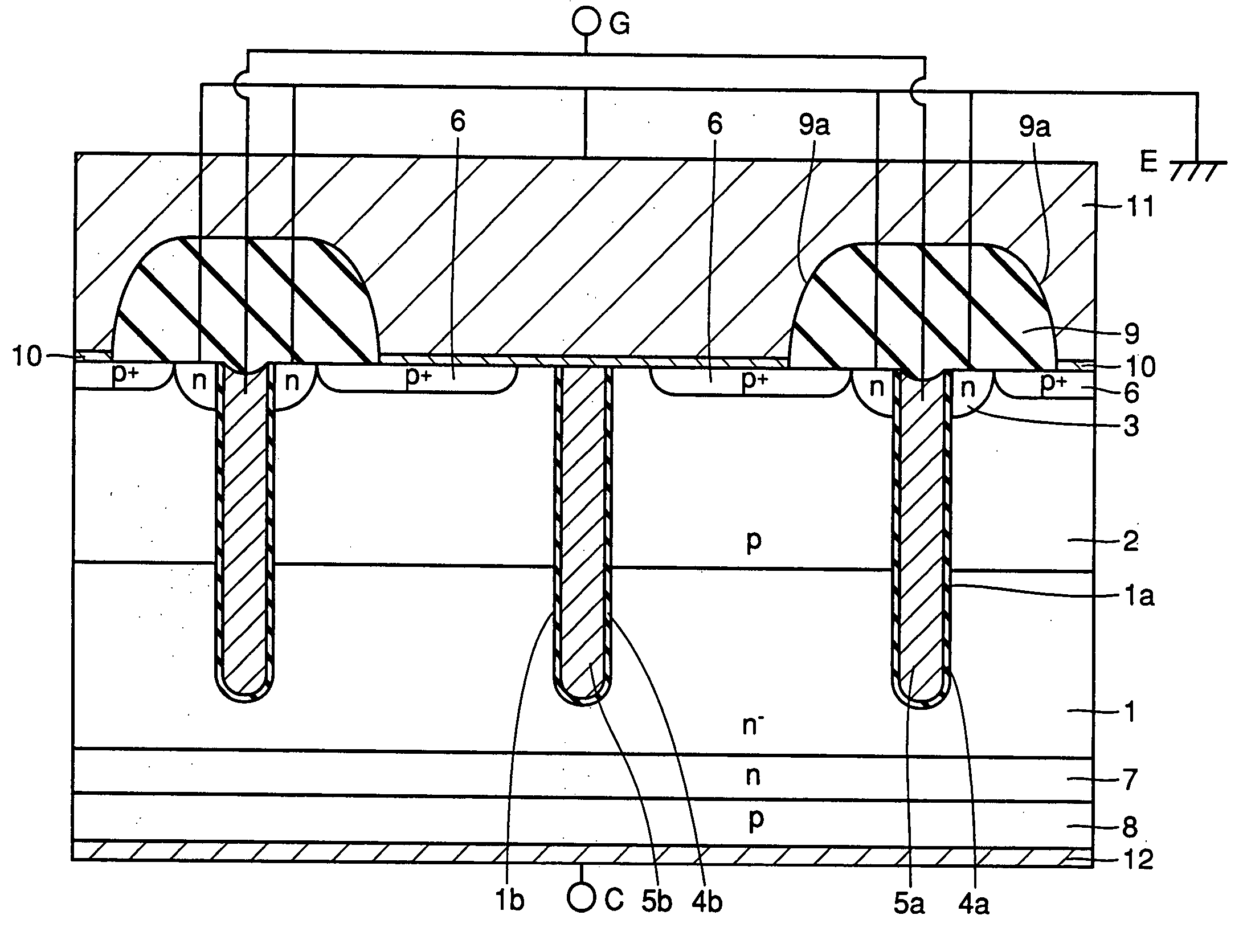
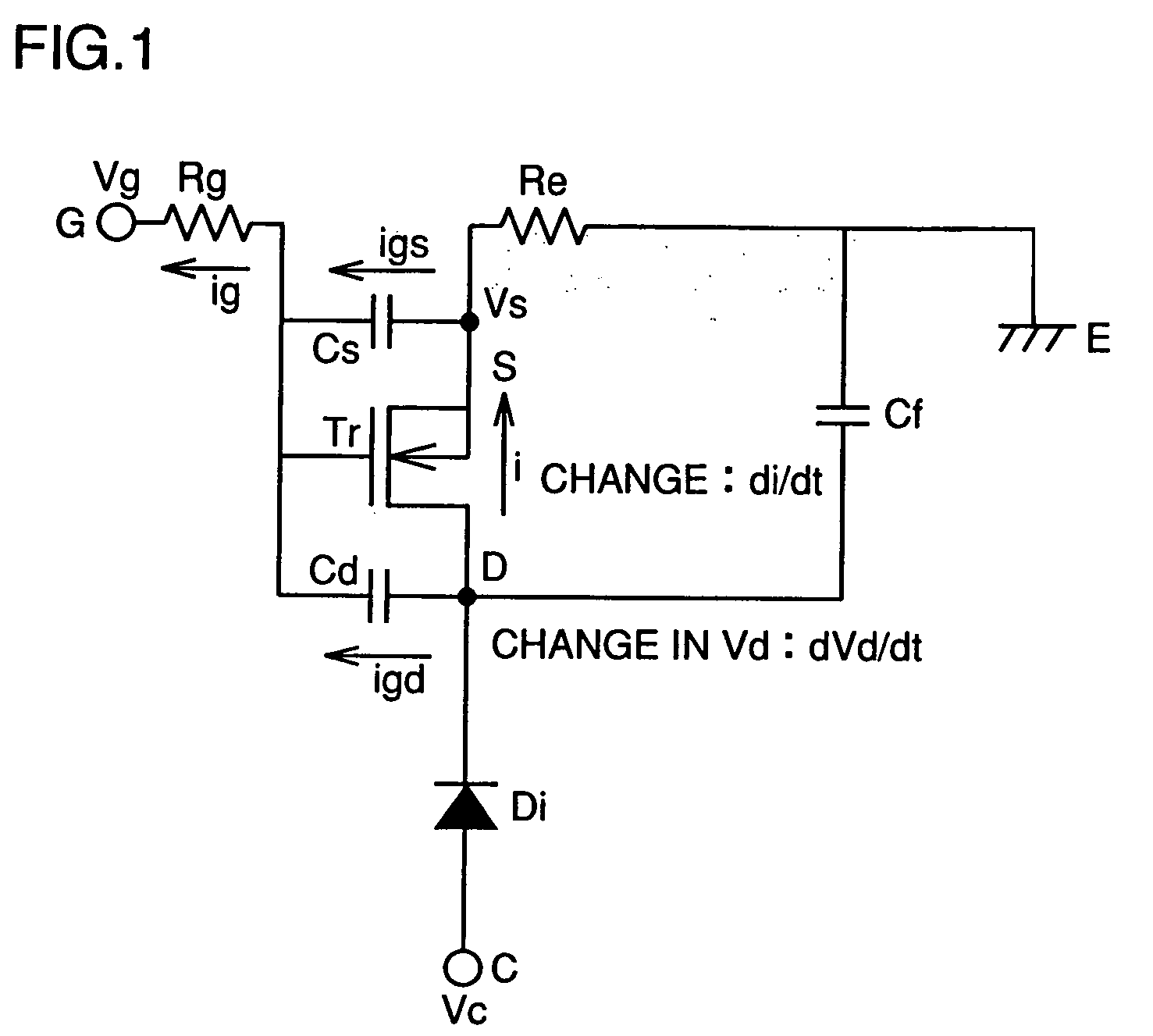
A semiconductor device of the present invention has an insulating gate type field effect transistor portion having an n-type emitter region (3) and an n− silicon substrate (1), which are opposed to each other sandwiching a p-type body region (2), as well as a gate electrode (5a) which is opposed to p-type body region (2) sandwiching a gate insulating film (4a), and also has a stabilizing plate (5b). This stabilizing plate (5b) is made of a conductor or a semiconductor, is opposed to n− silicon substrate (1) sandwiching an insulating film (4, 4b) for a plate, and forms together with n− silicon substrate (1), a capacitor. This stabilizing plate capacitor formed between stabilizing plate (5b) and n− silicon substrate (1) has a capacitance greater than that of the gate-drain capacitor formed between gate electrode (5a) and n− silicon substrate (1).
Owner:MITSUBISHI ELECTRIC CORP
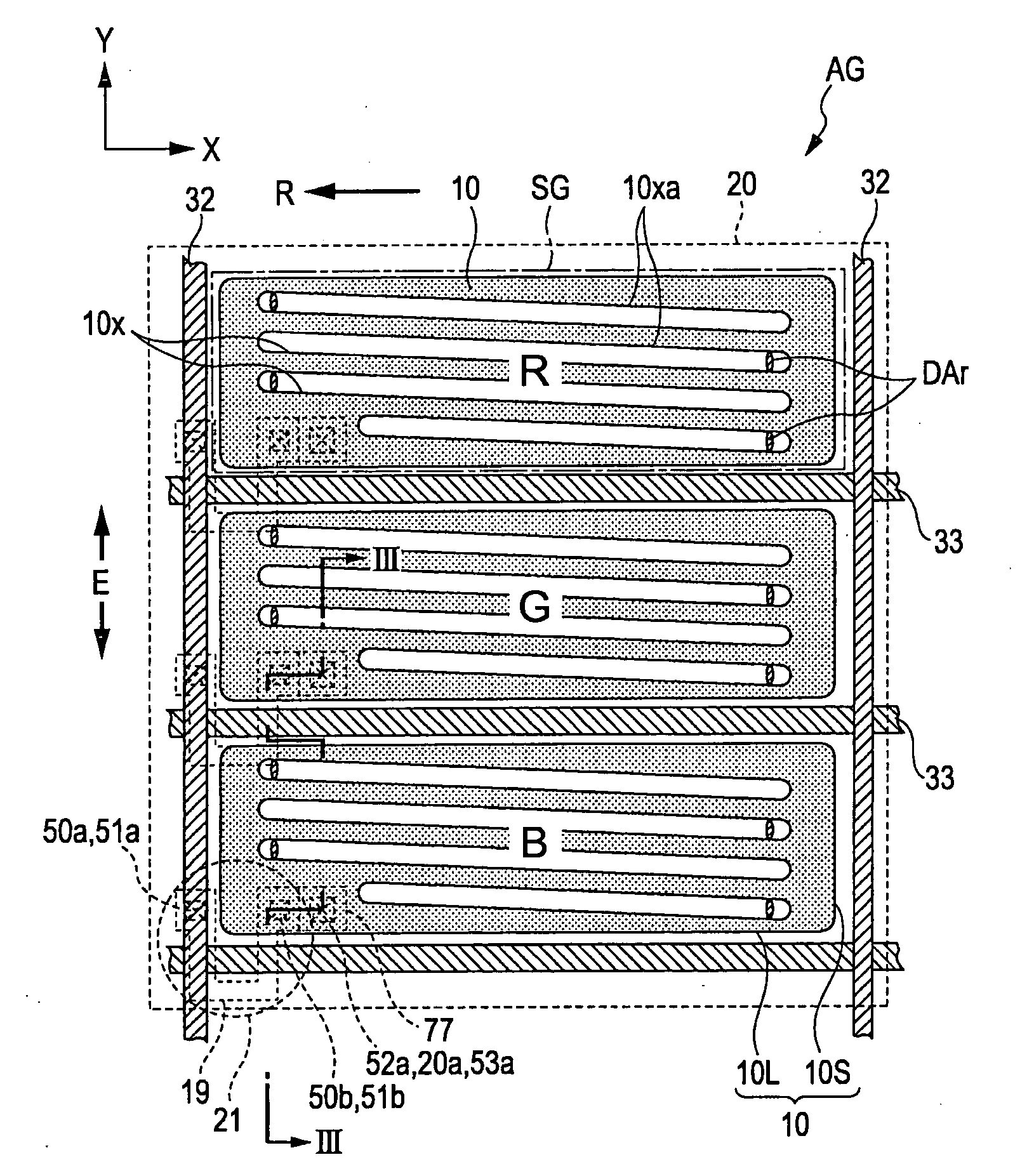
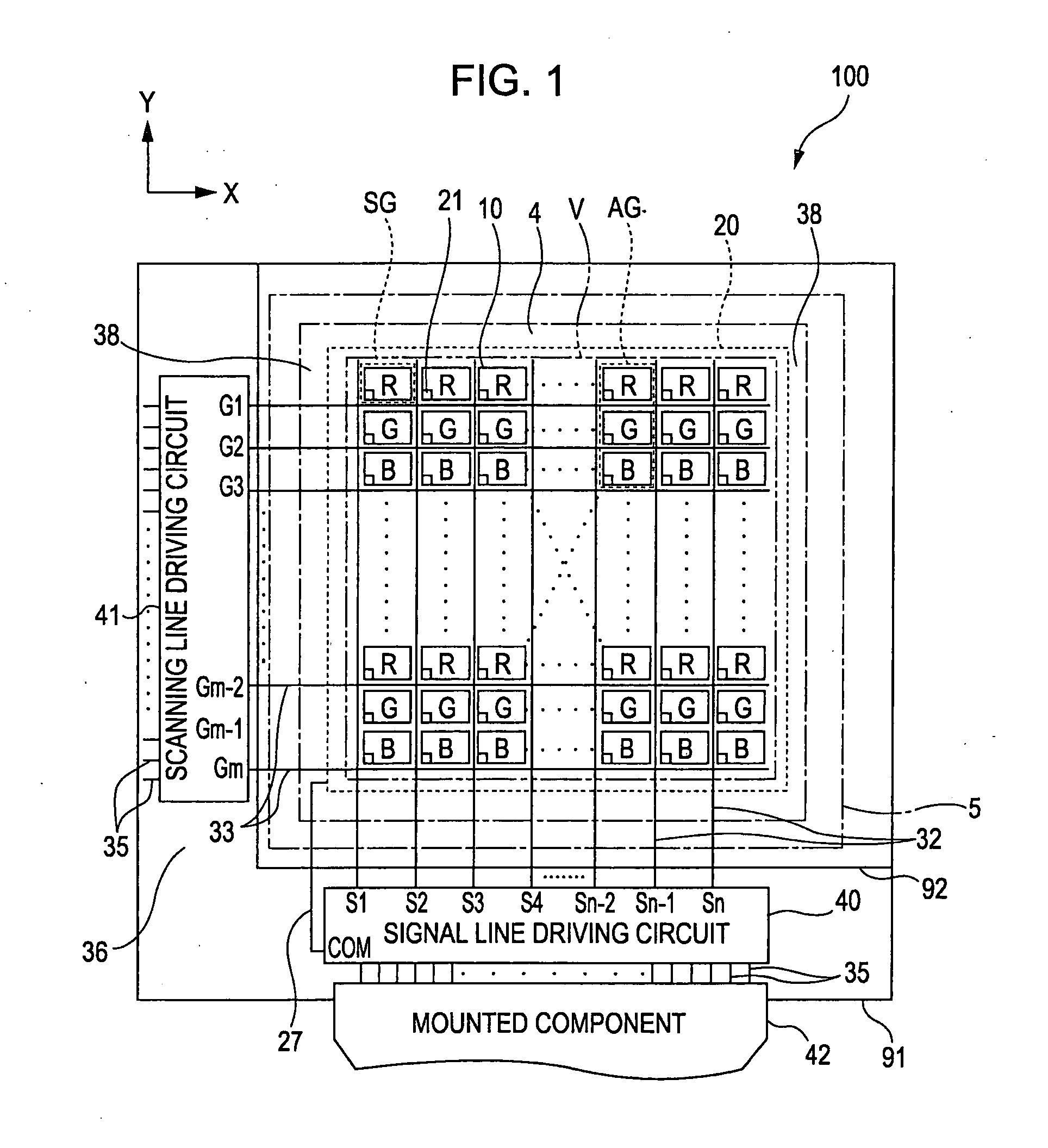
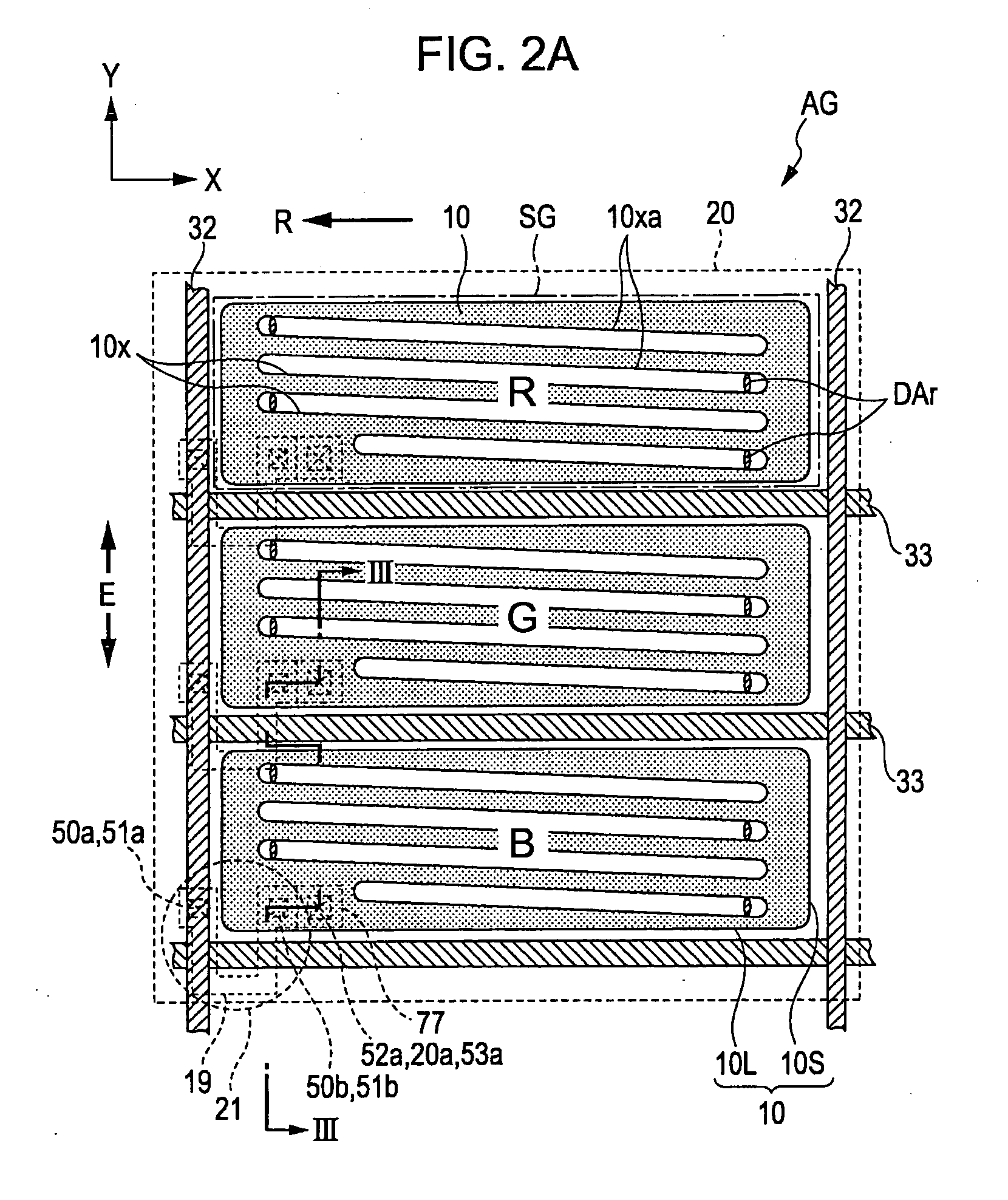
Liquid crystal device and electronic apparatus
1. A liquid crystal device includes a substrate including unit pixels each composed of a plurality of sub pixels arranged in a plurality of rows and one column and having a long side in the row direction and a short side in the column direction, wherein the substrate includes switching elements, an insulating film provided on at least the upper side of each of the switching elements, at least one first transparent electrodes provided on the upper side of the insulating film, other insulating film provided on the upper side of the first transparent electrodes, and at least one second transparent electrode formed on the upper side of the other insulating film and having a plurality of slits formed for each of the sub pixels and generating an electric field, the electric field being generated between the first transparent electrode and the second transparent electrode through each of the slits, and the extending direction of the long side of each of the slits is defined in a direction not the same as the extending direction of the short side of the sub pixels.
Owner:EPSON IMAGING DEVICES CORP
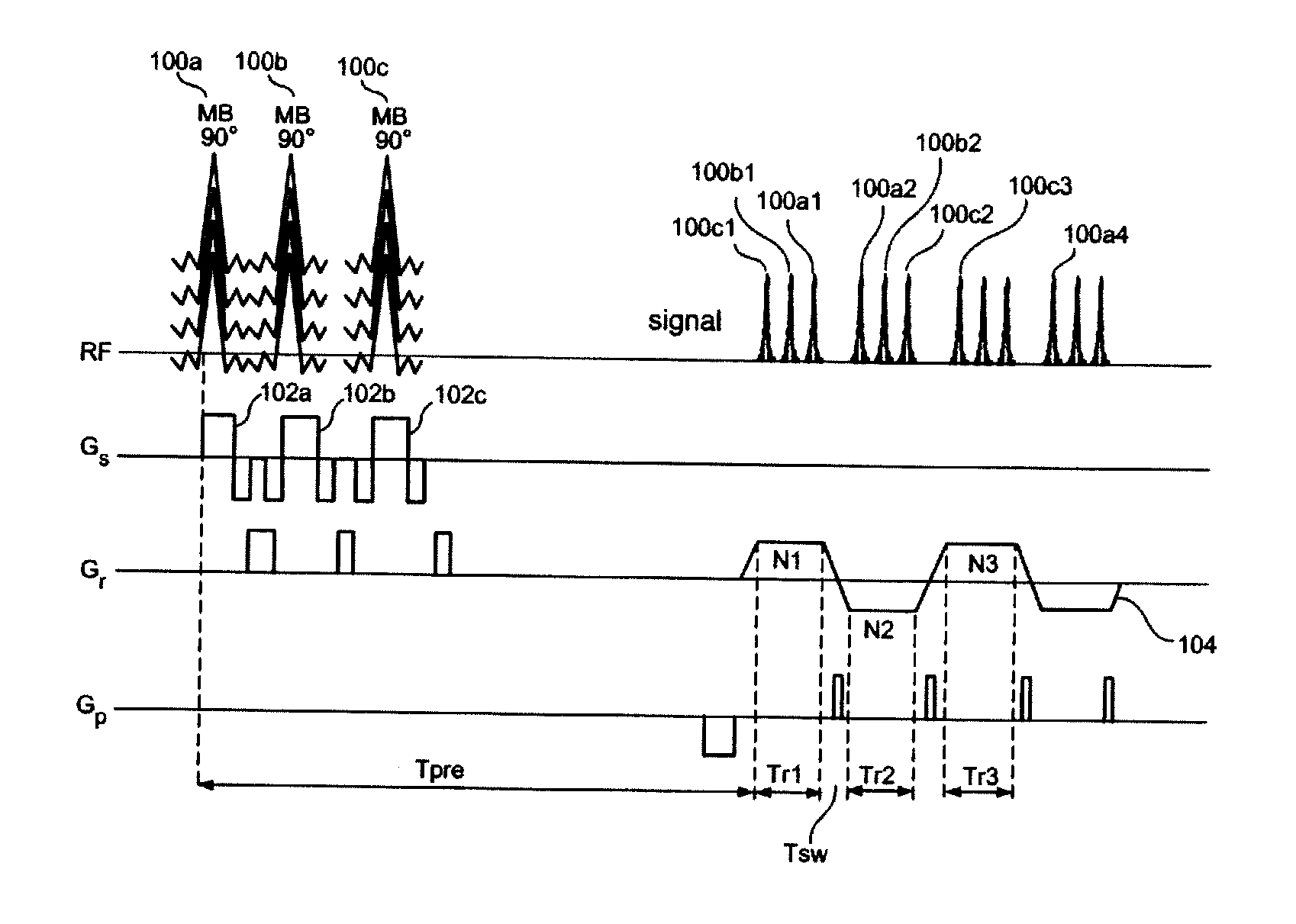
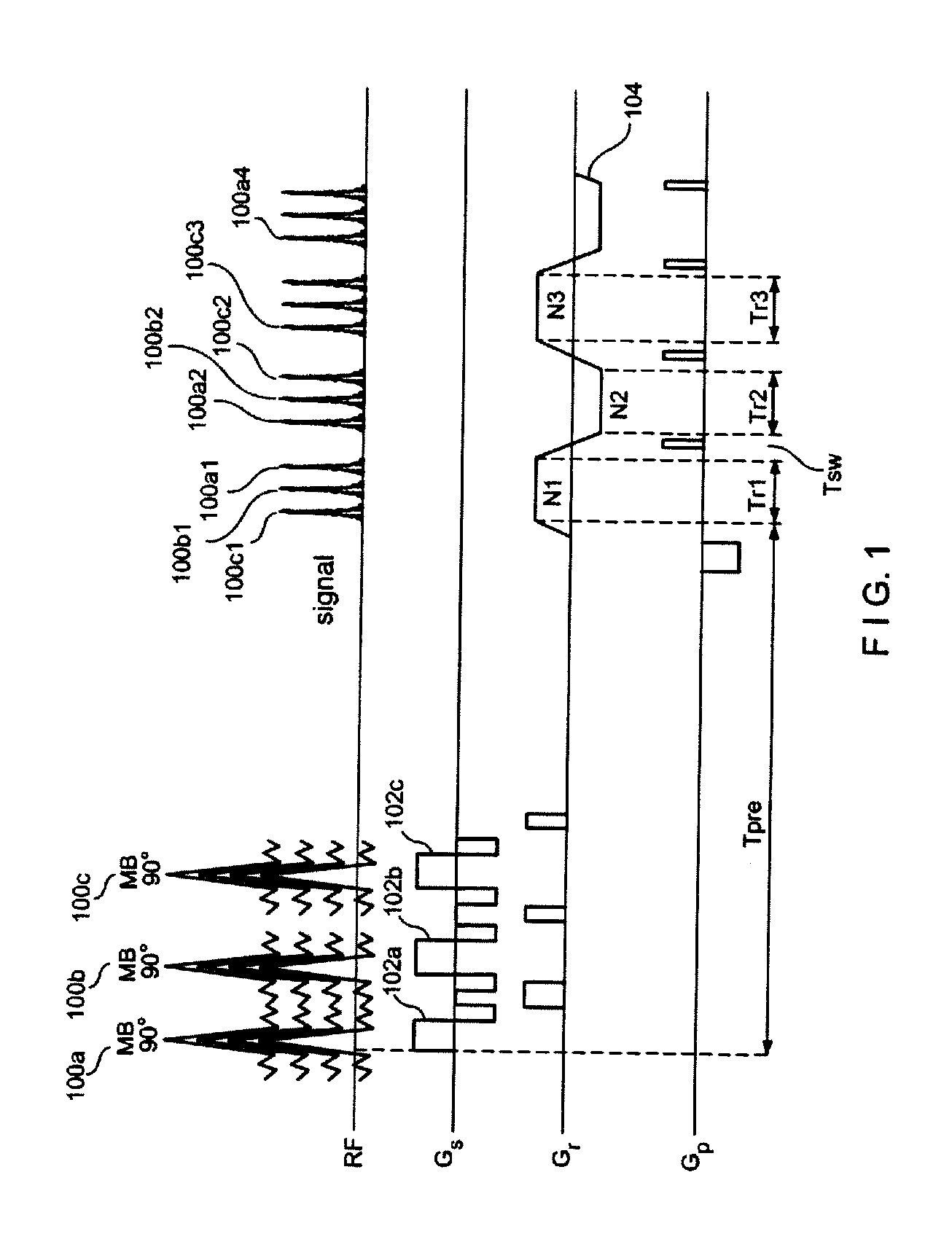
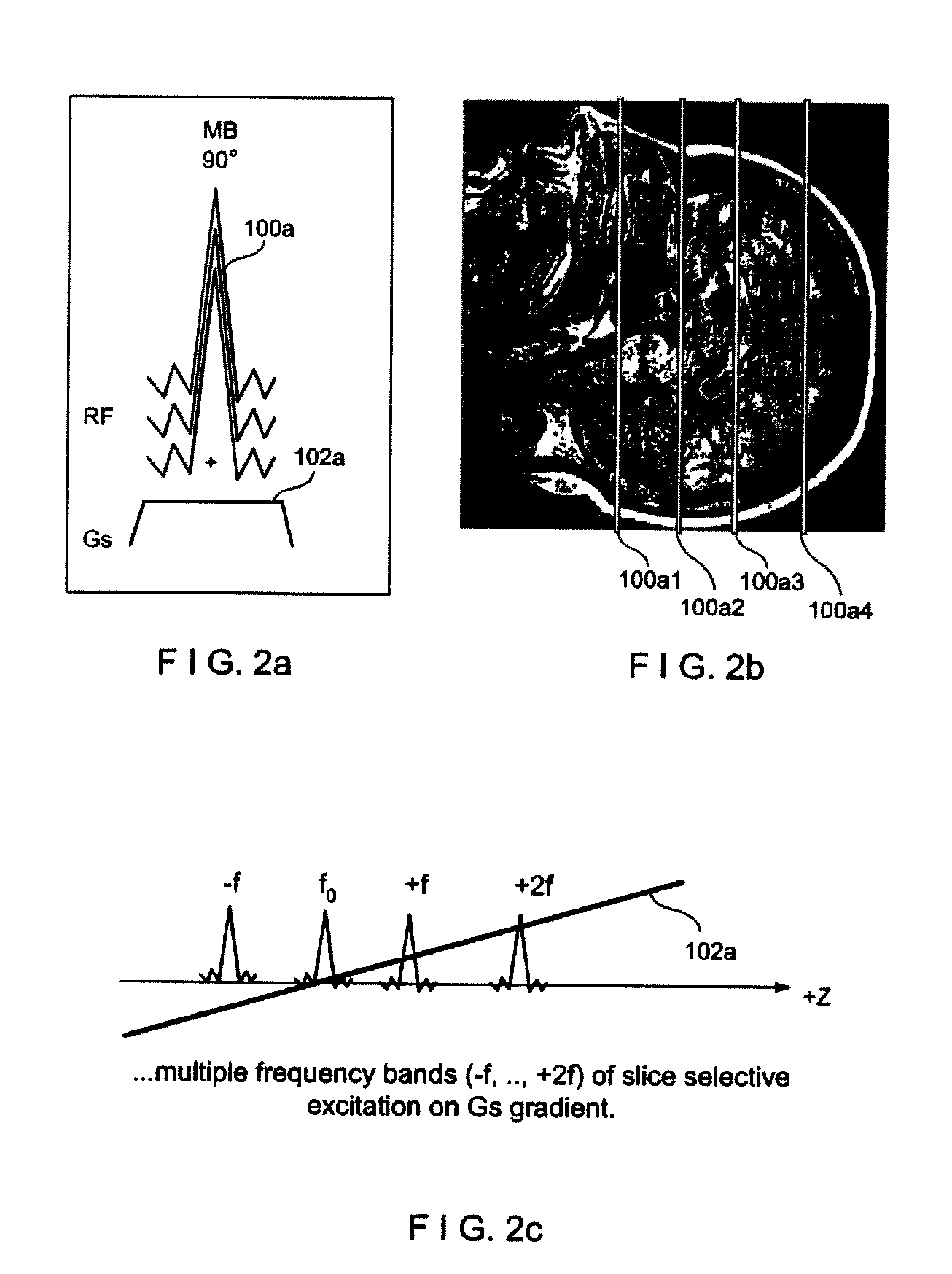
Multiplicative increase in MRI data acquisition with multi-band RF excitation pulses in a simultaneous image refocusing pulse sequence
ActiveUS20120056620A1Reduce acquisition timePromote absorptionMeasurements using NMR imaging systemsElectric/magnetic detectionMulti bandMultiplexing
Disclosed are methods and systems for carrying out super-multiplexed magnetic resonance imaging that entwines techniques previously used individually and independently of each other in Simultaneous Echo (of Imaging) Refocusing (SER or SIR) and Multi-Band (MB) excitation, in a single pulse sequence that provides a multiplication rather than summation of desirable effects while suppressing undesirable effects of each of the techniques that previously were used independently.
Owner:FEINBERG DAVID +1
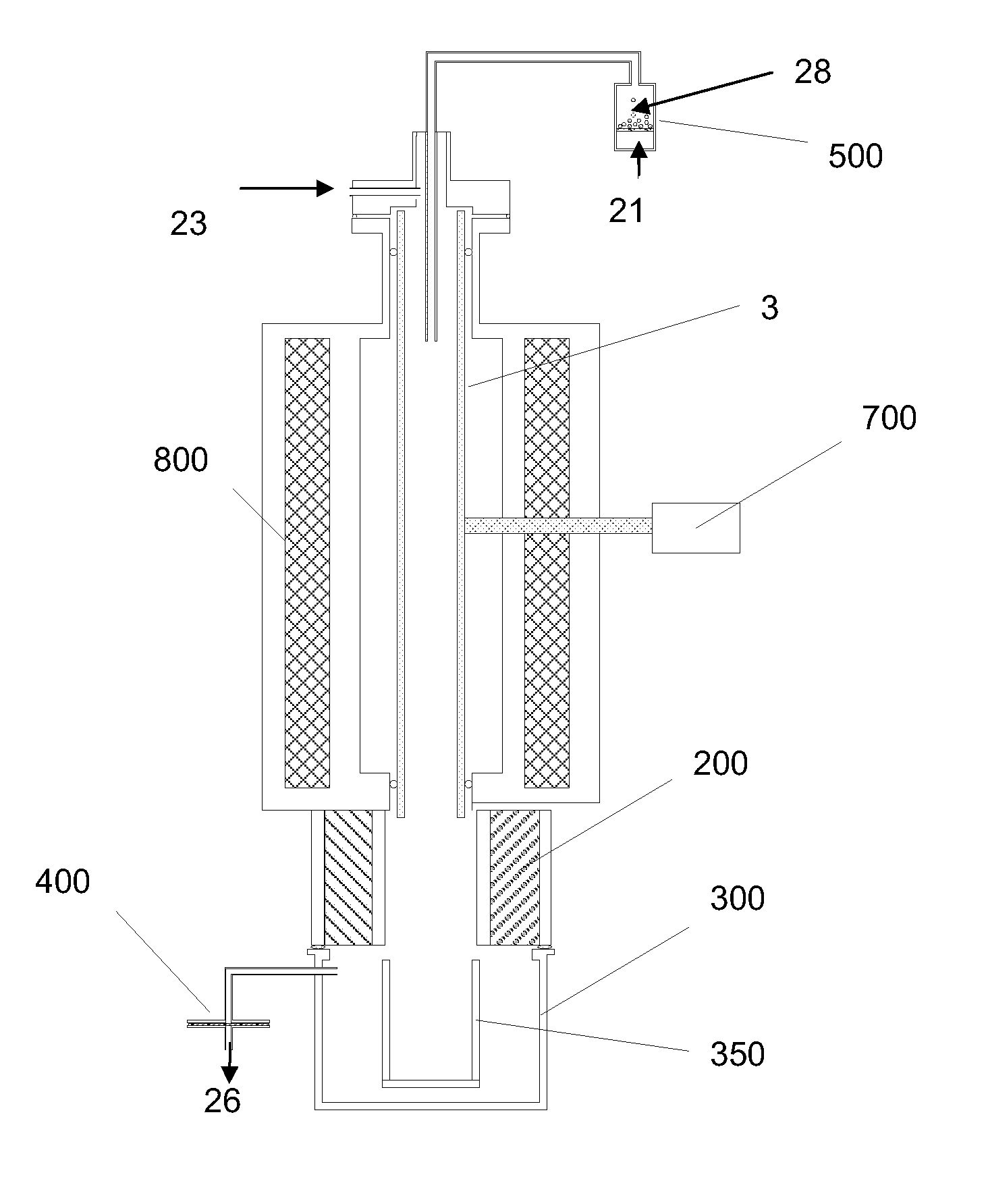
System and process for hydrodesulfurization, hydrodenitrogenation, or hydrofinishing
ActiveUS20090000989A1Increase ratingsLow costShaking/oscillating/vibrating mixersFlow mixersHydrogenFixed bed
A method for hydrodesulfurization by forming a dispersion comprising hydrogen-containing gas bubbles with a mean diameter of less than 1 micron dispersed in a liquid phase comprising sulfur-containing compounds. Desulfurizing a liquid stream comprising sulfur-containing compounds by subjecting a fluid mixture comprising hydrogen-containing gas and the liquid to a shear rate greater than 20,000 s−1 to produce a dispersion of hydrogen in a continuous phase of the liquid and introducing the dispersion into a fixed bed hydrodesulfurization reactor from which a reactor product is removed. Systems of apparatus for hydrodesulfurization are also presented.
Owner:HRD CORP
Acid gas management in liquid fuel production process
ActiveUS20150005398A1Promote formationLow costGas modification by gas mixingLiquid hydrocarbon mixture productionSyngasSteam reforming
A process and method for producing liquid fuel product from hydrogen and carbon monoxide containing streams produced by gasifying solid carbonaceous feedstock and steam reforming of light fossil fuels. The gasifier syngas is treated to preferentially remove at least 99% of sulfur containing impurities and less than 50% of the CO2 to produce a treated gasifier syngas and a CO2 enriched gas. The treated gasifier syngas and the light fossil fuel conversion unit product gas are combined to form a mixed syngas that is converted into a liquid fuel product. The CO2 enriched gas is used in the gasification unit.
Owner:PRAXAIR TECH INC
Apparatus and methods relating to collecting and processing human biological material containing adipose
ActiveUS20130164731A1Easy to transportCutting portionMedical devicesDead animal preservationLipoplastiesFat grafting
A portable apparatus for collection and processing of human biological material containing adipose, such as extracted during a lipoplasty procedure, is useful for multi-step processing to prepare a concentrated product (e.g., stromal vascular fraction) or a fat graft composition. The apparatus has a container with a containment volume with a tissue retention volume and a filtrate volume separated by a filter and with a tapered portion to a collection volume for collecting concentrate product. Inlet and suction ports provide access to the tissue retention volume and filtrate volume, respectively, and an extraction port provides versatile access for removal of target processed concentrate material or fat graft material, which access may be via a lumen through a rotatable mixer shaft. Access ports may be configured for access only from above the container. The apparatus is incorporatable into a variety of assemblies, systems, kits, methods and uses.
Owner:THE GID GROUP
Protein modeling tools
InactiveUS20030130797A1Rapid and computationally efficient generationEfficient representationDepsipeptidesPeptide preparation methodsProtein modellingSide chain
The invention provides a new, efficient method for the assembly of protein tertiary structure from known, loosely encoded secondary structure constraints and sparse information about exact side chain contacts. The method is based on a new method for the reduced modeling of protein structure and dynamics, where the protein is described by representing side chain centers of mass rather than alpha-carbons. The model has implicit, built-in multi-body correlations that simulate short- and long-range packing preferences, hydrogen bonding cooperativity, and a mean force potential describing hydrophobic interactions. Due to the simplicity of the protein representation and definition of the model force field, the Monte Carlo algorithm is at least an order of magnitude faster than previously published Monte Carlo algorithms for three-dimensional structure assembly. In contrast to existing algorithms, the new method requires a smaller number of tertiary constraints for successful fold assembly; on average, one for every seven residues as compared to one for every four residues. The reliability and robustness of the invention make it useful for routine application in model building protocols based on various (and even very sparse) experimentally-derived structural constraints.
Owner:SKOLNICK JEFFREY +1
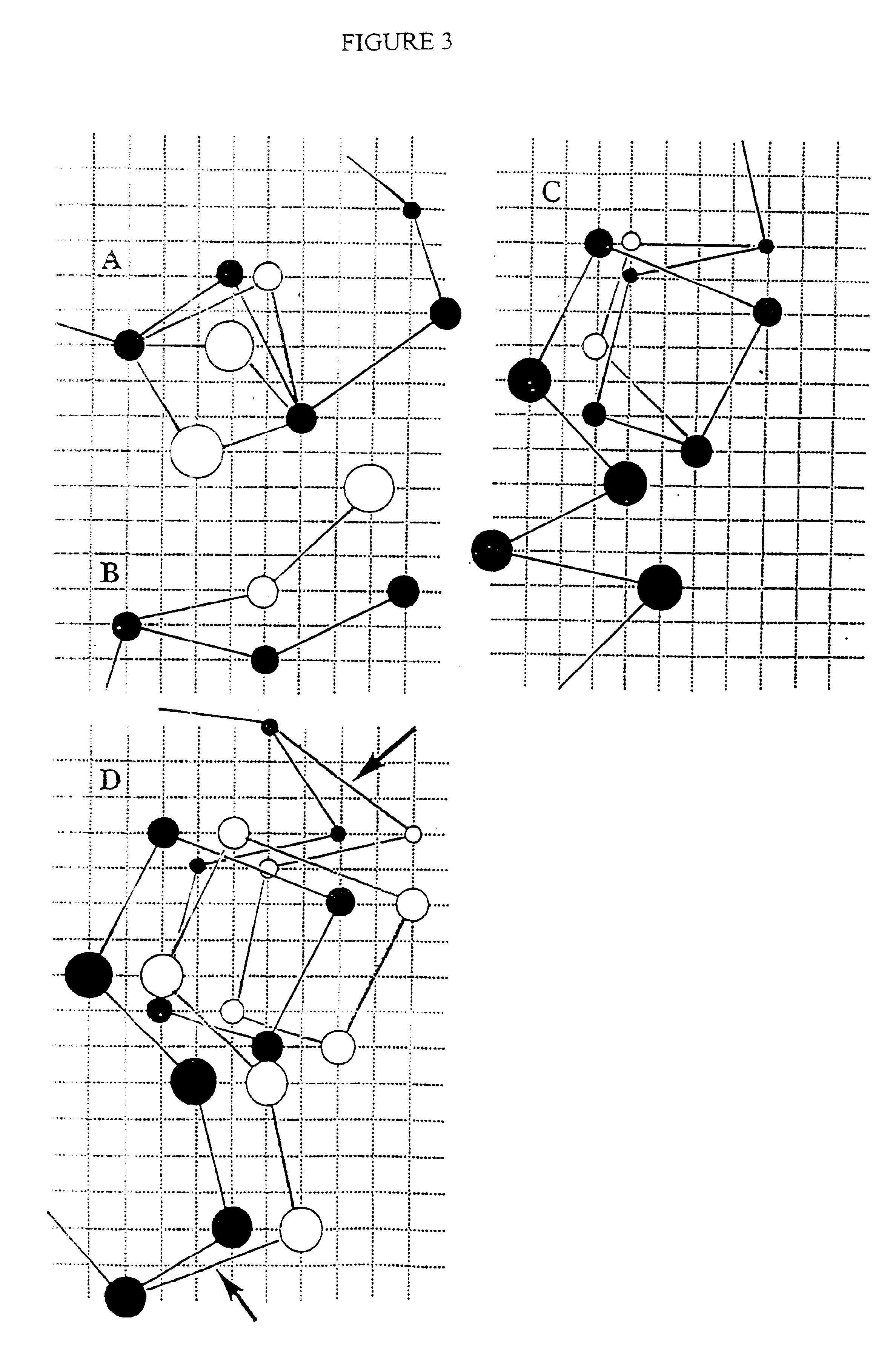
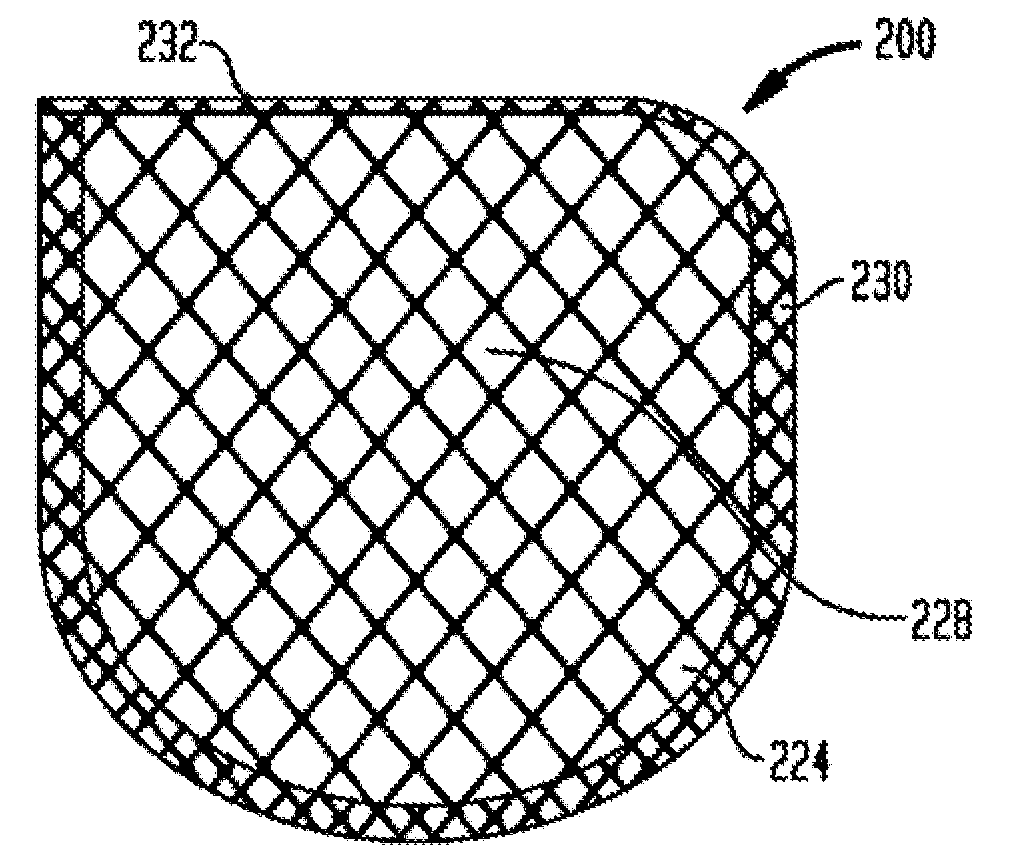
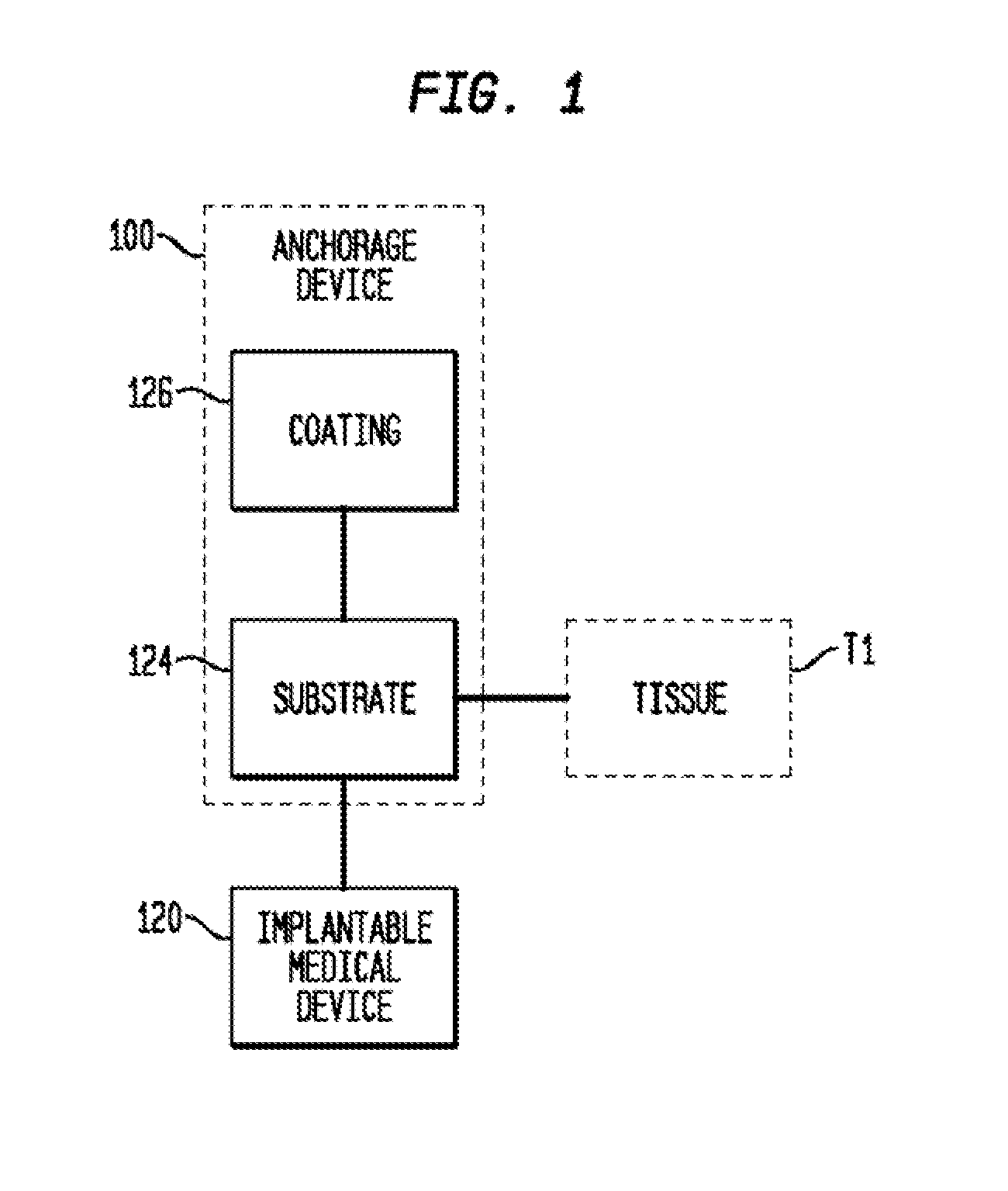
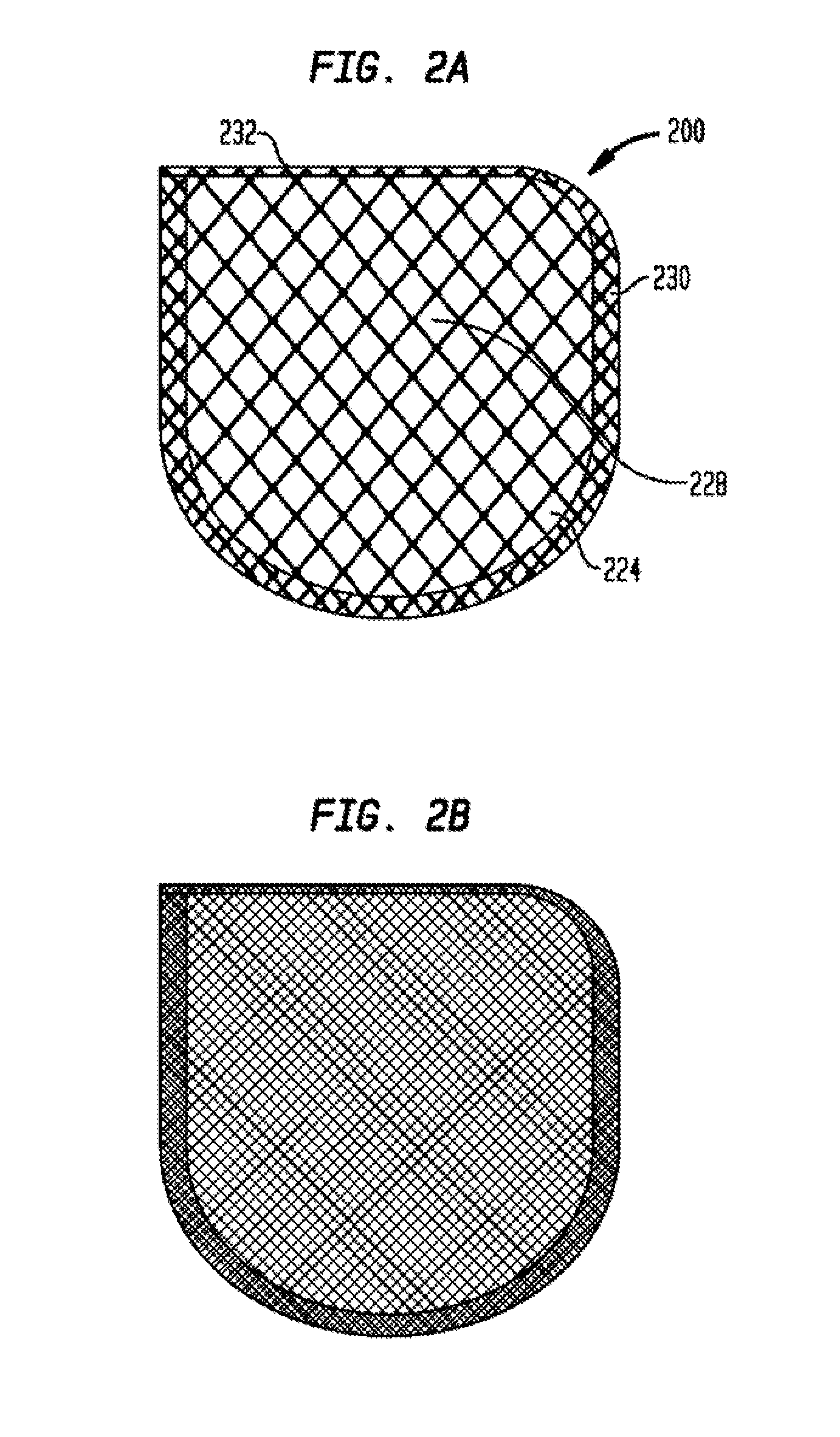
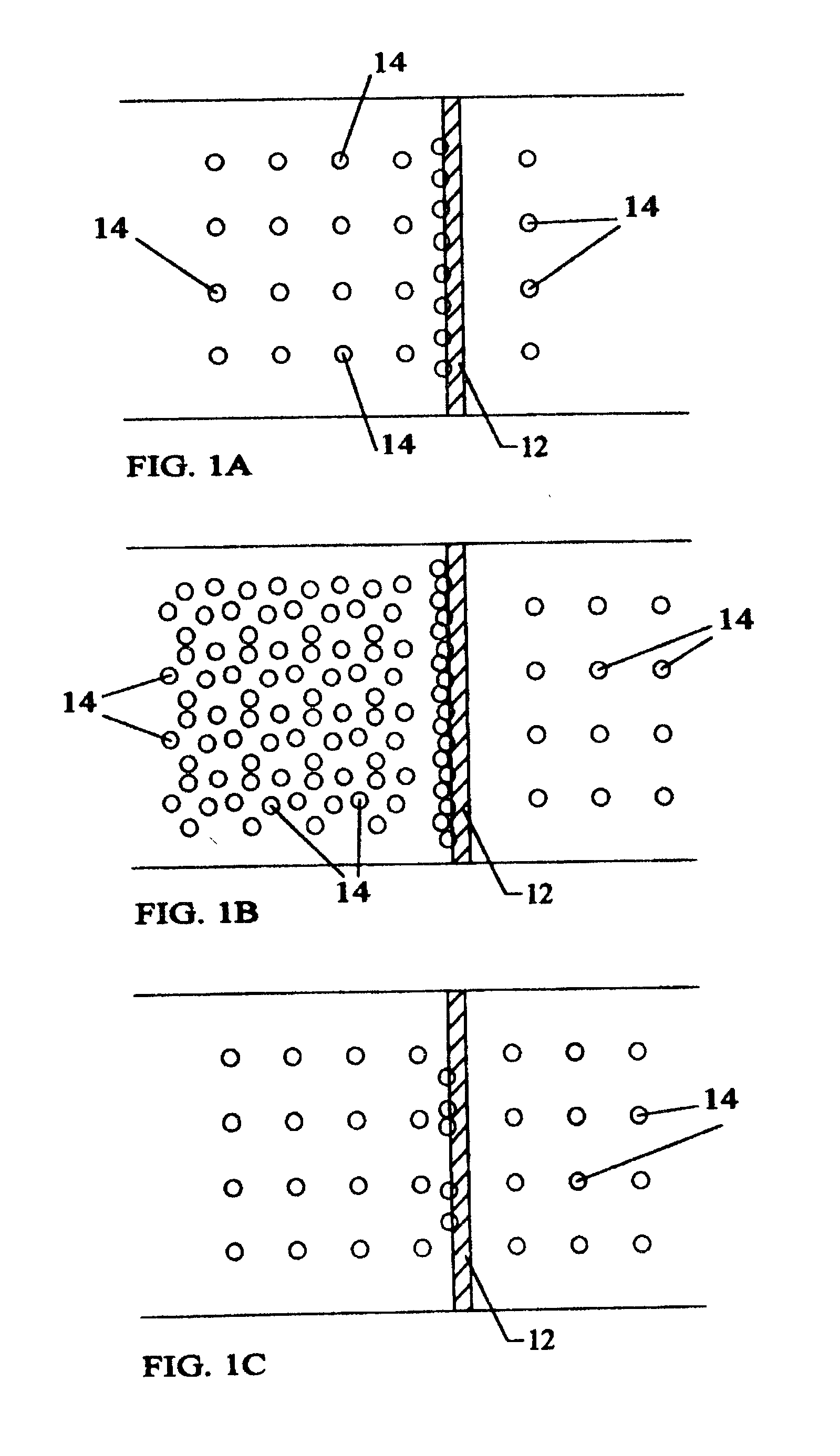
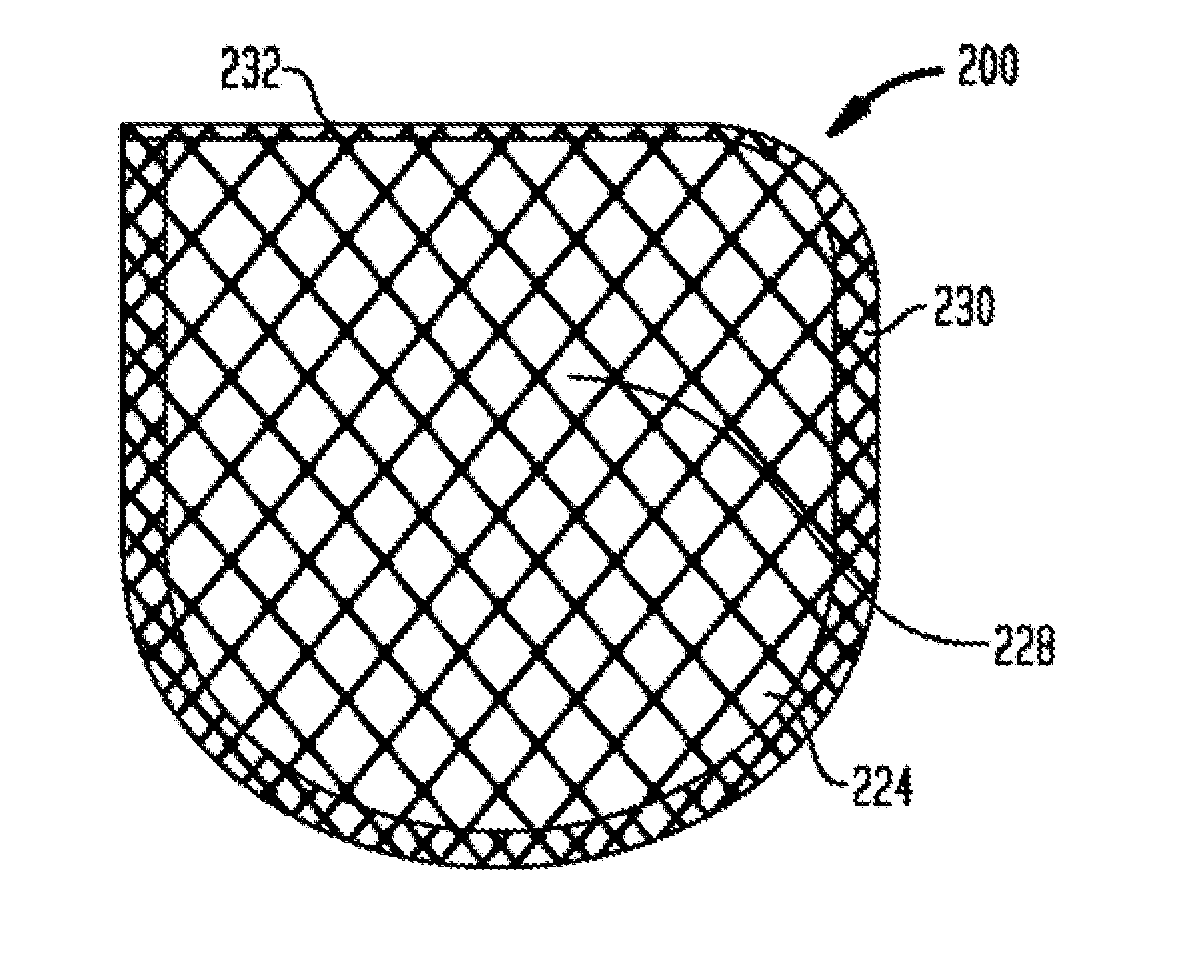
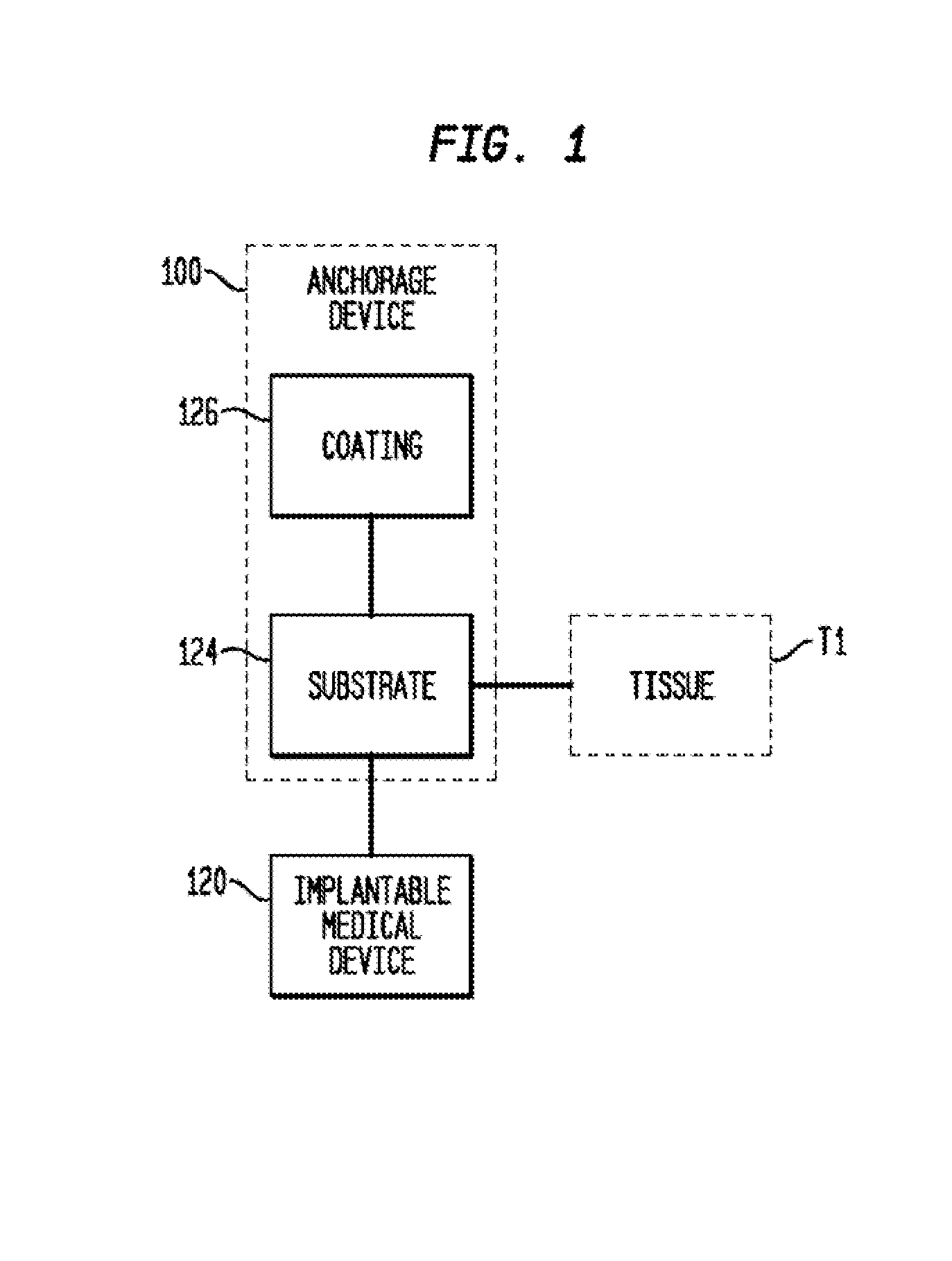
Anchorage devices comprising an active pharmaceutical ingredient
ActiveUS20120185004A1Incidence can be reduced and preventedSmall surface areaEpicardial electrodesHeart defibrillatorsBiologyMesh grid
An anchorage device comprising a mesh substrate coupled to an implantable medical device is disclosed, where the mesh substrate has a coating comprising a polymer, and the mesh further comprises at least one active pharmaceutical ingredient. The active pharmaceutical agent is designed to elute from the mesh over time. The mesh substrate can be configured to reduce the mass of the anchorage device such that tissue in-growth and / or scar tissue formation at the treatment site is reduced. In some embodiments, the mesh substrate can be formed with a mesh having a low areal density. In some embodiments, the mesh substrate can include one or more apertures or pores to reduce the mass of the substrate.
Owner:MEDTRONIC INC
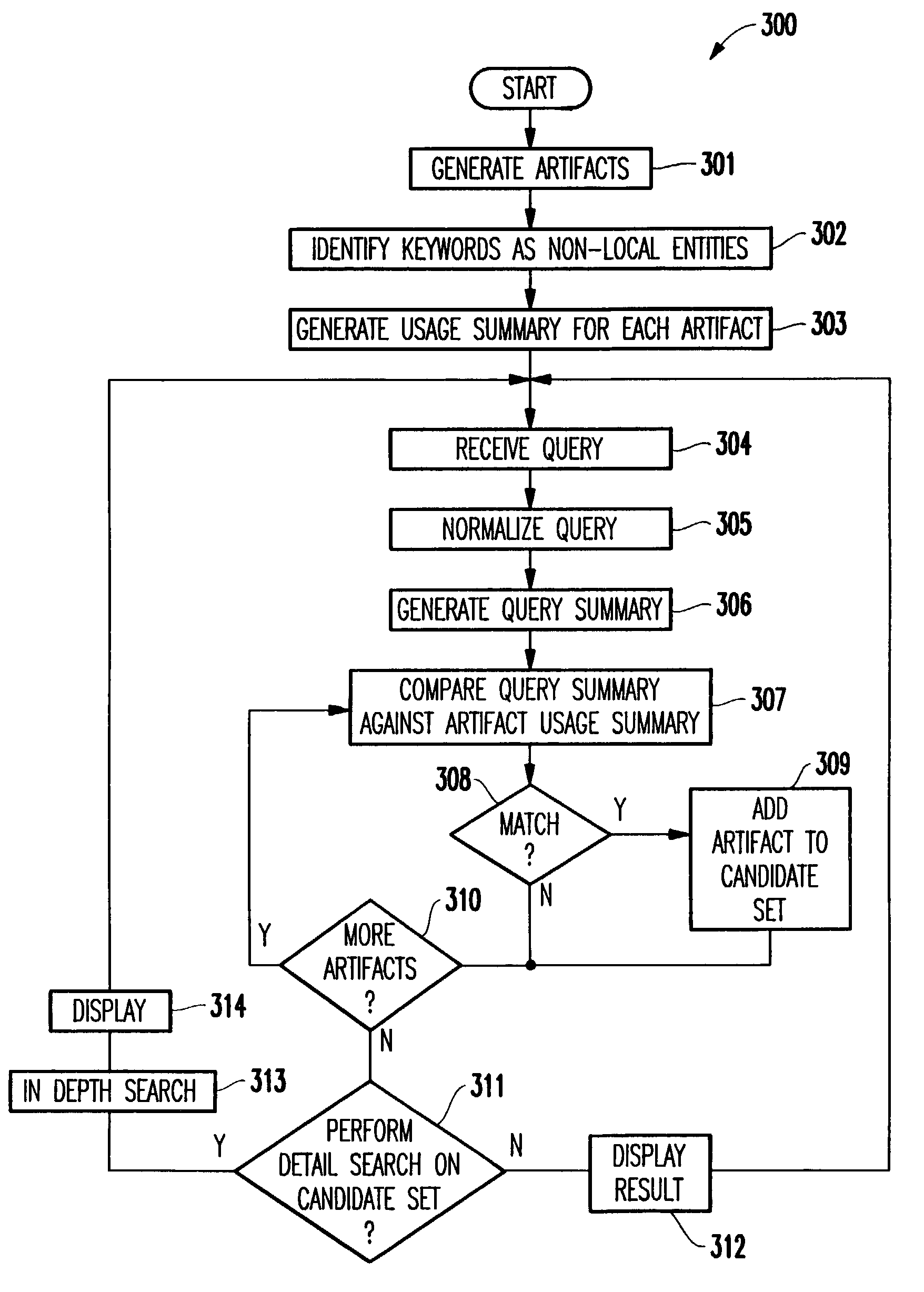
Method and structure for efficiently retrieving artifacts in a fine grained software configuration management repository
InactiveUS7146355B2Increase speedSmall setData processing applicationsSoftware reuseSoftware configuration managementFine grain
Owner:LINKEDIN
Moist wipe and method of making same
InactiveUS20030157856A1Eliminate needSurface chargingCosmetic preparationsToilet preparationsFiberAqueous medium
A moist wipe having a web of fibers stabilized as with a suitable binder, and the stabilized, dry web having an anionic surface charge not greater than about 1.2 meq / Kg. A cationic functional agent in an aqueous imbuement is added to the web which is partially adsorbed by the web and a portion of the agent remaining free. Because the anionic surface charge on the substrate is relatively low, there remains in the free aqueous medium a sufficient quantity of the functional agent deliverable to the surface to achieve the desired efficacy. The resulting web will adsorb a limited amount of the cationic functional agent in the aqueous imbuement, and thereby an adequate amount of the agent remains in the solution free of the web for deliverance to the surface, thereby obviating high loadings of the imbuement and the active functional agent.
Owner:GEORGIA PACIFIC CONSUMER PRODS LP
Anchorage Devices Comprising an Active Pharmaceutical Ingredient
ActiveUS20140031912A1Reduce swellingReduce inflammationEpicardial electrodesSurgeryMedical deviceScar tissues
An anchorage device comprising a mesh substrate coupled to an implantable medical device is disclosed, where the mesh substrate has a coating comprising a polymer, and the mesh further comprises at least one active pharmaceutical ingredient. The active pharmaceutical agent is designed to elute from the mesh over time. The mesh substrate can be configured to reduce the mass of the anchorage device such that tissue in-growth and / or scar tissue formation at the treatment site is reduced. In some embodiments, the mesh substrate can be formed with a mesh having a low areal density. In some embodiments, the mesh substrate can include one or more apertures or pores to reduce the mass of the substrate.
Owner:MEDTRONIC INC
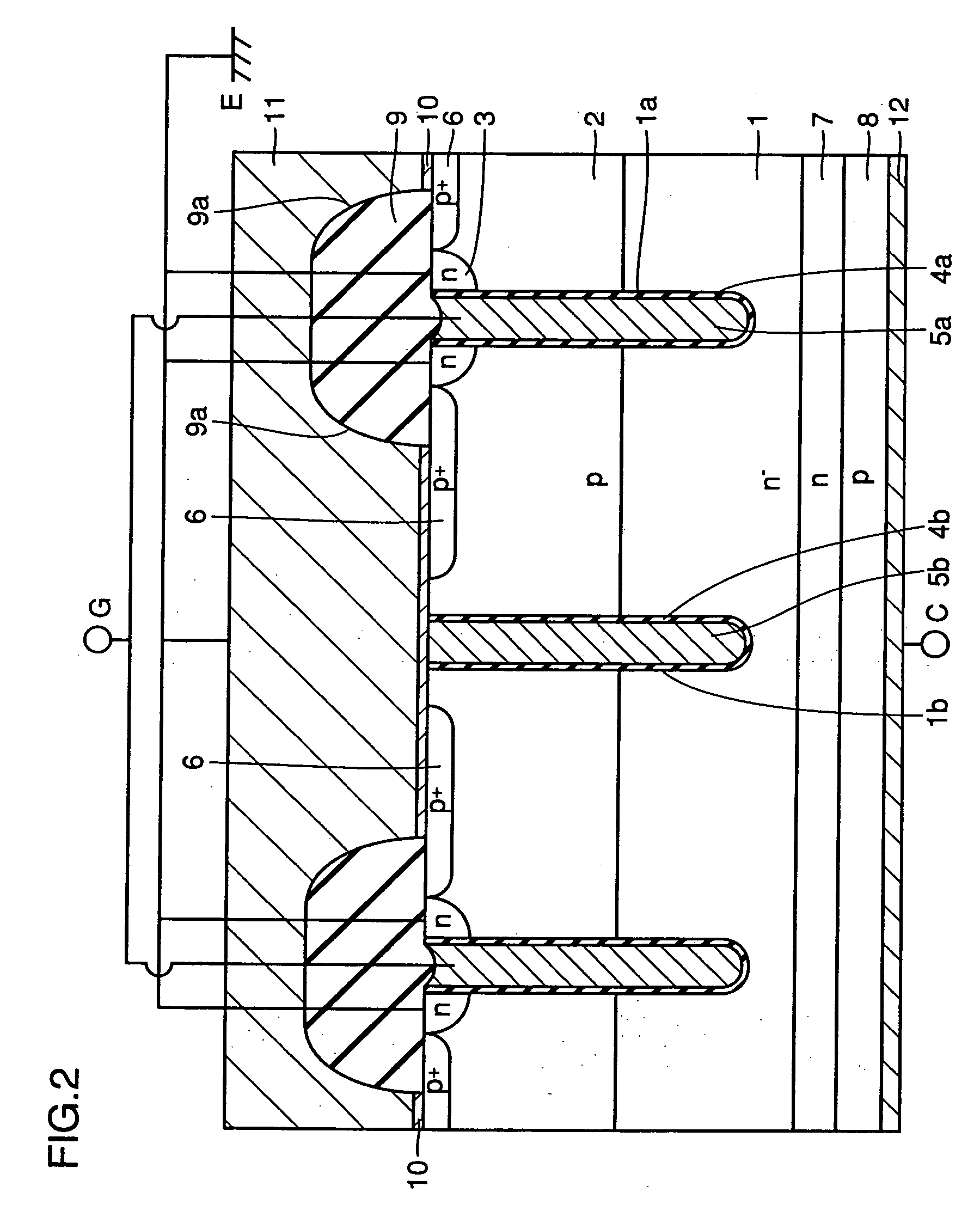
Semiconductor device
A semiconductor device of the present invention has an insulating gate type field effect transistor portion having an n-type emitter region (3) and an n− silicon substrate (1), which are opposed to each other sandwiching a p-type body region (2), as well as a gate electrode (5a) which is opposed to p-type body region (2) sandwiching a gate insulating film (4a), and also has a stabilizing plate (5b). This stabilizing plate (5b) is made of a conductor or a semiconductor, is opposed to n− silicon substrate (1) sandwiching an insulating film (4, 4b) for a plate, and forms together with n− silicon substrate (1), a capacitor. This stabilizing plate capacitor formed between stabilizing plate (5b) and n− silicon substrate (1) has a capacitance greater than that of the gate-drain capacitor formed between gate electrode (5a) and n− silicon substrate (1).
Owner:MITSUBISHI ELECTRIC CORP
Sensing display
ActiveUS20060098004A1Cutting portionCathode-ray tube indicatorsInput/output processes for data processingDisplay deviceEngineering
A display and method for making the same are provided that can sense a force applied thereto. The display comprises a substrate, an array of individually controllable image-forming elements arranged on the substrate; and a support located proximate to the substrate. The support has an arrangement of light paths formed therein to transmit light from a light source to a light sensor. The light paths are arranged in an area that is, at least in part, co-extensive with the array. The support is elastically deformable so that an application of force to the support causes the light paths to deform in a manner that reduces the portion of light passing through light paths elastically deformed by the application of force.
Owner:GLOBAL OLED TECH
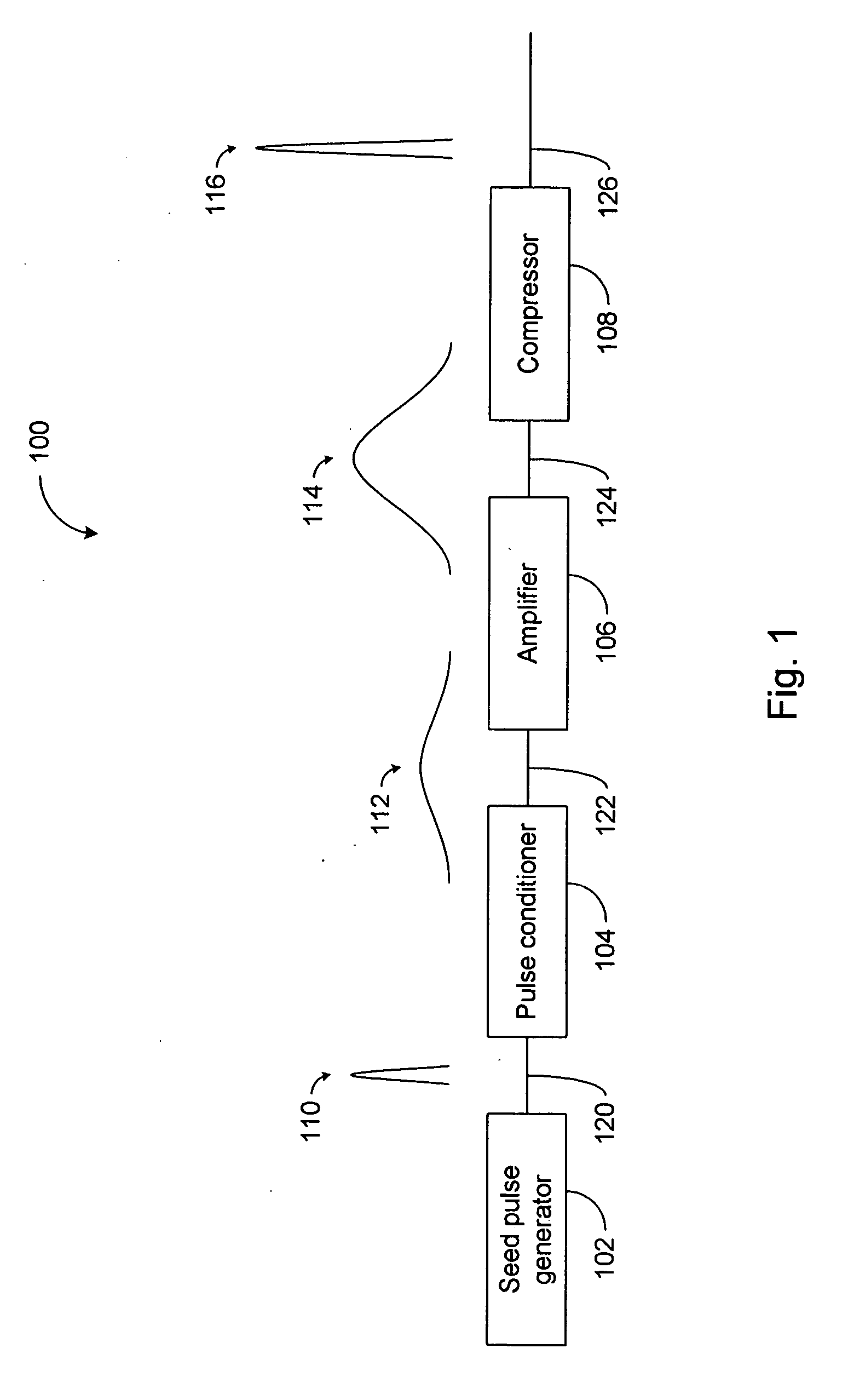
High power short pulse fiber laser
InactiveUS7804864B2Reduce widthWeaken energyLaser using scattering effectsLaser arrangementsAudio power amplifierPigtail
A pulsed laser comprises an oscillator and amplifier. An attenuator and / or pre-compressor may be disposed between the oscillator and amplifier to improve performance and possibly the quality of pulses output from the laser. Such pre-compression may be implemented with spectral filters and / or dispersive elements between the oscillator and amplifier. The pulsed laser may have a modular design comprising modular devices that may have Telcordia-graded quality and reliability. Fiber pigtails extending from the device modules can be spliced together to form laser system. In one embodiment, a laser system operating at approximately 1050 nm comprises an oscillator having a spectral bandwidth of approximately 19 nm. This oscillator signal can be manipulated to generate a pulse having a width below approximately 90 fs.
Owner:IMRA AMERICA
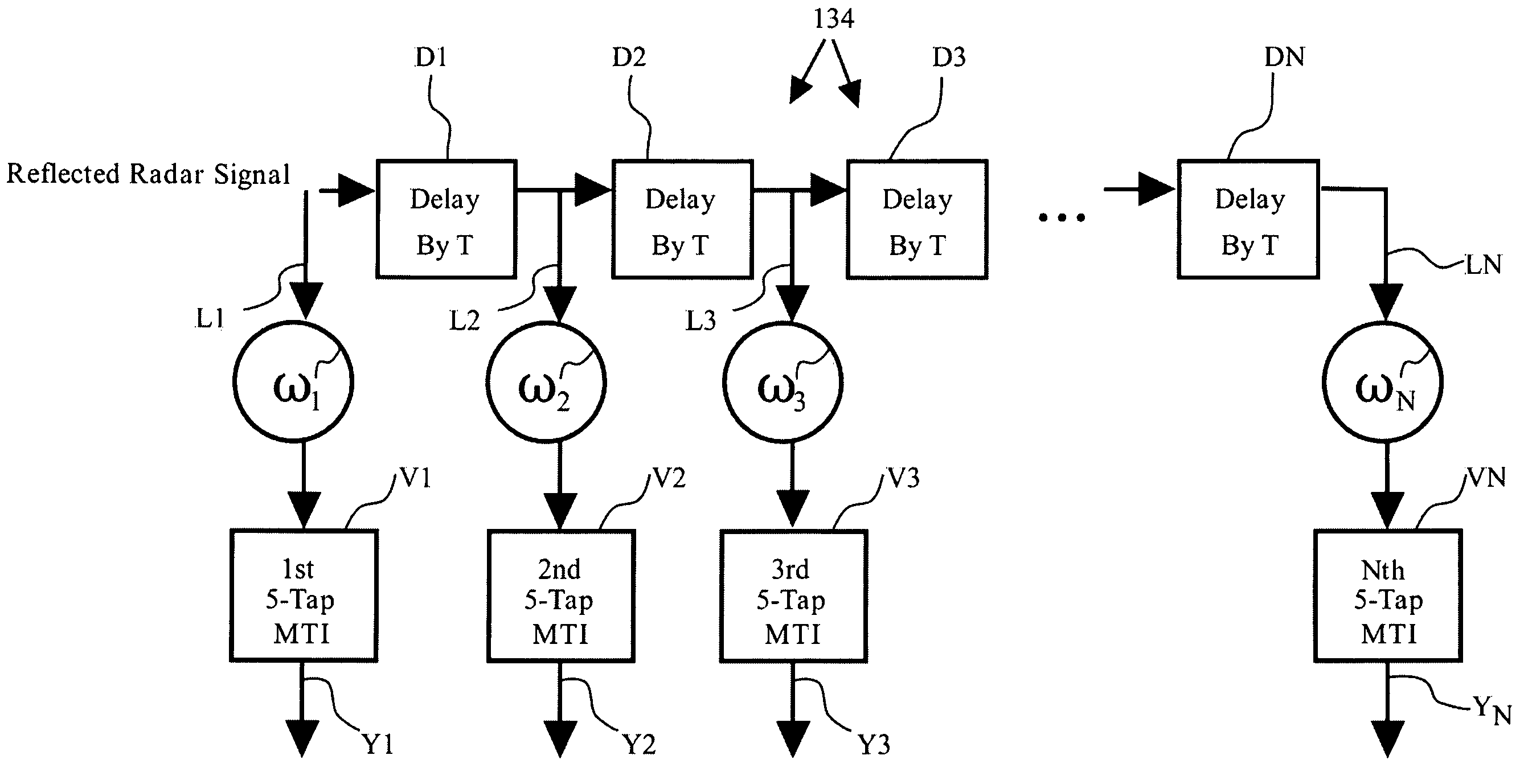
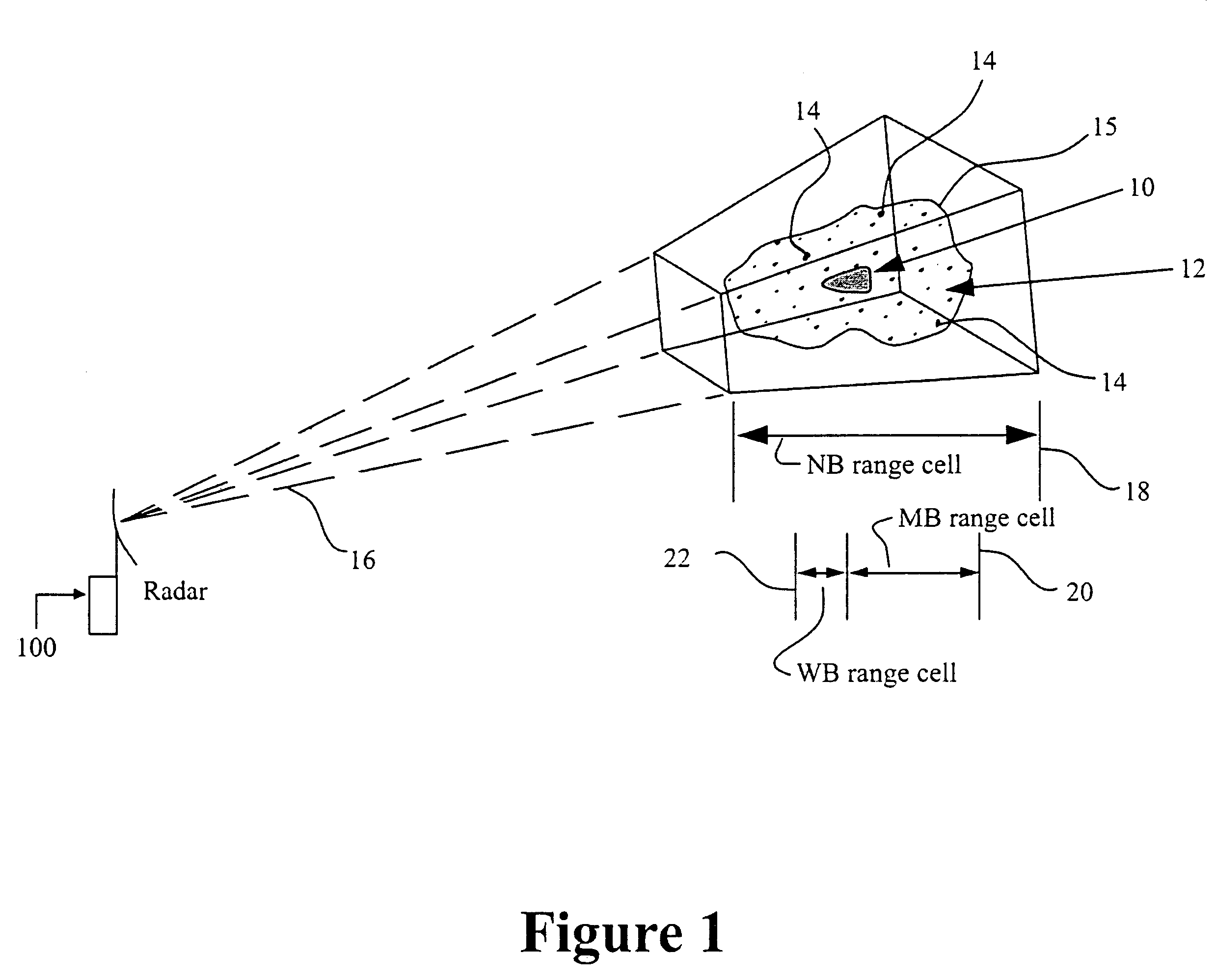
Method and device for the detection and track of targets in high clutter
InactiveUS7154433B1Effectively detectsEffective trackingRadio wave reradiation/reflectionImage resolutionAcoustics
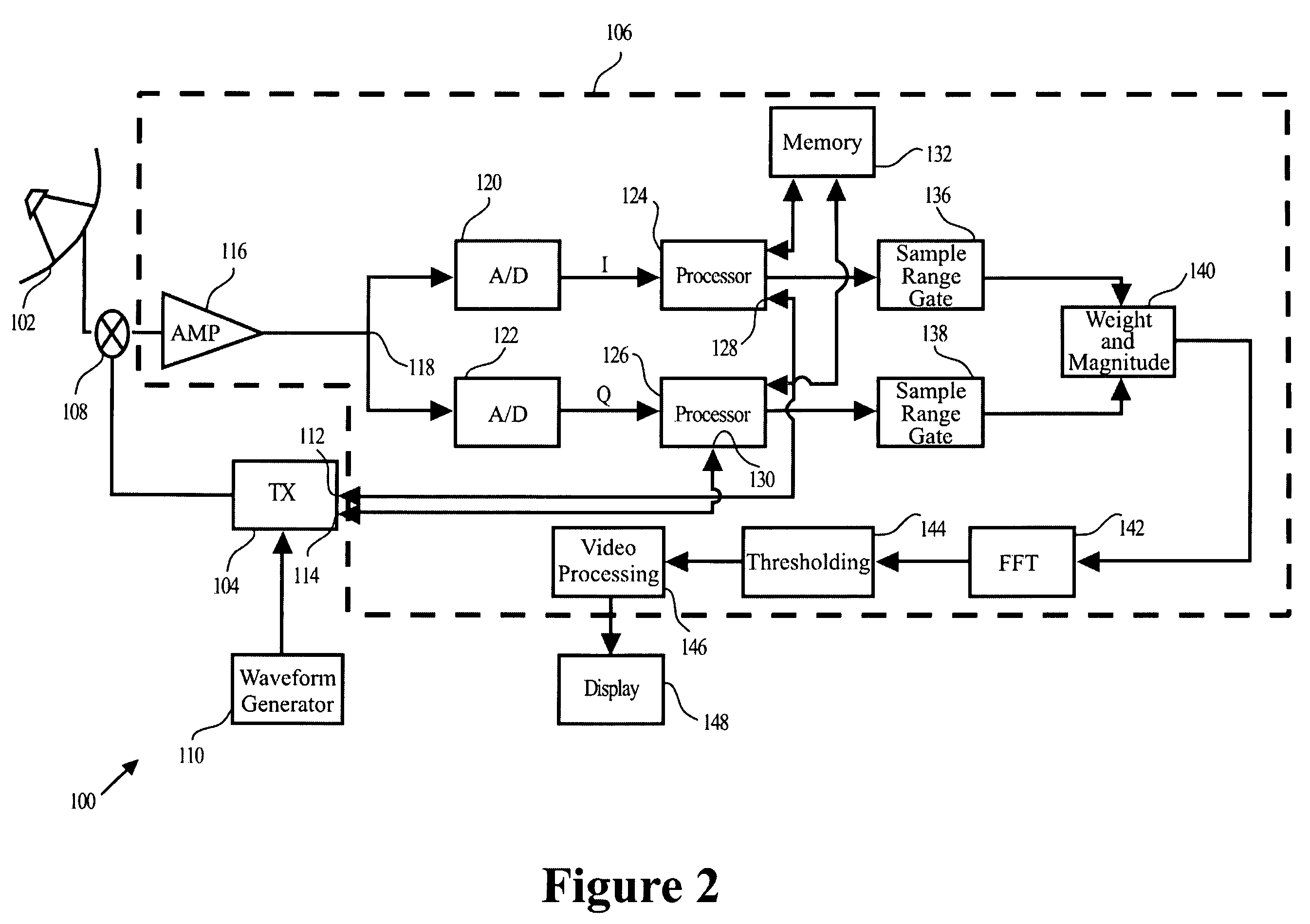
A method for discriminating and tracking a target in a clutter cloud includes transmitting a radar signal at a signal bandwidth to: identify a range extent of a clutter cloud; determine a centroid and a velocity growth rate of the clutter cloud; and identify a direction of movement of the centroid of the clutter cloud. The method may also include locking a another radar signal having a greater signal bandwidth onto the centroid of the clutter cloud whereby the centroid is tracked within one radar range resolution bin; providing a delay line that includes at least two Doppler filters and is configured to cover a Doppler frequency range corresponding to a velocity growth rate of the clutter cloud; and processing a reflected radar signal corresponding to the greater signal bandwidth. The processing of the reflected radar signal may comprise passing the reflected radar signal through the delay line to mitigate a portion of the reflected signal that is reflected by the clutter cloud. A system and apparatus for performing the method is also provided.
Owner:US OF AMER REPT BY SEC OF THE ARMY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com