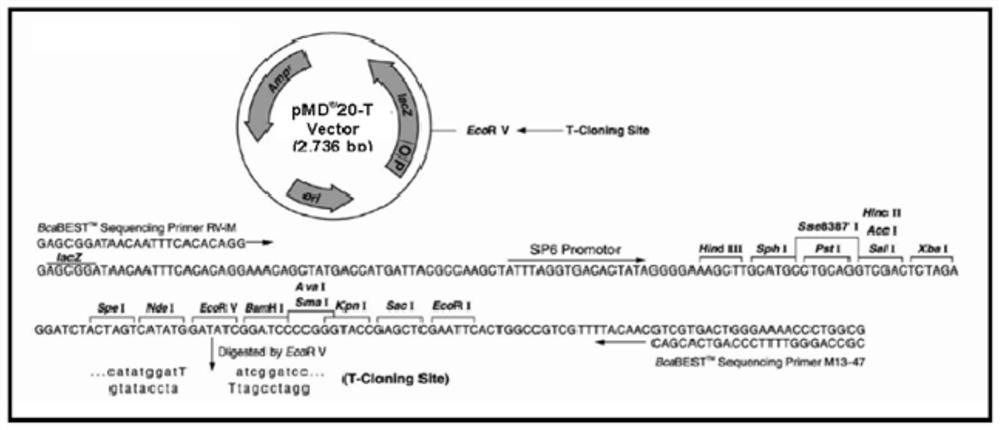
Patents
Literature
41 results about "Full length genome" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
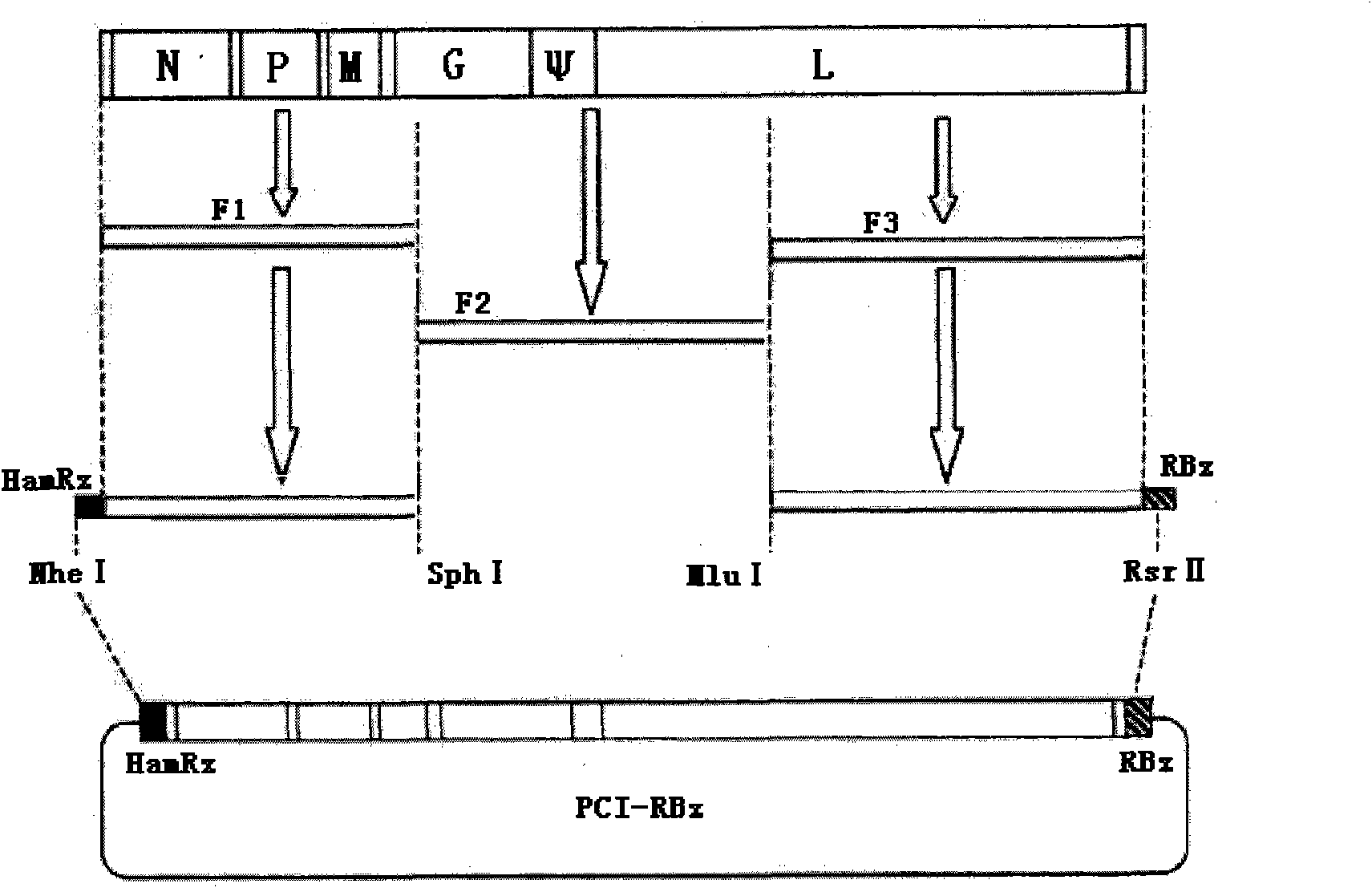
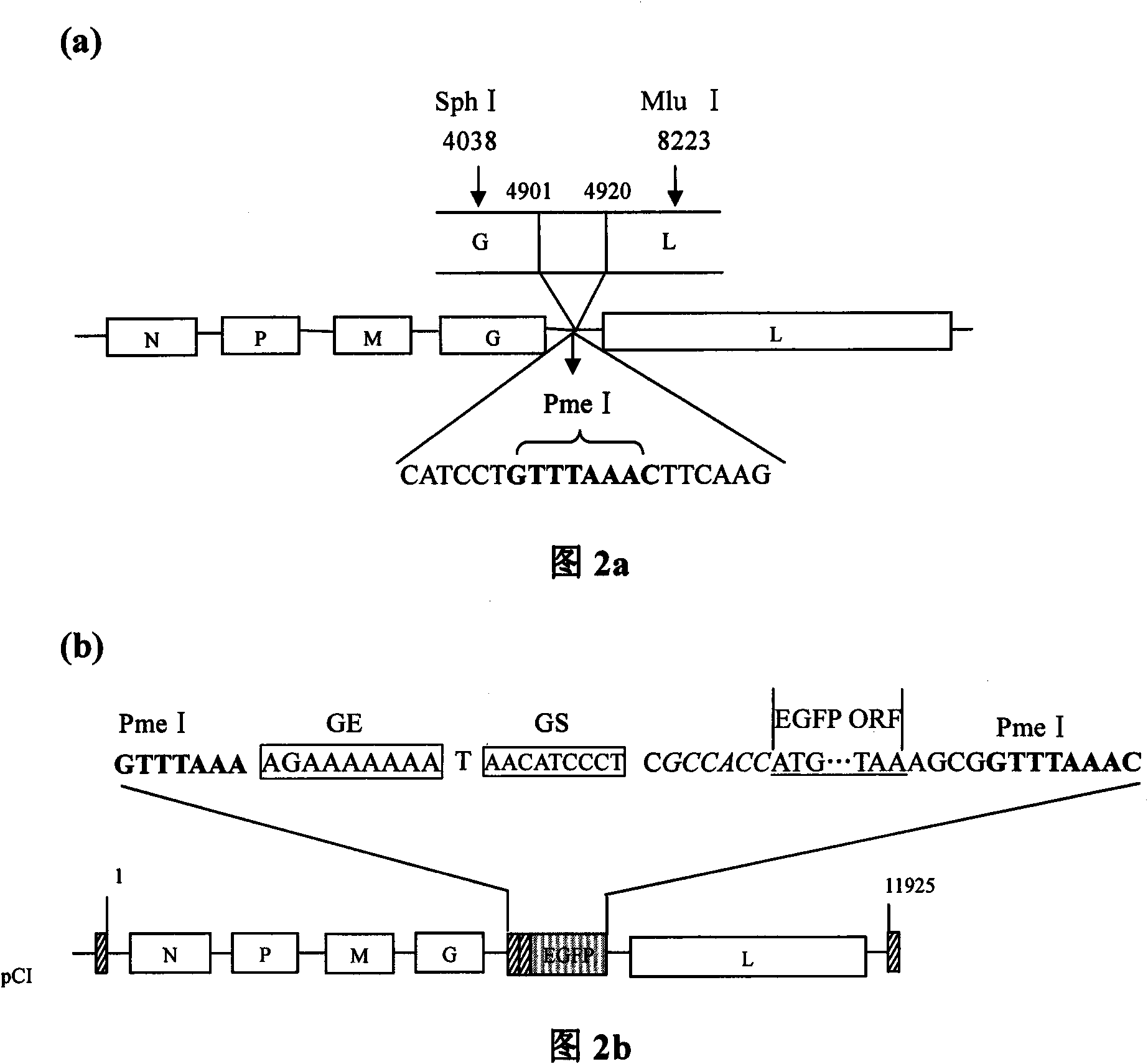
Rabies virus Flury-LEP vaccine strain reverse genetic operating system and LEP green fluorescent protein recombination viral vector
InactiveCN101586120AMicrobiological testing/measurementMicroorganism based processesPhosphoric acidViral vector
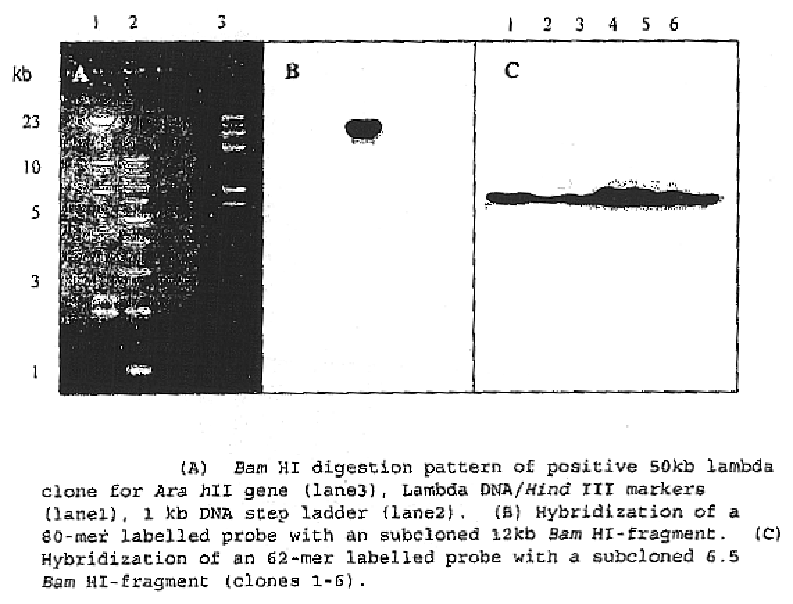
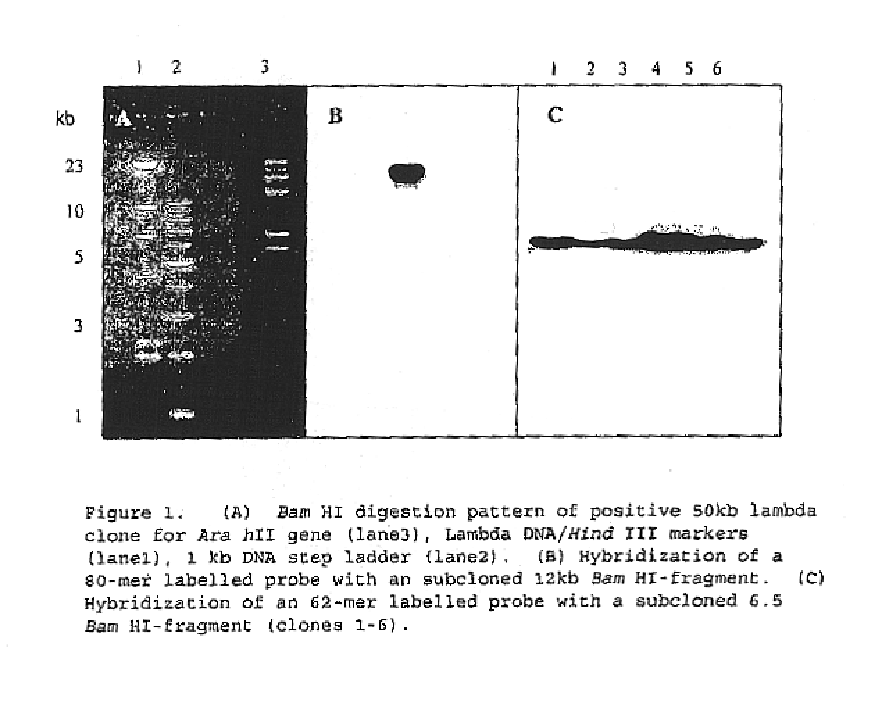
The invention relates to a rabies virus Flury-LEP vaccine strain reverse genetic operating system comprising a recombination vector for coding LEP full length genome cDNA and an auxiliary plasmid system thereof, wherein, the auxiliary plasmid system respectively codes nucleoprotein of LEP, phosphoric acid protein P and polyase protein L. the invention also relates to LEP green fluorescent protein recombination viral vector built by the reverse genetic operating system, concretely, the rabies virus Flury-LEP vaccine strain reverse genetic operating system is pCI-LEP and auxiliary plasmid systems pCAGG-N, pCAGG-P and pCAGG-L, the LEP green fluorescent protein recombination viral vector is pCI_LEP_EGEP. The invention also relates to recombination viruses rescued by the rabies virus Flury-LEP vaccine strain reverse genetic operating system and the LEP green fluorescent protein recombination viral vector, and applications thereof.
Owner:HARBIN VETERINARY RES INST CHINESE ACADEMY OF AGRI SCI
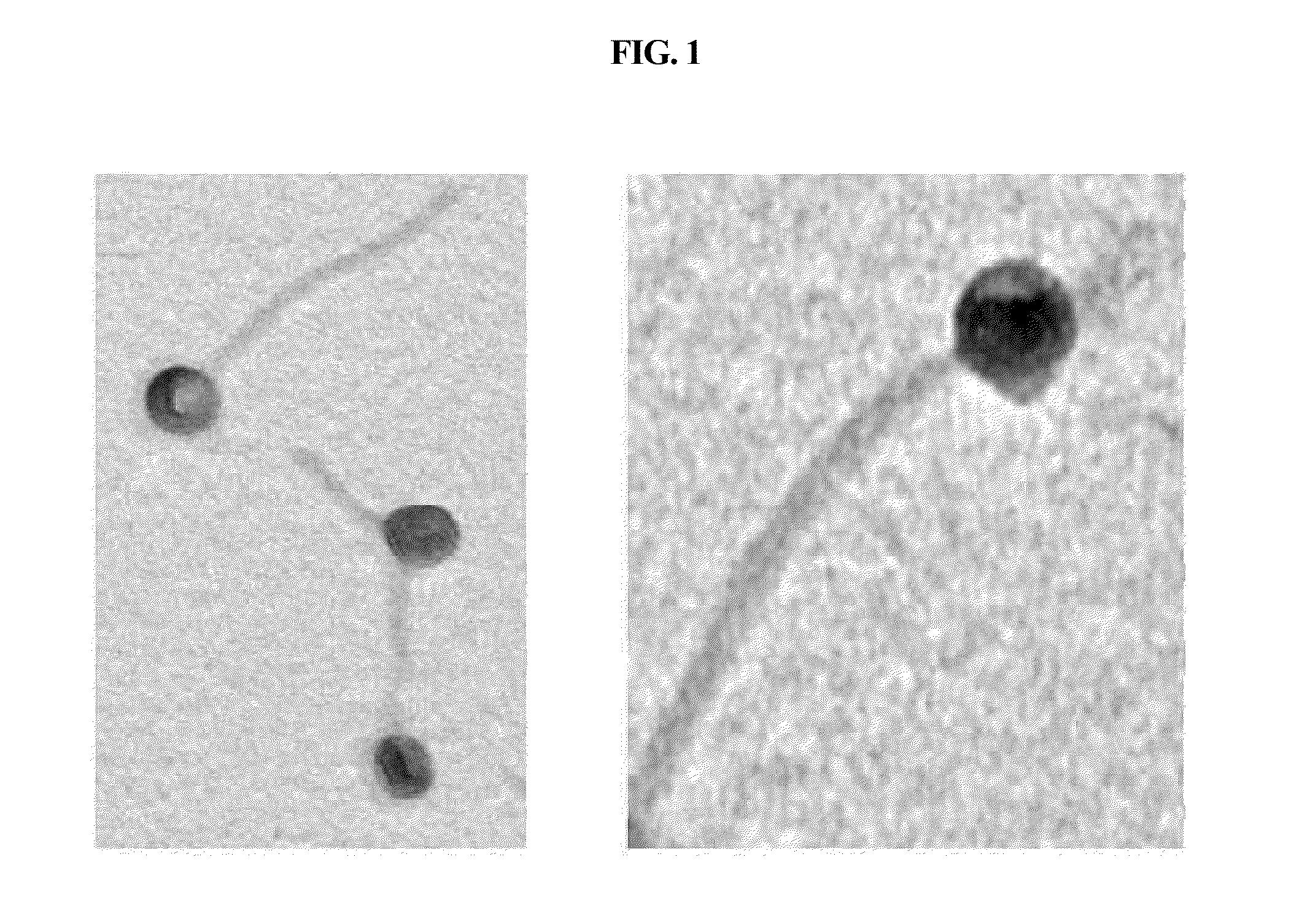
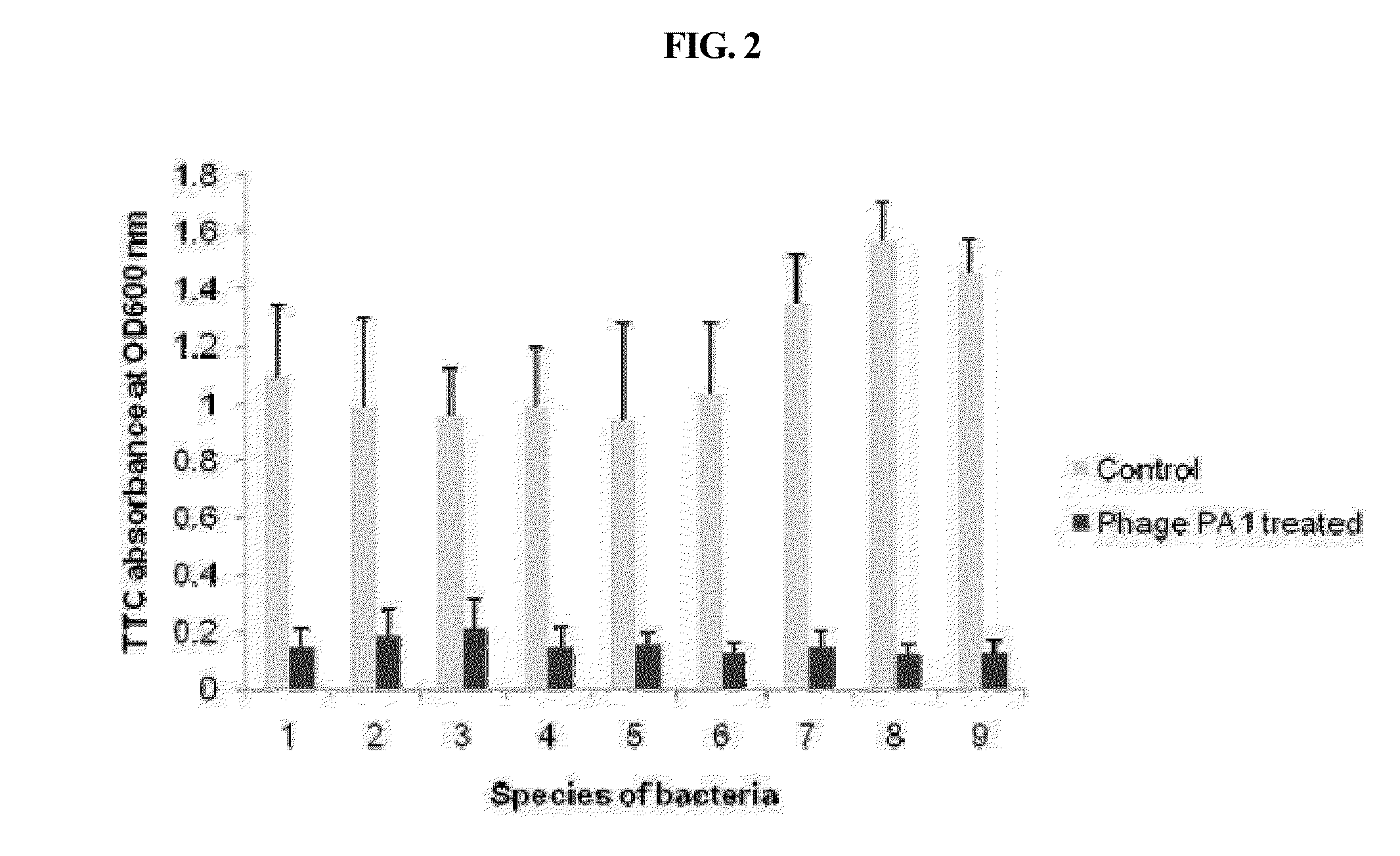
Bacteriophage killing pseudomonas aeruginosa and staphylococcus aureus
InactiveUS20130273635A1Causative bacteriaDisadvantageous to contaminate environmentViral/bacteriophage medical ingredientsBacteriophagesBacteroidesStaphylococcus cohnii
The present invention relates to a bacteriophage PA1Φ belonging to family Siphoviridae, characterized that it is capable of killing one or more bacteria strains selected from a group comprising Pseudomonas aeruginosa, Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus homonis, Shigella sonnei, Listeria monocytogenes and Streptococcus pneumonia, and contains a full-length genome of SEQ ID NO: 1.According to the present invention, the bacteriophage PA1Φ can be used to kill said bacteria and reduce the same effectively. Also, it can be used to remove biofilms generated by said bacteria. Especially, this bacteriophage is applicable for medical industry, food industry, biotechnology and the like, because it is a sort of virus that kills host bacteria without any adverse effect on human, animals and so on. In addition, this bacteriophage can kill noxious bacteria on target sites or target supports without any problem related with resistance development of bacteria.
Owner:INTRON BIOTECHNOLOGY INC
Method for constructing coronavirus infectious clone and application thereof
ActiveCN110079541AAvoid toxicityAvoid stabilitySsRNA viruses positive-senseViral antigen ingredientsInfectious bronchitisPlasmid Vector
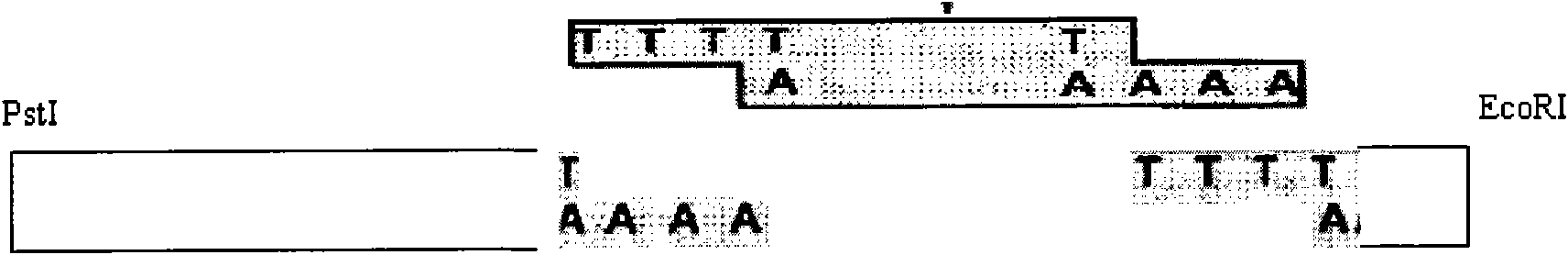
The invention discloses a method for constructing a coronavirus infectious clone and an application thereof. The method comprises the following steps: firstly, acquiring a cDNA segment covering a selected full-length genome of coronavirus; performing segmental cloning; adopting a RED / ET recombination technology for assembling the cDNA segment containing the full-length genome of coronavirus onto aplasmid vector; screening, thereby acquiring the infectious clone containing the full-length cDNA of coronavirus. The invention has the beneficial effects that a medium / low copy plasmid vector is firstly utilized to successfully construct the infectious bronchitis coronavirus infectious clone; a new method is established for the coronavirus infectious clone; the method is capable of solving the problems of difficulty in selecting coronavirus vector and instability of viral macro-genome in bacteria; the positive cloning efficiency is high; the acquired reverse genetic vaccine strain cloning has integrity, passage stability, infectivity and high rescuing efficiency; an effective tool is supplied for researching nosogenesis of coronavirus, developing a novel vaccine, and the like.
Owner:SOUTH CHINA AGRI UNIV
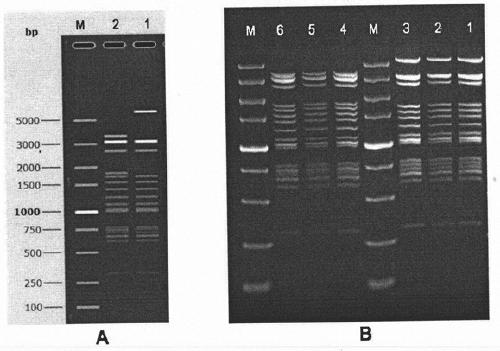
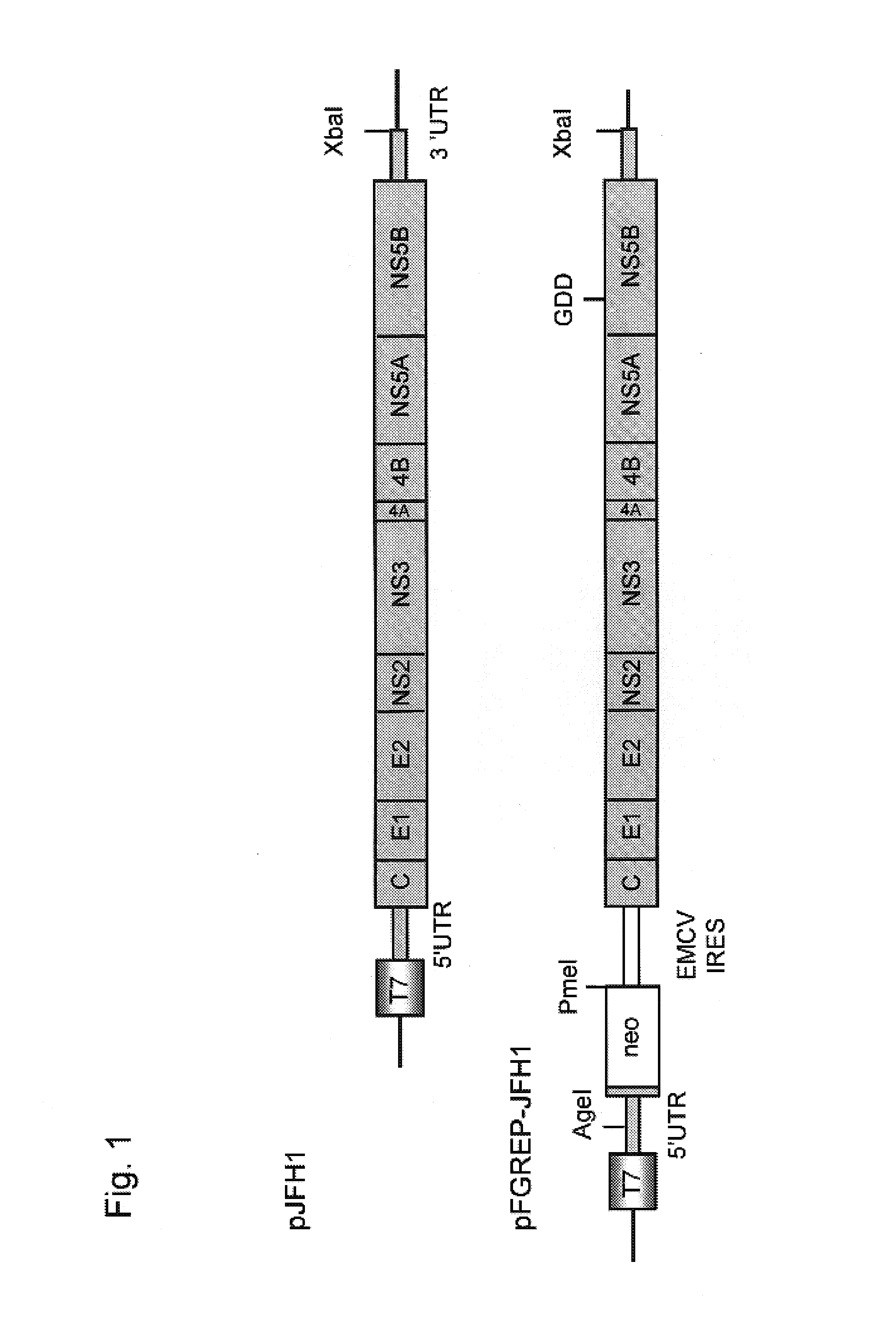
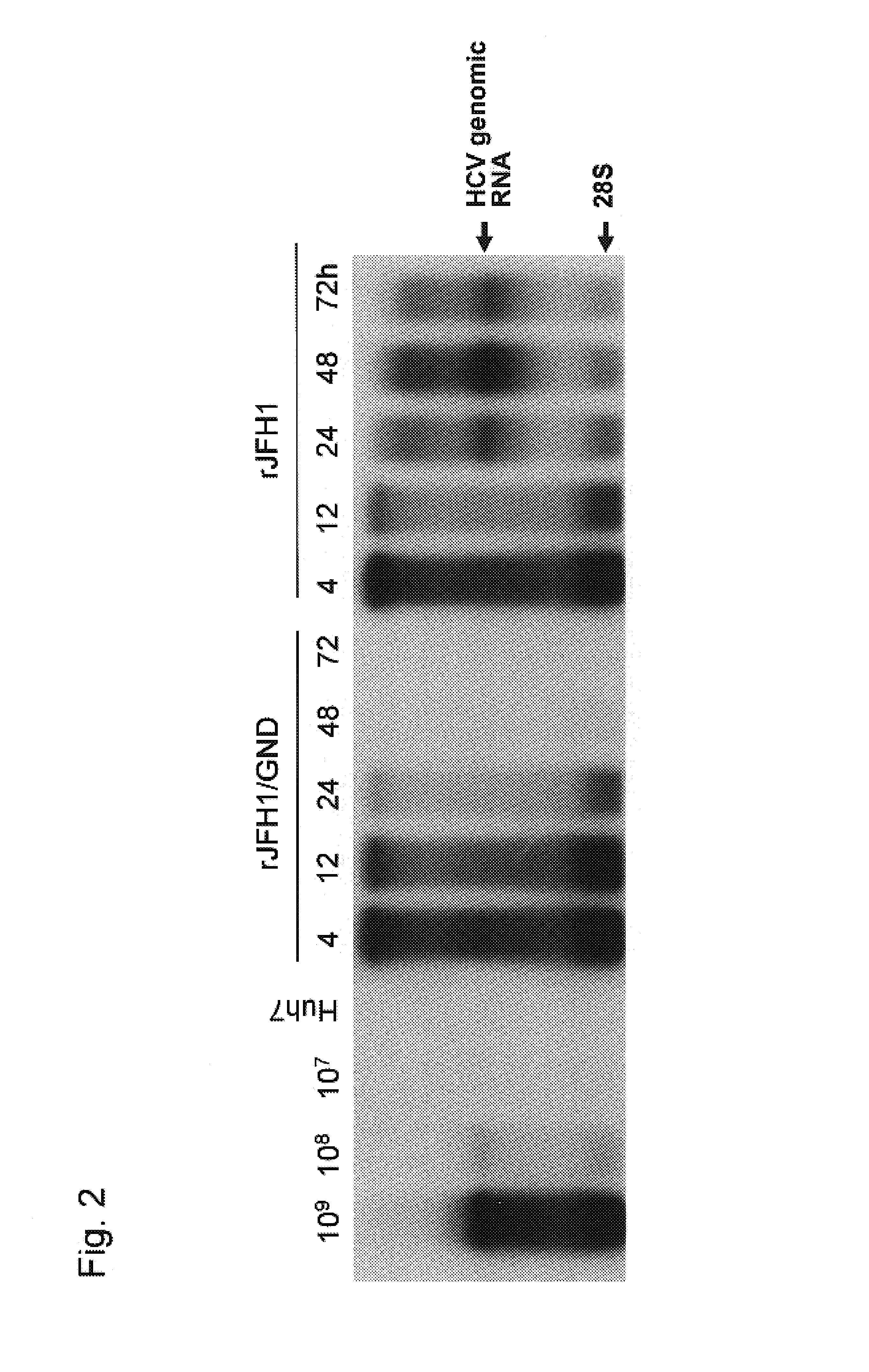
Nucleic acid construct containing fulllength genome of human hepatitis C virus, recombinant fulllength virus genome-replicating cells having the nucleic acid construct transferred thereinto and method of producing hepatitis C virus Particle
The present invention provides a method for replicating efficiently an RNA containing fulllength HCV genomic sequence and a method for producing HCV virus particles containing fulllength HCV replicon RNA or fulllength HCV genomic RNA by using a cell culture system. Further, the present invention relates to a method for producing hepatitis C virus particles which comprises culturing a cell, into which a replicon RNA comprising a nucleotide sequence comprising a fulllength genomic RNA sequence of hepatitis C virus of the genotype 2a, at least one selectable marker gene and / or at least one reporter gene and at least one IRES sequence or the fulllength genomic RNA of hepatitis C virus of the genotype 2a is introduced, and generating virus particles in the culture medium. Still further the present invention relates also to a hepatitis C vaccine and an antibody against hepatitis C virus particles.
Owner:TORAY IND INC +1
Preparation method, amplification primers and detection reagent for HPV full-length genome quality control product
ActiveCN108929869AAvoid connection inefficienciesSolve the problem of uncertain concentrationMicrobiological testing/measurementVector-based foreign material introductionHuman DNA sequencingCompetent cell
The invention relates to a preparation method, amplification primers and detection reagent for a HPV full-length genome quality control product. The preparation method of the HPV full-length genome quality control product comprises: (1) acquiring a DNA template, (2) carrying out PCR amplification through amplification primers, (3) connecting a target DNA fragment to a vector by a HD Cloning linkage technology, (4) introducing the plasmid into a competent cell and then extracting the plasmid and (5) diluting the plasmid with a human genome. The preparation method of the human papillomavirus full-length genome quality control product can successfully acquire the HPV gene detection quality control product which has no risk of biological infection, can simulate a clinical sample, can be mass-produced, has a controllable concentration and can be stably preserved. The preparation method has good applicability.
Owner:亚能生物技术(深圳)有限公司
Down-regulation and silencing of allergen genes in transgenic peanut plants
InactiveUS20050114924A1Reduced and undetectable allergen protein contentReduced protein contentOther foreign material introduction processesPlant peptidesTransgeneAmino acid
An allergen-free transgenic peanut seed is produced by recombinant methods. Peanut plants are transformed with multiple copies of each of the allergen genes, or fragments thereof, to suppress gene expression and allergen protein production. Alternatively, peanut plants are transformed with peanut allergen antisense genes introduced into the peanut genome as antisense fragments, sense fragments, or combinations of both antisense and sense fragments. Peanut transgenes are under the control of the 35S promoter, or the promoter of the Ara h2 gene to produce antisense RNAs, sense RNAs, and double-stranded RNAs for suppressing allergen protein production in peanut plants. A full length genomic clone for allergen Ara h2 is isolated and sequenced. The ORF is 622 nucleotides long. The predicted encoded protein is 207 amino acids long and includes a putative transit peptide of 21 residues. One polyadenilation signal is identified at position 951. Six additional stop codons are observed. A promoter region was revealed containing a putative TATA box located at position−72. Homologous regions were identified between Ara h2, h6, and h7, and between Ara h3 and h4, and between Ara h1P41B and Ara h1P17. The homologous regions will be used for the screening of peanut genomic library to isolate all peanut allergen genes and for down-regulation and silencing of multiple peanut allergen genes.
Owner:DODO HORTENSE +3
Hydrophobia-canine distemper-canine parvovirus genetic recombination virus strain, construction method and application thereof
InactiveCN107988174AHigh expressionExpress influenceSsRNA viruses negative-senseViral antigen ingredientsRabiesCanine parvovirus
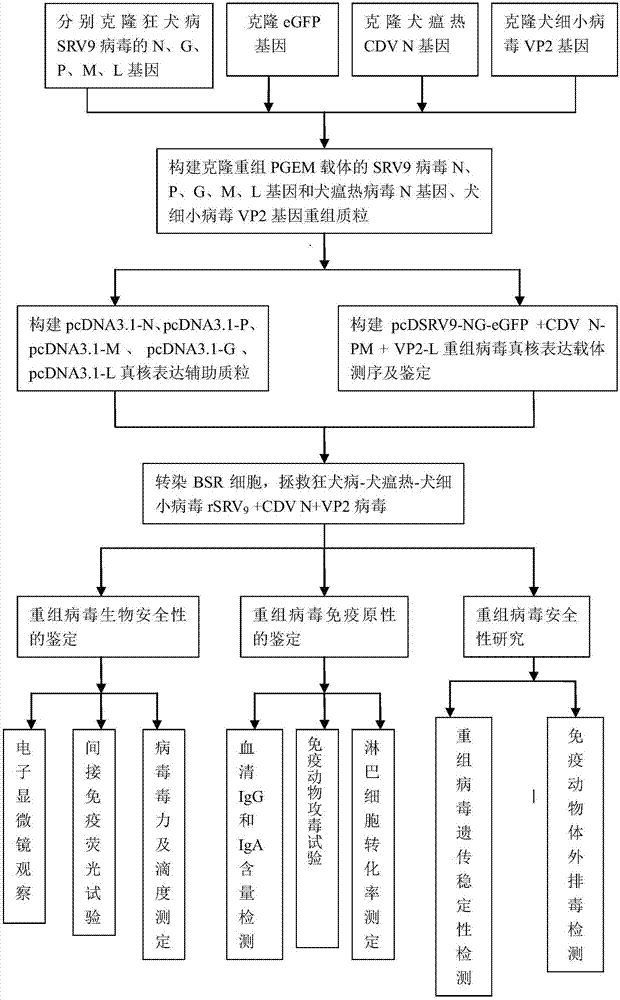
The invention discloses a hydrophobia-canine distemper-canine parvovirus genetic recombination virus strain, a construction method and application thereof. Through a homologous recombination method, agene replacement method and a gene insertion method, a full-length genome of a virus is subjected to gene rearrangement, and a rearranged hydrophobia virus genome plasmid with a canine distemper G gene and a canine parvovirus VP2 gene is constructed and is named as a hydrophobia-canine distemper-canine parvovirus, which is pcDSRV9-NG-eGFP+CDV N-PM+VP2-L genome plasmid; the hydrophobia-canine distemper-canine parvovirus is rescued through a reverse genetic method, and is an rSRV9+CDV N+VP2 gene recombinant virus; a gene nucleotide sequence of a recombination hydrophobia virus is SEQ ID NO:1. The recombination gene virus provided by the invention can be applied to researching a trigeminy oral vaccine for hydrophobia, canine distemper and canine parvovirus of canine and other wild animals soas to prevent three infectious diseases.
Owner:XINJIANG AGRI UNIV
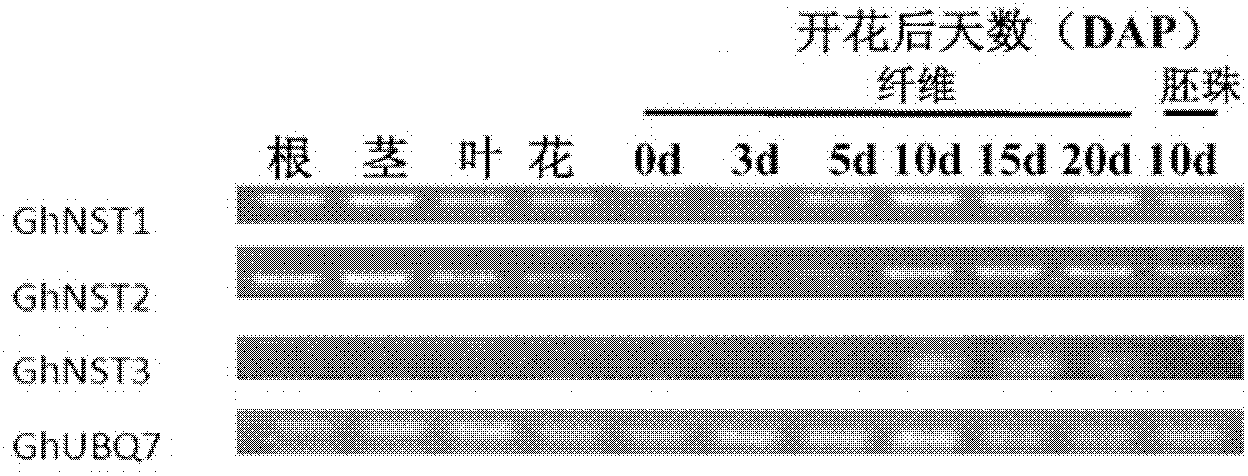
Cotton acetylcysteine (NAC) transcription factor NST sub-family and application thereof
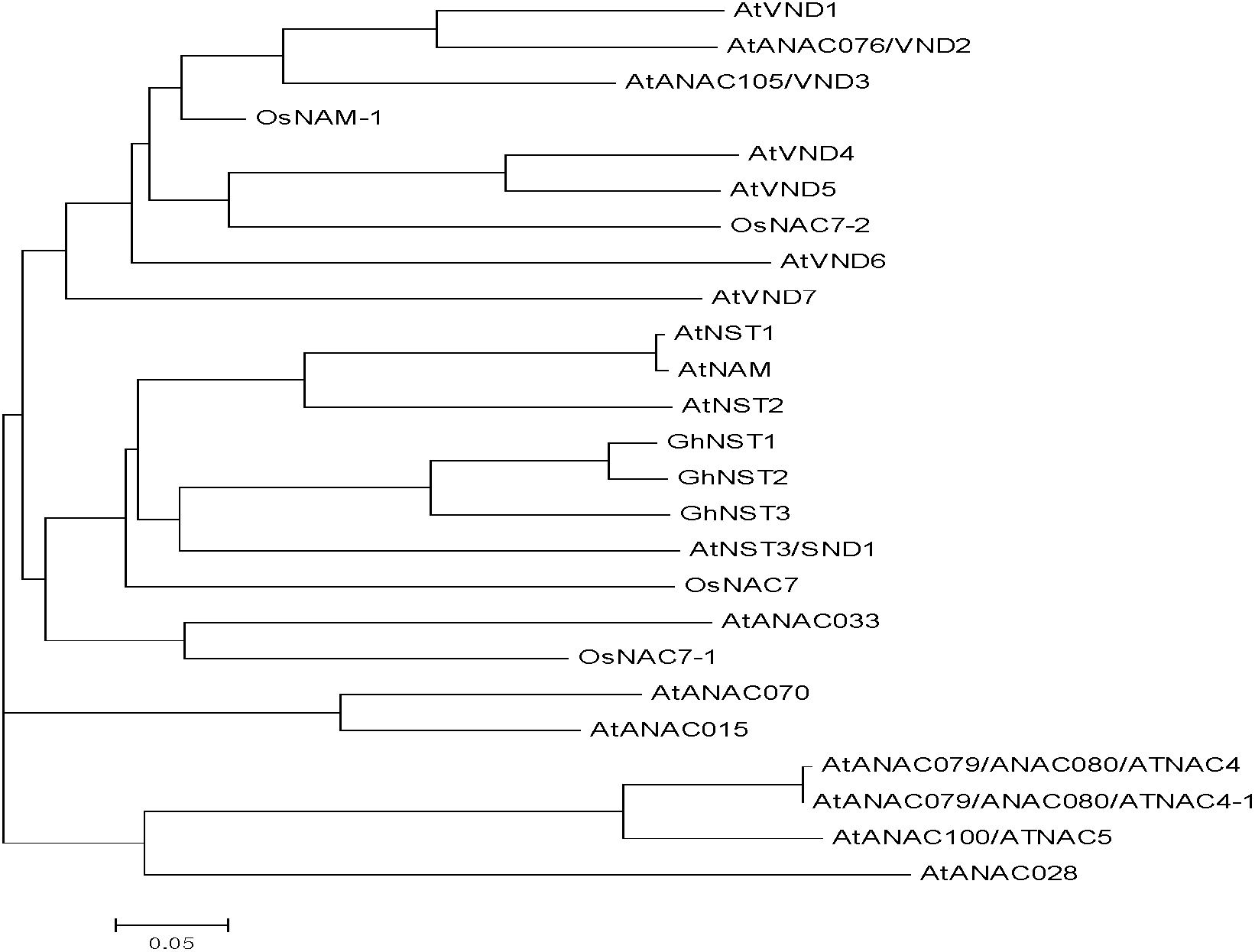
The invention utilizes bioinformatics and 3'-race to successfully clone cotton fiber blooming for 10 days and obtain three full-length genomes. The three full-length genomes obtained through bioinformatic comparison and cladogram analysis belong to an acetylcysteine (NAC) NST sub-family and are named as GhNST1, GhNST2 and GhNST3 respectively. Amino acid sequences of the three full-length genomes are shown as Seq ID No. 1-3. By analyzing growth and stage transcription level expression profile of the cotton fiber, the fact that the GhNST3 is expressed specifically at secondary wall synthesis stage of the cotton fiber is found, and the GhNST3 is related with synthesis of cell wall is suspected, thereby providing reliable theoretical basis for further study on the effect of the GhNST3 in the aspects of plant growth and development, hormonal regulation, stress resistance and the like.
Owner:BEIJING NORMAL UNIVERSITY
Hepatitis C virus and its external cell culture method
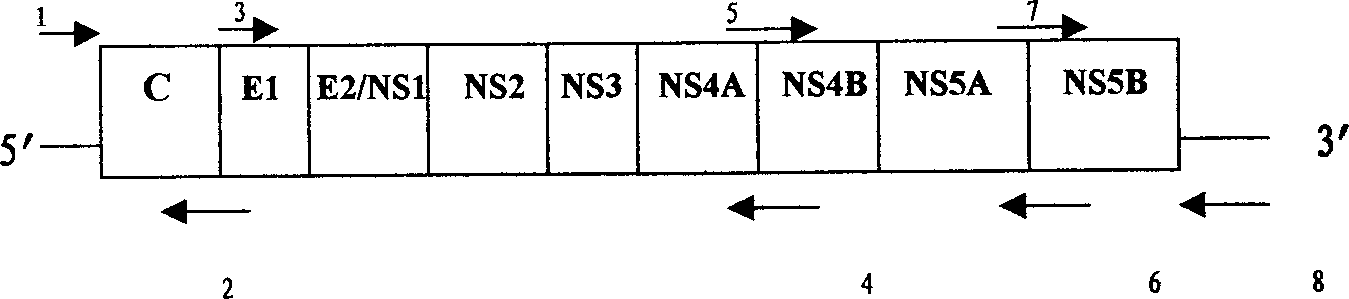
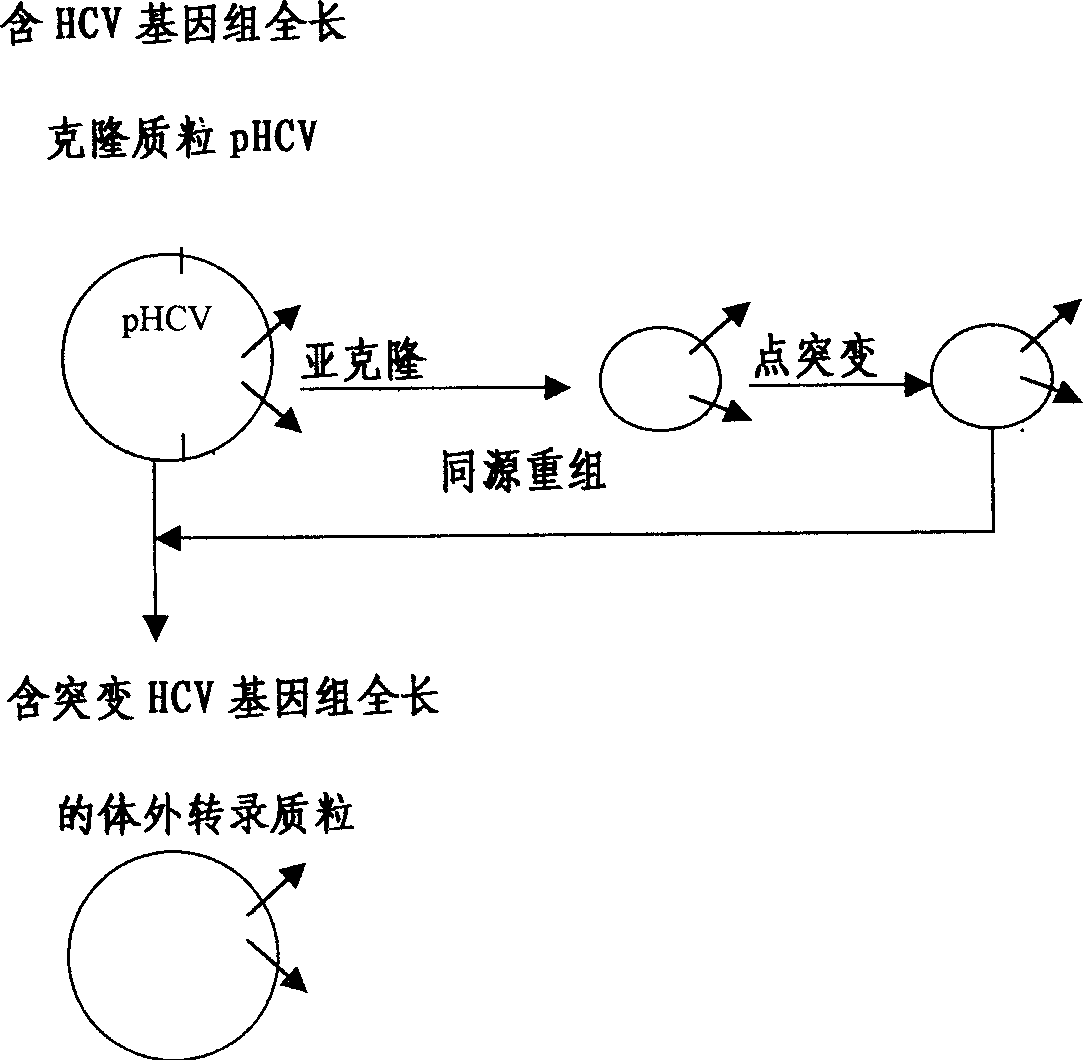
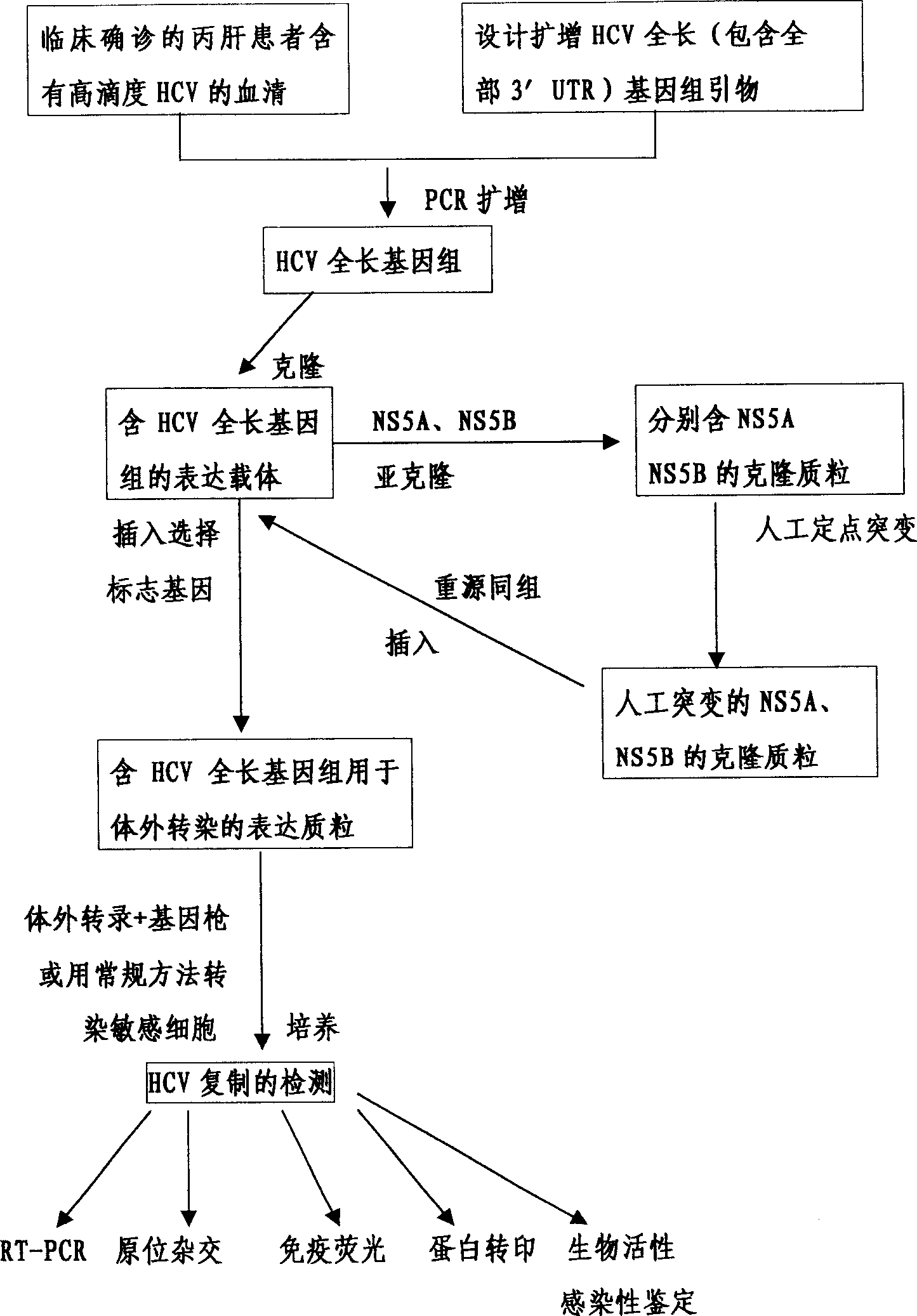
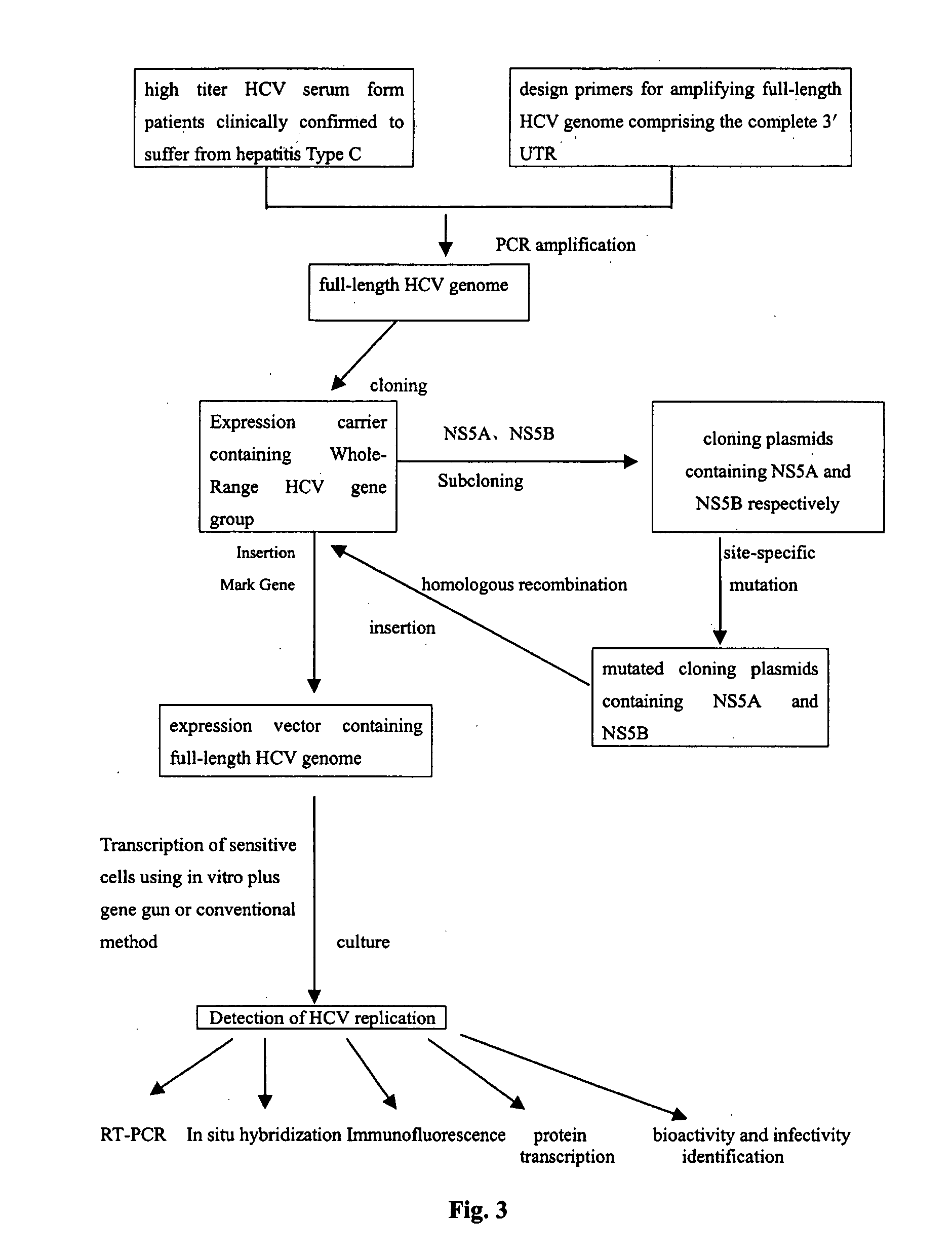
An external culture system of hepatitis C totivirus for culturing complete multiplicative HCV virus by the molecular biology and gene recombination technique is disclosed. Its culture steps include amplifying the full-length genom containing 98 nucleotides after 3' PolyA in the serum of hepatitis C patient; artificial site-directed mutation to NS5A and NS5B in HCV genon; inserting the expression box of marker gene IRES-GFP at NS5B3' terminal in the mutated HCV genom; transfecting sensitive cell and culturing to obtain progeny HCV virus with infection activity.
Owner:西安金昊科技投资管理有限公司
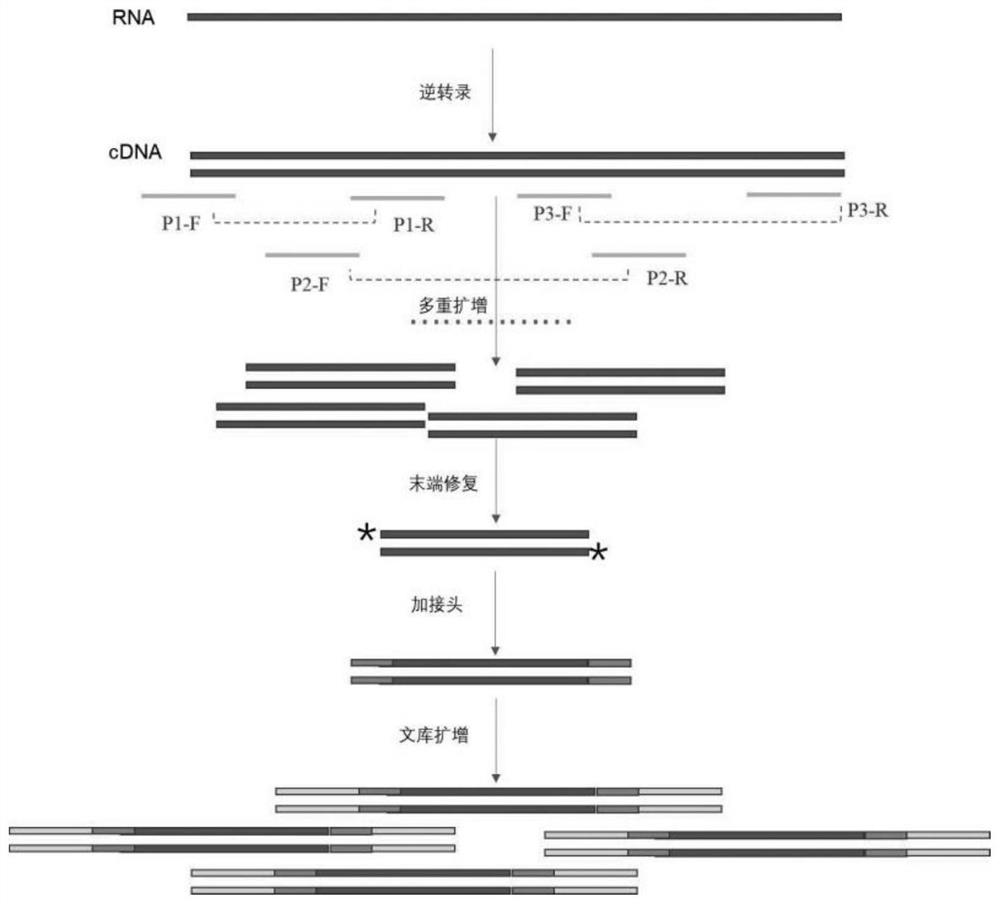
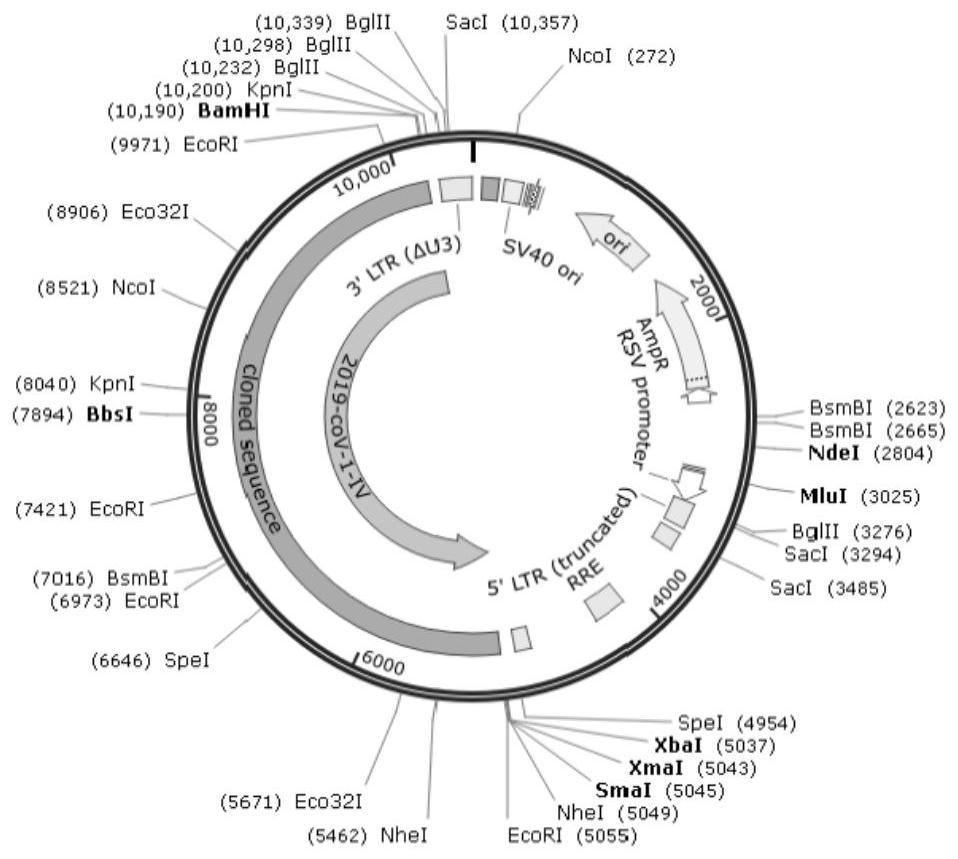
Method for constructing library for full-length genome sequencing of novel coronavirus SARS-CoV-2
InactiveCN111979353AGet satisfiedMeet high-throughput sequencingMicrobiological testing/measurementLibrary creationGenomic sequencingNucleic acid sequencing
The present invention relates to a method for constructing a library for full-length genome sequencing of novel coronavirus SARS-CoV-2. A complete mutation information genome library is established for gene sequencing to meet the requirements of an NGS technology, and solve the problems that some existing novel coronavirus nucleic acid sequencing library conducting kits have low detection, sensitivity poor specificity and more false positives, and the detected gene regions are not comprehensive, and the virus mutation condition cannot be further detected and analyzed.
Owner:上海融享生物科技有限公司
Rabies viruses CTN strain complete genome infectious cDNA cloning, and preparation and use thereof
InactiveCN101475943APreserve weak toxicitySafeMicroorganism based processesAntiviralsCdna cloningViral vector
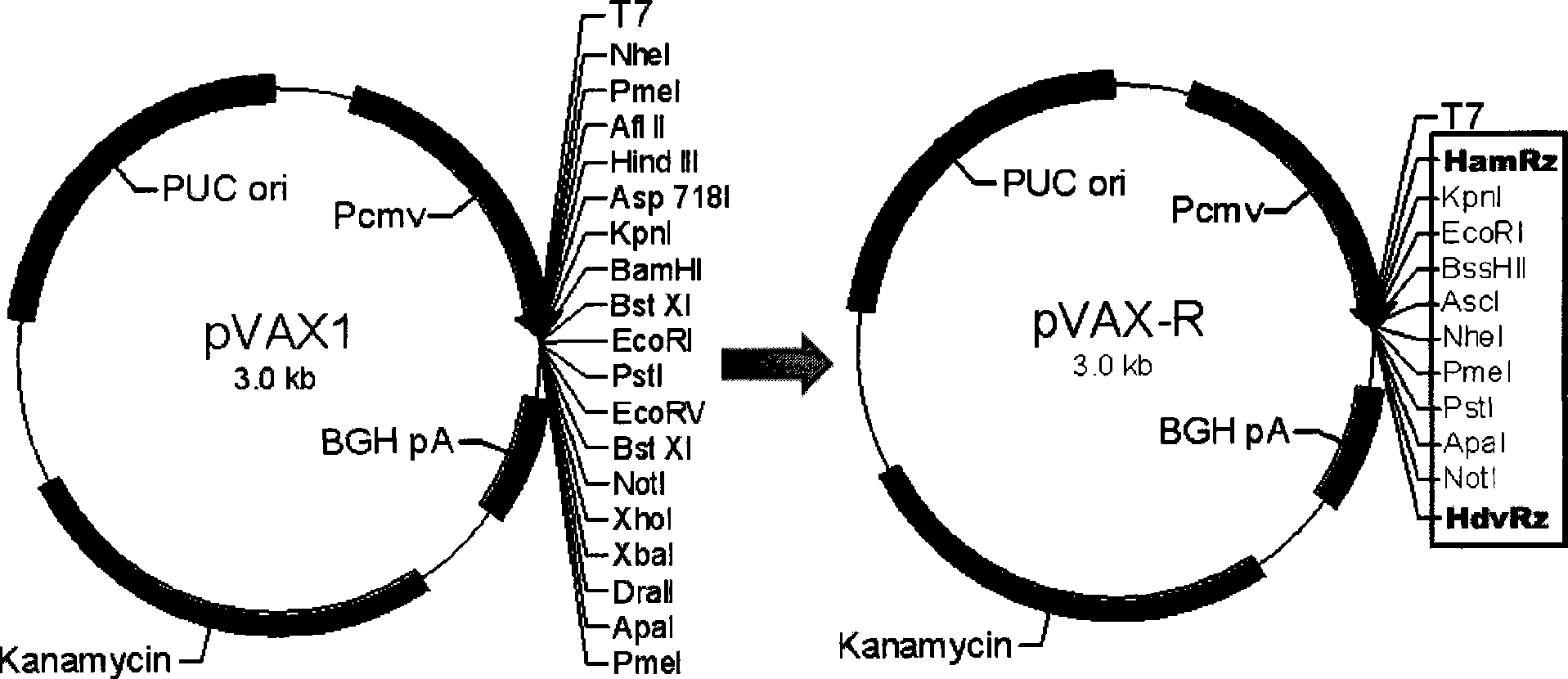
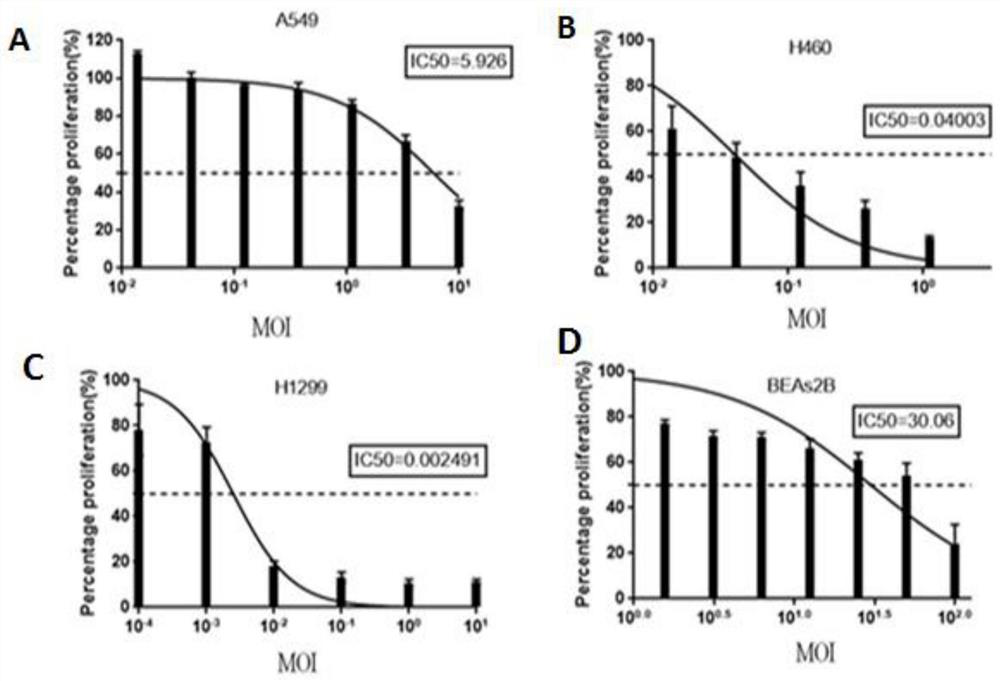
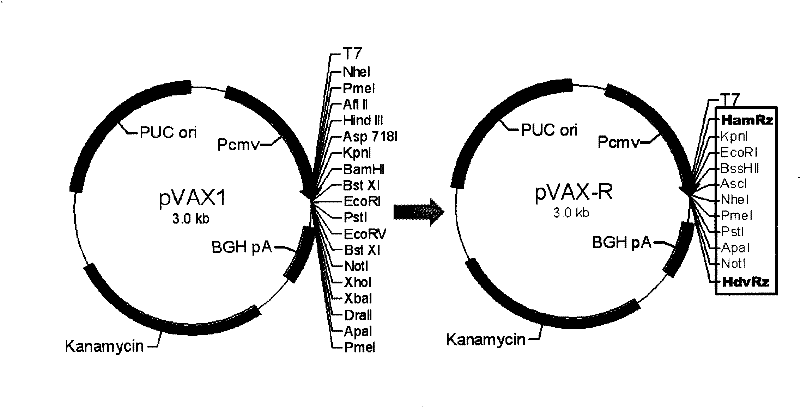
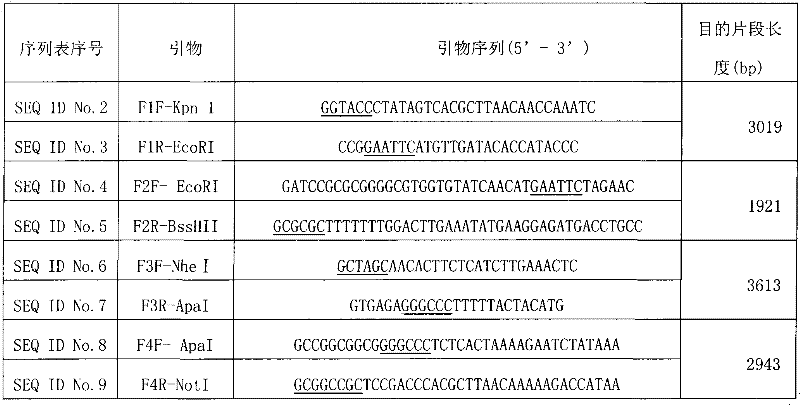
The present invention provides a rabies virus CTN strain whole genome cDNA infectious clone, as well as a method of using reverse genetics technology to save the active rabies virus. The present inventive method includes: using the single restriction enzyme sites in the CTN strain of rabies virus full-length genome to divide the full-length virus into four segments to amplify, simultaneously using the green fluorescent protein gene to substitute 423bp base at the pseudo-gene point, cloning in the pVAX-R expression vector, to construct a recombinant full-length plasmid of the virus genome; performing PCR amplification of nucleoprotein, phosphoprotein, glycoprotein and transcription large protein gene sequence of the CTN strain of the rabies virus, and cloning the amplified fragment into a vector pVAX1, to constitute four auxiliary plasmids in the reverse heredity system; and then using the five plasmids to jointly transfect cells, to rescue the rabies virus. The present invention successfully saves the rabies virus, and has a great significance in the study of the pathogenic mechanism of rabies virus, development of new vaccines, viral vectors and the like areas.
Owner:中国疾病预防控制中心病毒病预防控制所
Full-Length Infectious Cdna Clone for Porcine Reproductive and Respiratory Syndrome Virus(Prrsv) and Uses Thereof
The present invention relates to a novel full-length infectious cDNA clone for Porcine Reproductive and Respiratory Syndrome Virus (PRRSV), derivatives therefrom and uses thereof Particularly, the present invention relates to a full-length PRRSV genomic RNA represented by SEQ. ID. No 27 and the genetically stable full-length infectious PRRSV cDNA clone thereof. The PRRSV genomic RNA and infectious PRRSV cDNA clone of the present invention can be used not only for the identification of the PRRSV viral genes, but also for the molecular biological studies including viral replication, transcription and translation. Moreover, it can also be applied to the development of the therapeutic agents, vaccines, diagnostic reagents and diagnostic devices for porcine reproductive and respiratory syndrome and can be used as a novel expression vector for a variety of heterologous genes of interest.
Owner:LEE SEUNG HAN +1
Intact hepatitis c virus and the method for culturing it in a vitro cell culture
InactiveUS20040166488A1Easy to transcribeImprove efficiencySsRNA viruses positive-senseMicrobiological testing/measurementNucleotideCell
The present invention relates to the establishment of an in vitro cell culture system for culturing the whole HCV proliferating virus by using molecular biology and gene recombinant technology. Said method comprises the step of amplifying from the serum of HCV patients the full-length HCV genome comprising the 98 nucleotides at the 3' end of the genome; site-specific mutating the NS5A and NS5B of HCV genome; inserting a marker gene IRES-GFP expression cassette into the NS5B 3' end in the mutated HCV genome; and transfecting the sensitive cells and culturing, to obtain the infectious HCV offspring virus.
Owner:YANGLING DAIYING BIOLOGICAL ENG
Ribosomal RNA sequence extraction method and microorganism identification method using extracted ribosomal RNA sequence
InactiveUS20190119717A1High completenessMicrobiological testing/measurementBiostatisticsMicroorganismContig
The present disclosure relates to a method for obtaining rRNA sequence information, comprising the steps of calling an rDNA sequence from full-length genomic DNA sequence information, assembling the rDNA to form rDNA contigs; and extracting DNA sequence of 16S rRNA having high completeness from the rDNA contigs and correcting an assembly error, and a method for identifying a microorganism, using the rDNA information.
Owner:CHUNLAB INC
Construction method of HBV (Hepatitis B Virus) isolating strain replicating-form plasmid
InactiveCN101993893AImprove digestion efficiencyInefficient digestionMicroorganism based processesVector-based foreign material introductionPhosphorylationHepatitis B virus
The invention discloses a construction method of HBV (Hepatitis B Virus) isolating strain replicating-form plasmid, comprising the following steps of: 1, extraction of HBV full-length genome, PCR (Polymerase Chain Reaction) amplification and clone construction; 2, construction of a recombinant transition vector; 3, enzyme cutting and dephosphorylation of the recombinant transition vector; and 4, construction of replicating-form plasmid, wherein the recombinant transition vector is synchronously and directionally connected by three connection segments, two reverse BspQI enzyme cutting sites are formed between two inserted HBV segments, and the clone of the HBV full-length genome and the bacterial colony cultivation and amplification of the replicating-form plasmid are carried out by an SOC culture medium. The invention applies a novel full-length genome clone vector, improves the enzyme cutting solution and adopts a novel bacterial colony culture medium. The invention has the advantages of simple performance, high efficiency and stability, can be applied to the comprehensive phenotype research on a China HBV clinic isolating strain whole genome and has very important application value.
Owner:RUIJIN HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
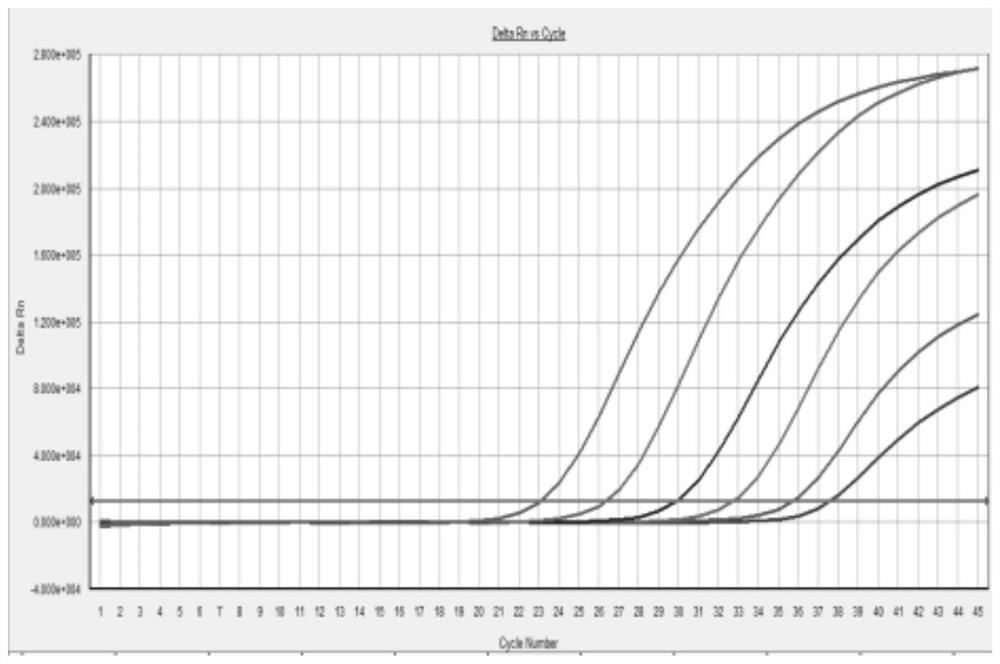
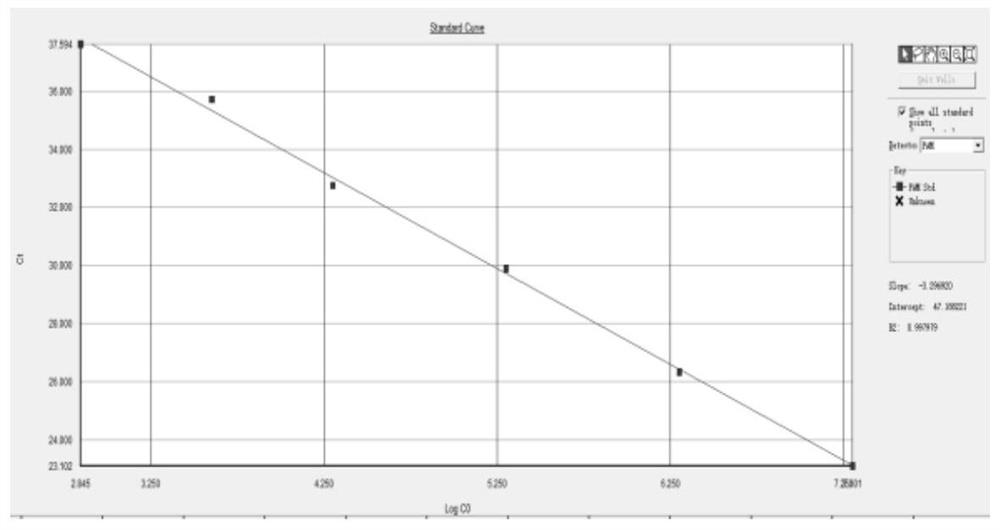
Quality control material for detecting respiratory tract pathogen nucleic acid and preparation method thereof
PendingCN112609023ANo Biosafety ConcernsNo risk of infectionMicrobiological testing/measurementMicroorganism based processesMycoplasmal pneumoniaPneumonitis
The invention provides a quality control material for detecting respiratory pathogen nucleic acid. The quality control material comprises any one or more of the following respiratory pathogens: coronavirus, influenza virus, adenovirus, mycoplasma pneumoniae, streptococcus pneumoniae, rhinovirus or / and legionella pneumophila. A full-length genome sequence of the coronavirus is divided into six target fragments, and the length of each fragment ranges from 4000 bp to 5500 bp. The quality control material disclosed by the invention covers all detection target sequences (or target spots) of the coronavirus, the coverage is wide, the detection target spots are comprehensive, and the phenomenon of missing detection is avoided. The quality control material provided by the invention contains main respiratory tract infection pathogens, has a wide coverage range, and has more accurate detection targets by taking a real virus sample and a slow virus sample as raw materials. According to the coronavirus quality control material prepared by the invention, a target gene sequence is integrated into a host genome through a lentiviral vector, an autonomously replicated gene is knocked out in the preparation process, and the coronavirus quality control material has a self-inactivation capability, so that the recombinant lentivirus cannot be replicated in a target cell and infects other cells.
Owner:GUANGZHOU BDS BIOLOGICAL TECH CO LTD
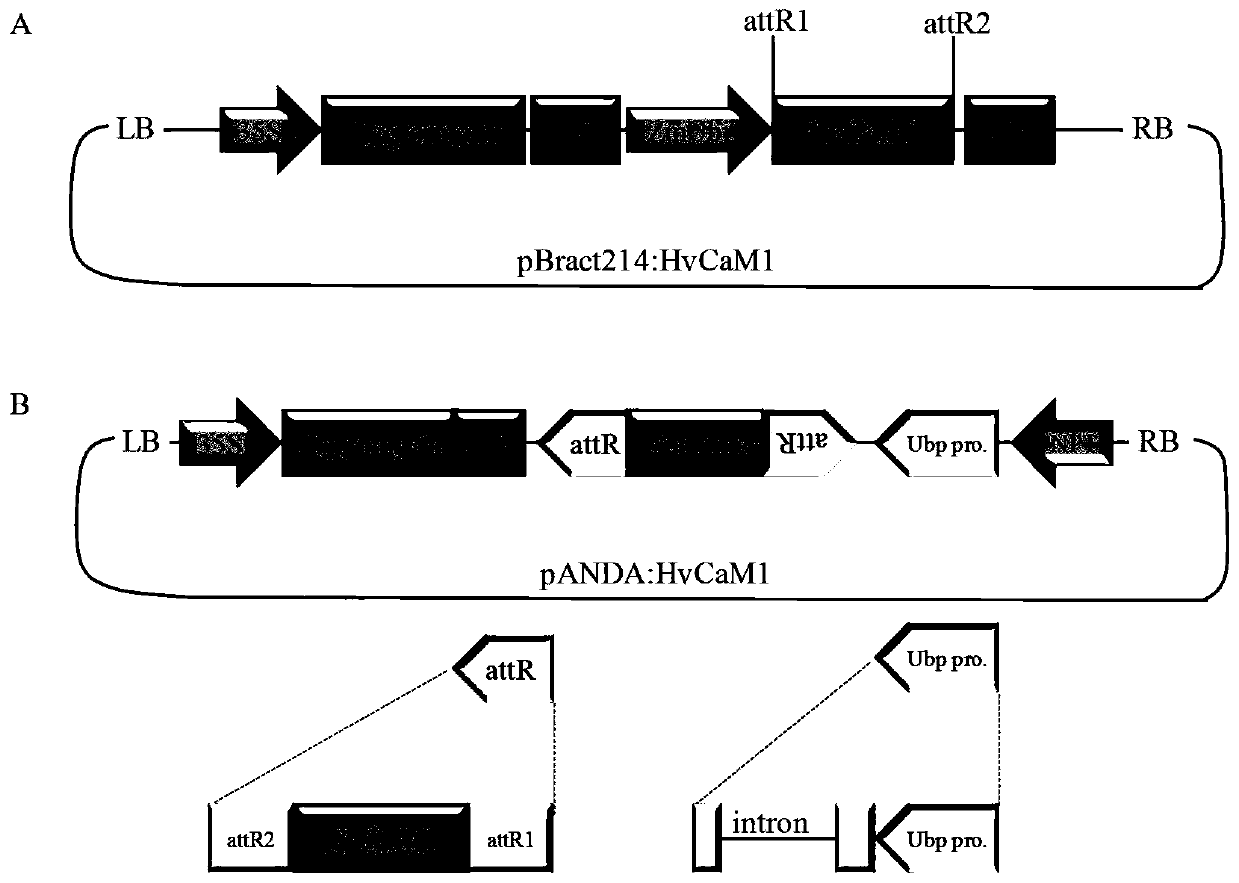
Hordeum vulgare calmodulin gene HvCAM1 and salt tolerant application thereof
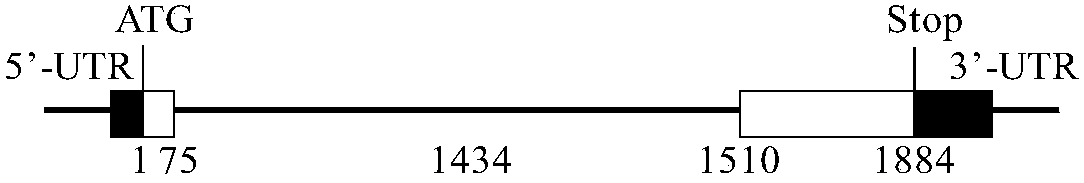
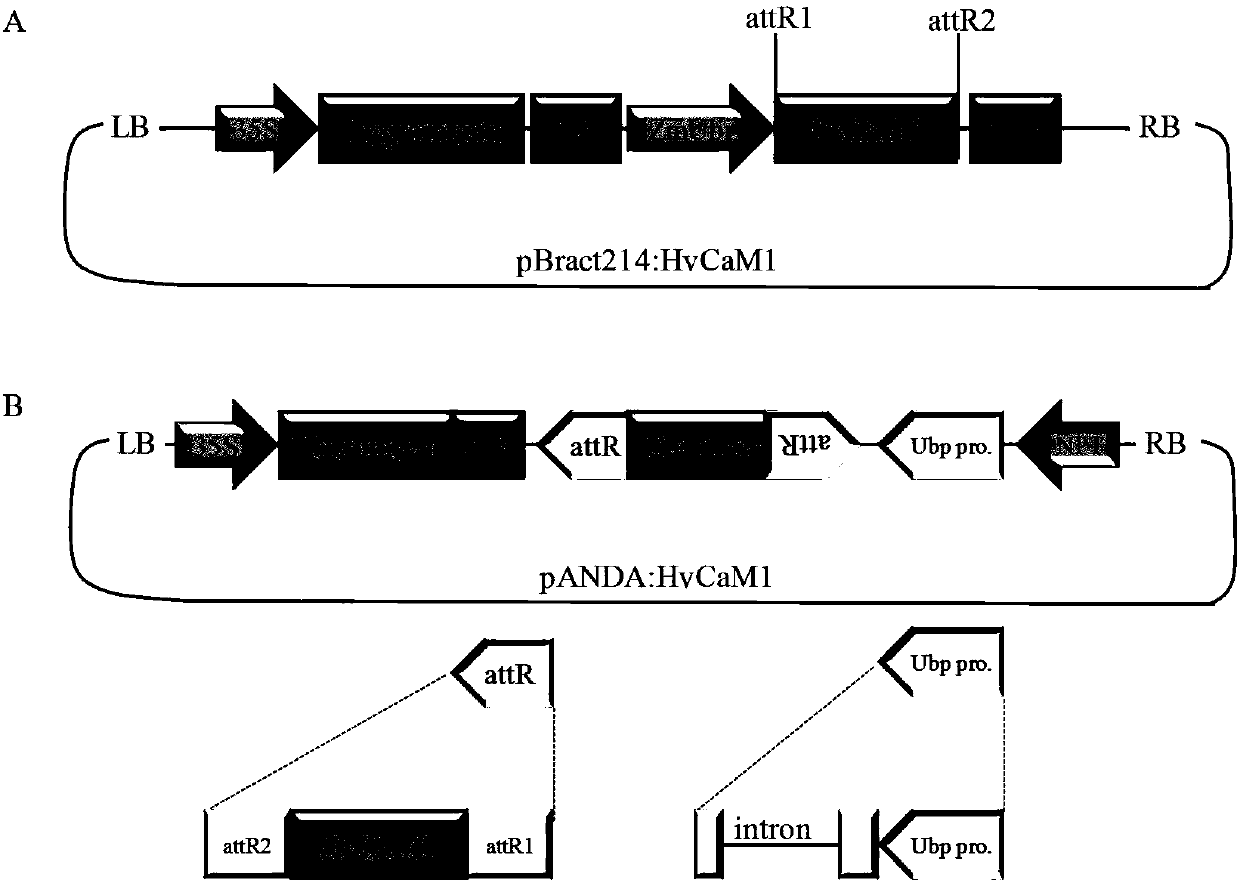
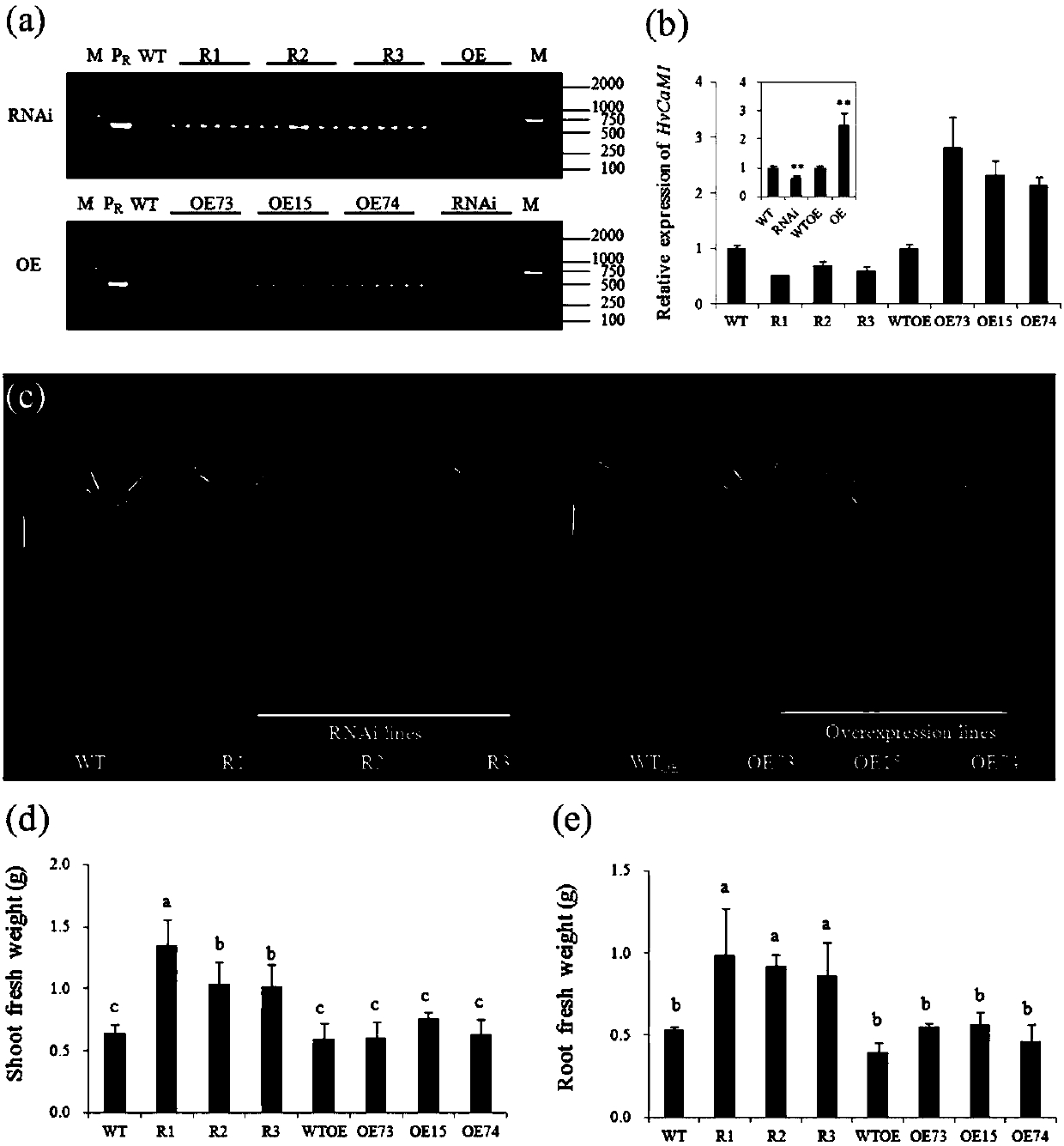
ActiveCN107653249AImprove salt toleranceGuaranteed normal growthPlant peptidesFermentationNucleotideFull length cdna
Belonging to the technical field of biological genetic engineering, the invention in particular relates to a hordeum vulgare calmodulin gene HvCAM1 and salt tolerant application thereof. The hordeum vulgare calmodulin gene HvCAM1 has a nucleotide sequence shown as SEQ ID No:1 and a full-length genome sequence 2124 bp. According to the invention, firstly a hordeum vulgare calmodulin coded gene is obtained by cloning and is named as HvCaM1, and the transcript cDNA full-length nucleotide sequence is shown as SEQ ID No:2, and the full-length cDNA sequence is 728 bp. The invention also utilizes RNAi interference method to silence the HvCAM1 gene successfully in hordeum vulgare Golden Promise, also utilizes the material to verify the negative regulation effect of HvCAM1 gene under salt stress can effectively enhance the salt tolerance of hordeum vulgare. At the same time, overexpression of the HvCAM1 gene in hordeum vulgare Golden Promise cannot enhance the salt tolerance of hordeum vulgare.
Owner:ZHEJIANG UNIV
Down-regulation and silencing of allergen genes in transgenic peanut seeds
InactiveUS20110004959A1Sugar derivativesOther foreign material introduction processesBiotechnologyNucleotide
An allergen-free transgenic peanut seed is produced by recombinant methods. Peanut plants are transformed with multiple copies of each of the allergen genes, or fragments thereof, to suppress gene expression and allergen protein production. Alternatively, peanut plants are transformed with peanut allergen antisense genes introduced into the peanut genome as antisense fragments, sense fragments, or combinations of both antisense and sense fragments. Peanut transgenes are under the control of the 35S promoter, or the promoter of the Ara h2 gene to produce antisense RNAs, sense RNAs, and double-stranded RNAs for suppressing allergen protein production in peanut plants. A full length genomic clone for allergen Ara h2 is isolated and sequenced. The ORF is 622 nucleotides long. The predicted encoded protein is 207 amino acids long and includes a putative transit peptide of 21 residues. One polyadenilation signal is identified at position 951. Six additional stop codons are observed. A promoter region was revealed containing a putative TATA box located at position −72. Homologous regions were identified between Ara h2, h6, and h7, and between Ara h3 and h4, and between Ara h1P41B and Ara h1P17. The homologous regions will be used for the screening of peanut genomic library to isolate all peanut allergen genes and for down-regulation and silencing of multiple peanut allergen genes.
Owner:NGATEGEN INC +1
Down-regulation and silencing of allergen genes in transgenic peanut seeds
Owner:NGATEGEN INC +1
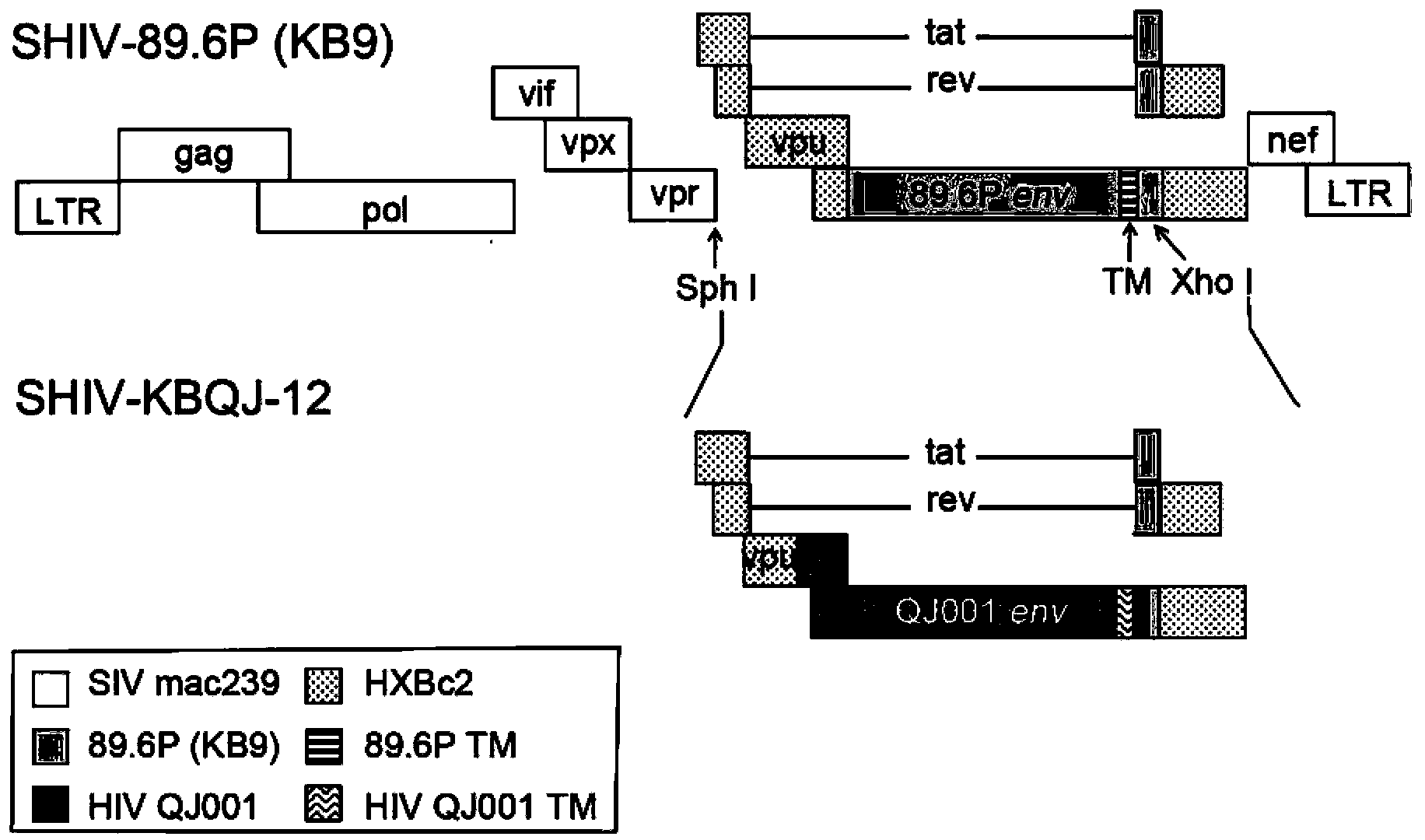
Construction and application of SHIV (simian/human immunodeficiency virus) infectious clone
InactiveCN103374550AGood replicationStable productive infectionMicroorganism based processesViruses/bacteriophagesNucleotideEnvelope Gene
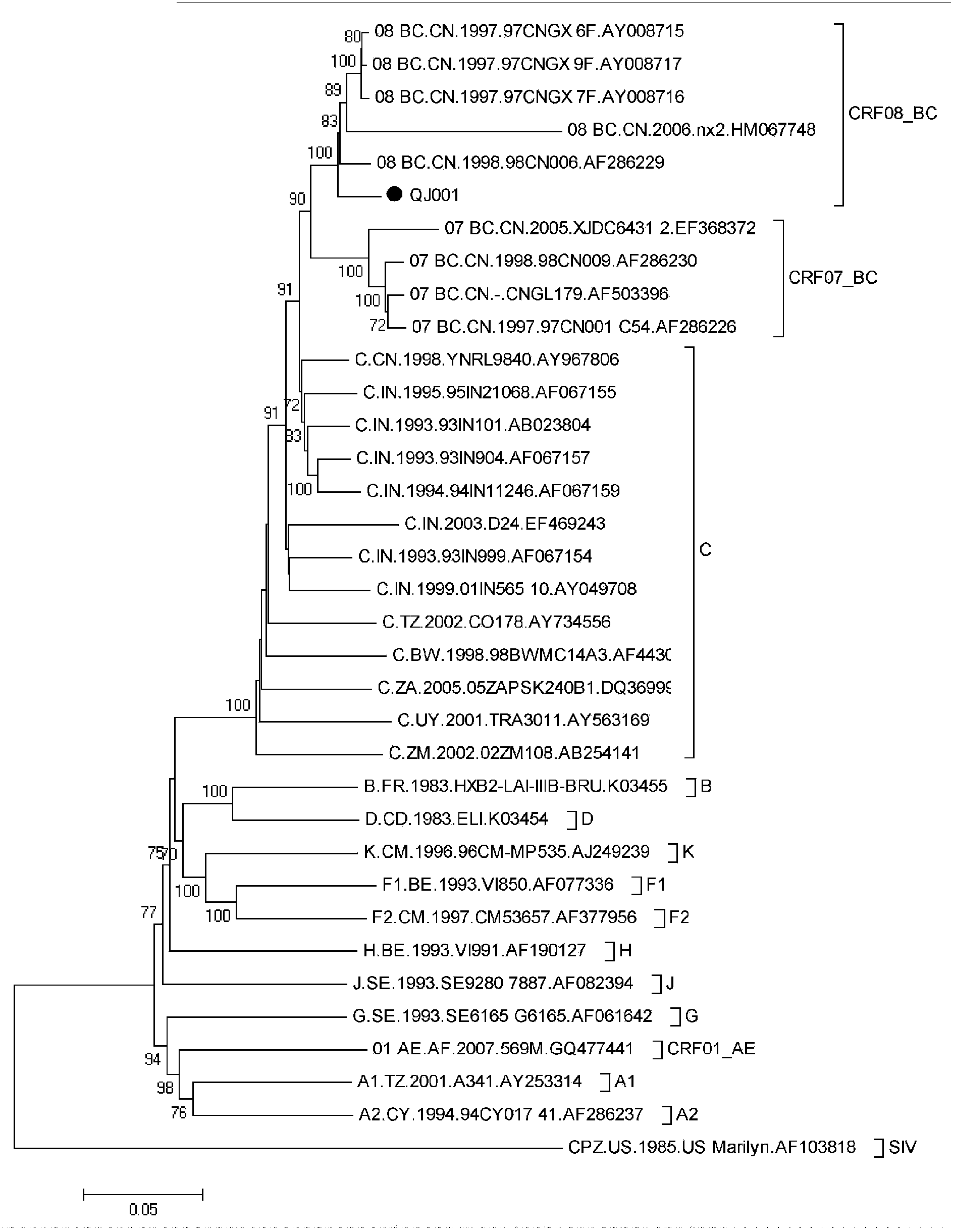
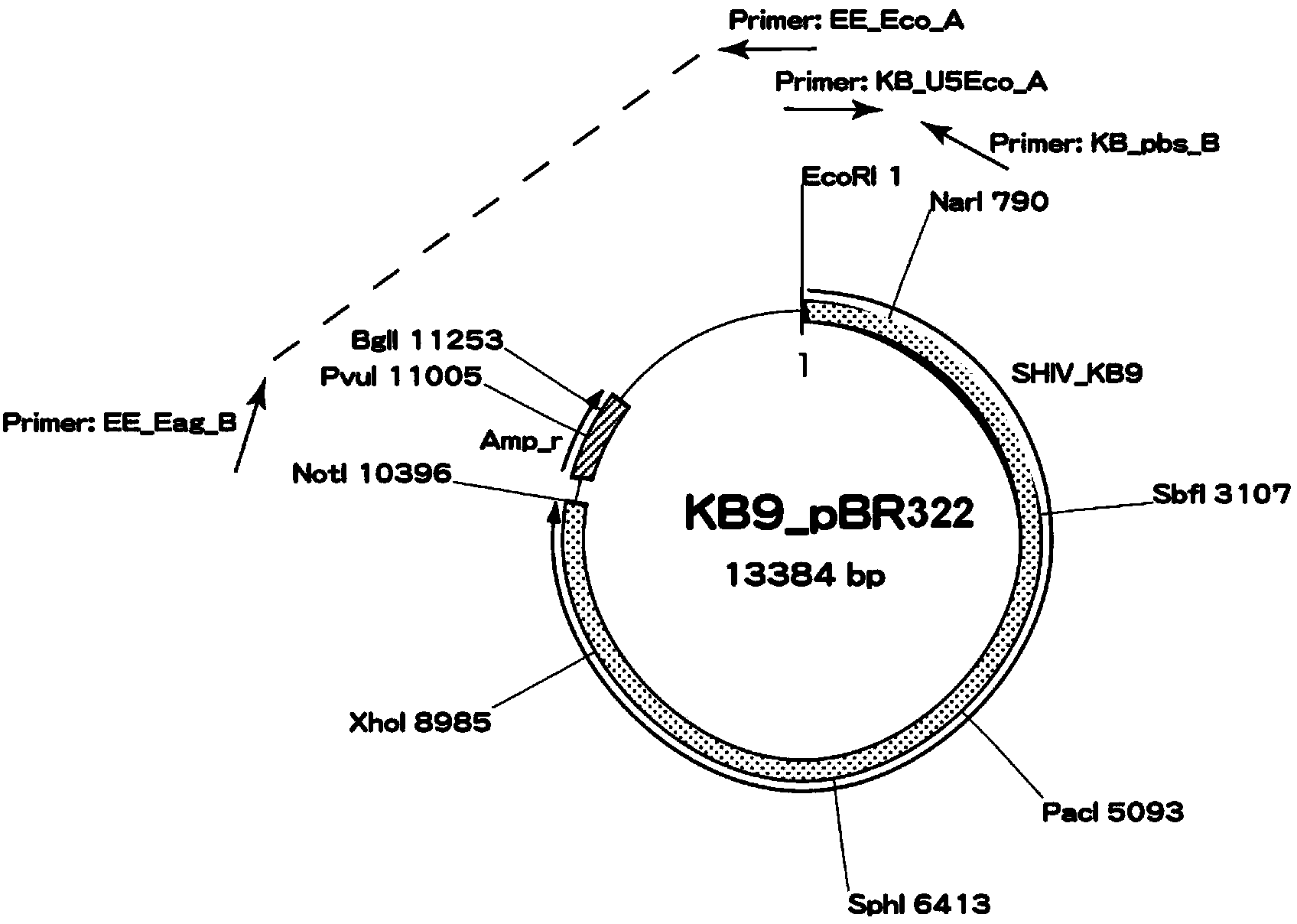
The invention relates to construction and application of SHIV (simian / human immunodeficiency virus) infectious clone. Firstly, the overall length genome of a virulent strain SHIV-KB9 embedded with a B subtype env gene is cloned to a low copy number carrier pBR322, the obtained plasmid SHIV-KB9 / pBR322 is taken as a genome skeleton, the env gene of Chinese main prevalent strain CRF08_BC of HIV-1 is used for substituting for corresponding gene segments so as to obtain the infectious clone SHIV-KBQJ-12, the replacing env gene is from the infectious clone strain QJ001 of HIV-1CRF08_BC and comprises the whole intracellular region, a transmembrane region and part of an extracellular region of gp120 and gp41, and the position of the replacing env gene, corresponding to the position of the nucleotide of a standard reference strain HXB2, is 6103-8249. The construction is simple and convenient to operate, has high success rate, can obtain the infectious clone in a short time, and can be used for establishing an SHIV / Chinese rhesus infection model.
Owner:WUHAN INST OF VIROLOGY CHINESE ACADEMY OF SCI
Agrobacterium strain carrying Yunnan tomato leaf curl virus infectious clones and application thereof
InactiveCN102690776AEfficient infectionEasy to operateBacteriaMicrobiological testing/measurementDiseasePathogenicity
The invention discloses a agrobacterium strain carrying Yunnan tomato leaf curl virus infectious clones and application thereof. 1.4 forward repetitive Yunnan tomato leaf curl virus full-length genomes of DNAs (deoxyribonucleic acids) are constructed to an efficient plant expression vector pBinPLUS by a molecular biology method, and an agrobacterium strain EHA105 is introduced by an electric shock method to obtain a virus infectious clone agrobacterium strain ToLCYnV-Y194 capable of efficiently infecting plants, wherein the collection number of the agrobacterium strain is CGMCC No.5931. The strain can generate massive Yunnan tomato leaf curl virus infectious clone which can efficiently infect multiple host plants, and provides technical and material supports for pathogenicity analysis and genome function research of Yunnan tomato leaf curl virus as well as tomato breeding for disease resistance.
Owner:ZHEJIANG UNIV
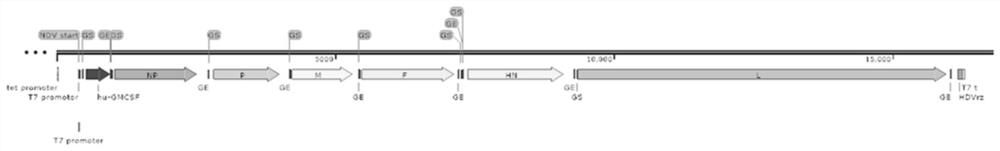
Recombinant oncolytic Newcastle disease virus for expressing human GM-CSF and use thereof
PendingCN113355295APromote differentiation and maturationEasy to processSsRNA viruses negative-sensePeptide/protein ingredientsTherapeutic effectTumor antigen
The invention relates to the field of biological virus construction and particularly discloses a recombinant oncolytic Newcastle disease virus for expressing human GM-CSF. The recombinant oncolytic Newcastle disease virus is obtained by inserting a human GM-CSF coding gene into a full-length genome plasmid of a Newcastle disease virus Italien strain and reviving the virus in vitro. The human GM-CSF gene is as shown in SEQ ID NO.1. A GM-CSF molecule is used for recruiting DC cells from mononuclear cells, activating neutrophil and promoting differentiation and maturation of the DC cells, especially has an important effect on promoting processing and presentation of antigens, combines a characteristic of selective replication of tumors of the oncolytic Newcastle disease virus, directly kills tumor cells, causes inflammation, recruits more DC cells, promotes maturation of the DC cells and presentation of tumor antigens, enhances local and whole-body anti-tumor immune responses of tumors, improves a treatment effect of single or combined use of the oncolytic virus in treating solid tumors, and brings a new treatment thought for tumor immunotherapy.
Owner:FOURTH MILITARY MEDICAL UNIVERSITY
Rabies viruses CTN strain complete genome infectious cDNA cloning, and preparation and use thereof
InactiveCN101475943BPreserve weak toxicitySafeMicroorganism based processesAntiviralsRabies virus strainCdna cloning
The present invention provides a rabies virus CTN strain whole genome cDNA infectious clone, as well as a method of using reverse genetics technology to save the active rabies virus. The present inventive method includes: using the single restriction enzyme sites in the CTN strain of rabies virus full-length genome to divide the full-length virus into four segments to amplify, simultaneously using the green fluorescent protein gene to substitute 423bp base at the pseudo-gene point, cloning in the pVAX-R expression vector, to construct a recombinant full-length plasmid of the virus genome; performing PCR amplification of nucleoprotein, phosphoprotein, glycoprotein and transcription large protein gene sequence of the CTN strain of the rabies virus, and cloning the amplified fragment into a vector pVAX1, to constitute four auxiliary plasmids in the reverse heredity system; and then using the five plasmids to jointly transfect cells, to rescue the rabies virus. The present invention successfully saves the rabies virus, and has a great significance in the study of the pathogenic mechanism of rabies virus, development of new vaccines, viral vectors and the like areas.
Owner:STATION OF VIRUS PREVENTION & CONTROL CHINA DISEASES PREVENTION & CONTROL CENT
Primers for cloning SPFMV (sweet potato feathery mottle virus) O strain and full length genome sequence of SPVC (sweet potato virus C) and cloning method
ActiveCN105695456AMethod saves timeMethod saves effortSsRNA viruses positive-senseMicrobiological testing/measurementAgricultural scienceSweet potato virus C
The invention discloses primers for cloning SPFMV (sweet potato feathery mottle virus) O strain and full length genome sequence of SPVC (sweet potato virus C). The primers are respectively a primer for cloning the SPFMV O strain, a primer for cloning the full length genome sequence of SPVC, and the primer for simultaneously cloning the SPFMV O strain and the full length genome sequence of SPVC. The SPFMV and the SPVC generally infect sweet potatoes in a mixed way, the separation and the purification are very difficult; the content of the virus in the sweet potatoes is low; the full length genome sequence is difficult to obtain by a conventional virus purification cloning method. The principle that SPCSV can promote the content rise of the SPFMV and the SPVC in the sweet potato plants is used; the sweet potato plants simultaneously infected with SPCSV, SPFMV-O and SPVC viruses are used as materials; the total RNA (ribonucleic acid) of the virus-infecting sweet potato plant leaves is extracted; the virus full length genome sequence cloning is performed; through a great amount of PCR (polymerase chain reaction) amplification and sequence testing, the genome full length sequence of SPVC and SPFMV is obtained through comparison and splicing.
Owner:INST OF PLANT PROTECTION HENAN ACAD OF AGRI SCI
Preparation method, amplification primer and detection reagent of hpv full-length genome quality control product
ActiveCN108929869BAvoid connection inefficienciesSolve the problem of uncertain concentrationMicrobiological testing/measurementVector-based foreign material introductionHuman DNA sequencingCompetent cell
The invention relates to a preparation method of a HPV full-length genome quality control product, an amplification primer and a detection reagent. Wherein, the preparation method of the HPV full-length genome quality control product comprises the following steps: (1) obtaining a DNA template; (2) using amplification primers to perform PCR amplification; (3) using HD Cloning connection technology to connect the target DNA fragment to the carrier ; (4) introducing the plasmid into competent cells, and then extracting the plasmid; (5) diluting the plasmid with human genome. The preparation method of the human papillomavirus full-length genome quality control product of the present invention can successfully obtain the HPV gene detection quality control product that has no biological infection risk type, can simulate clinical samples, can be produced in large quantities, has a controllable concentration, and is stably stored. Has good applicability.
Owner:亚能生物技术(深圳)有限公司
Full - length genomic RNA of papaya leaf - distortion mosaic virus
The purpose of the present invention is to determine the nucleotide sequence of the full-length genomic RNA of papaya leaf-distortion mosaic virus. The present invention provides the full-length genomic RNA of papaya leaf-distortion mosaic virus, a method for diagnosing infection with papaya leaf-distortion mosaic virus using the full-length genomic RNA, a method for producing a papaya leaf-distortion mosaic virus-resistant plant, and a method for producing a foreign protein in a plant body.
Owner:INDEPENDENT ADMINISTRATIVE INST JAPAN INT RES CENT FOR AGRI SCI
Recombinant Infectious Cloning Plasmid of Japanese Encephalitis Virus Attenuated Strain and Its Application
InactiveCN104152476BEasy to solveEasy to controlNervous disorderViral antigen ingredientsEncephalitis VirusesImmune protection
Owner:SHANGHAI VETERINARY RES INST CHINESE ACAD OF AGRI SCI
Adenovirus vector and its application in preparing HCV cell and mouse models
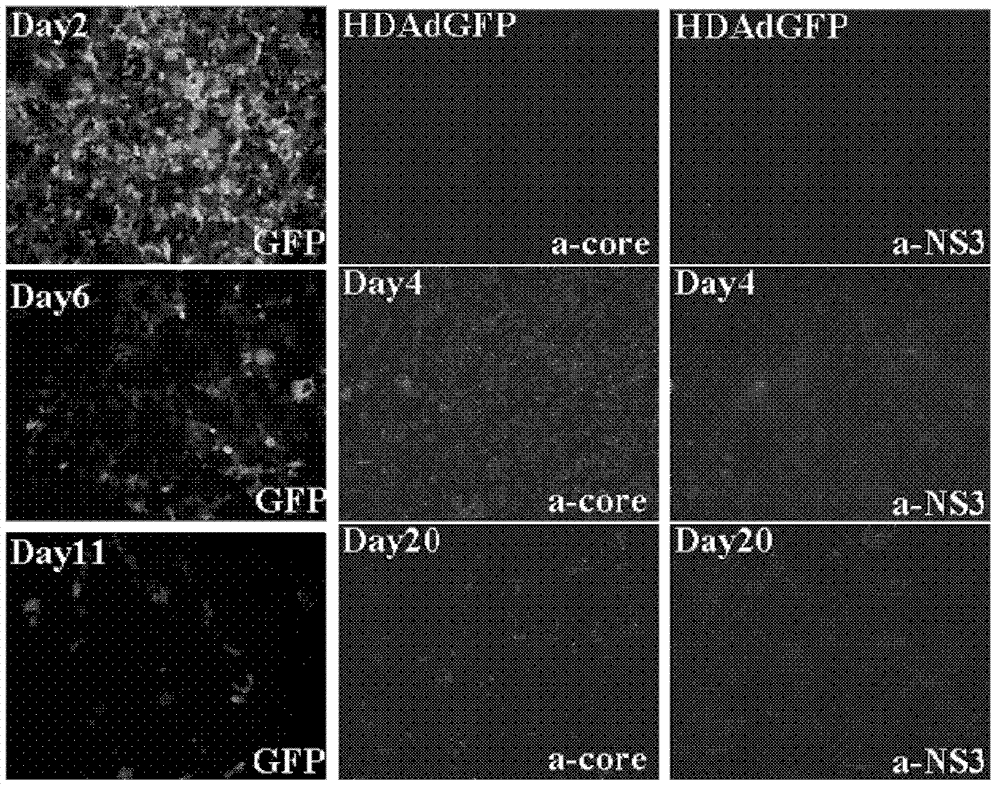
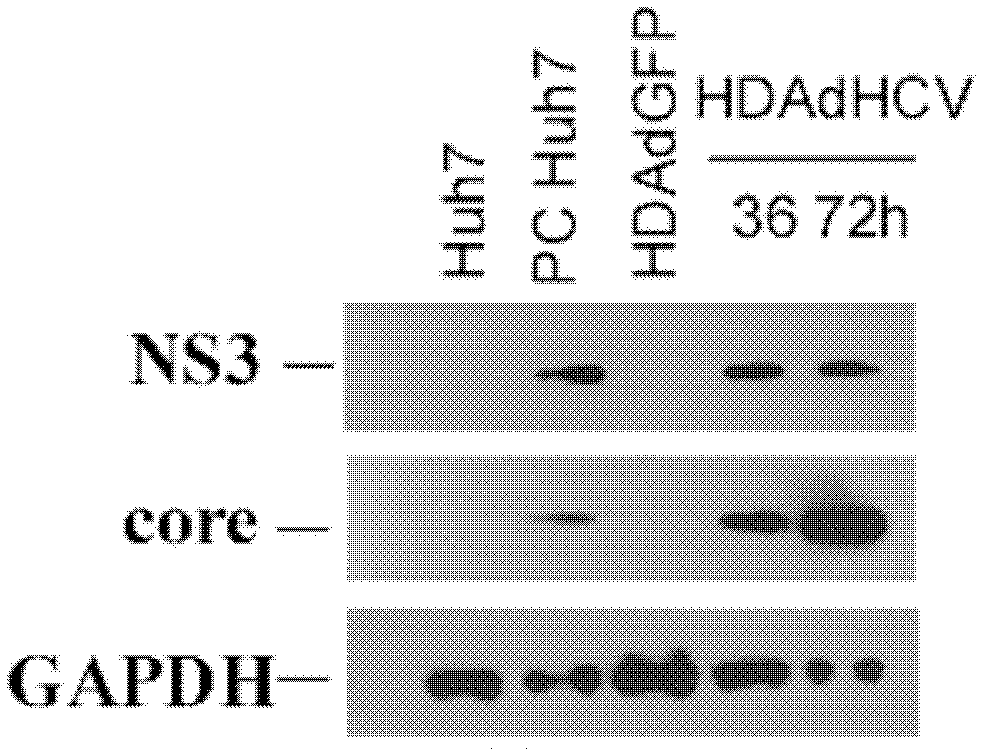
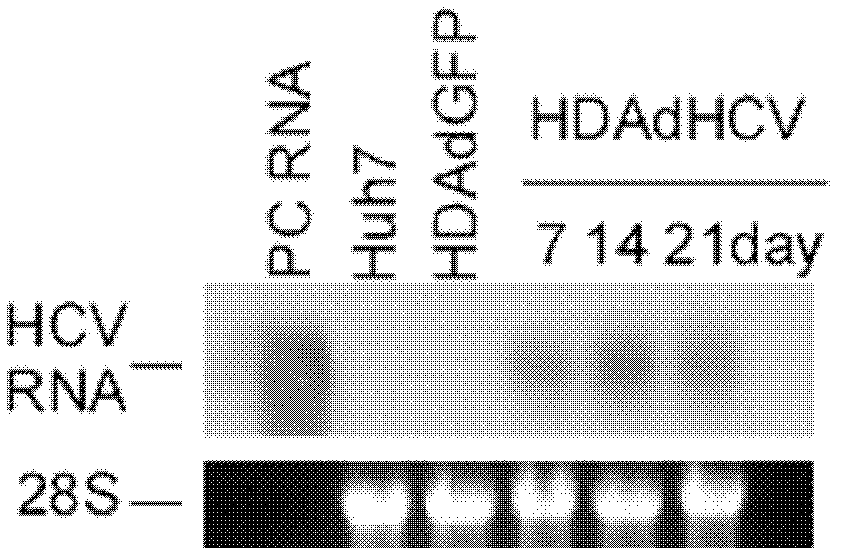
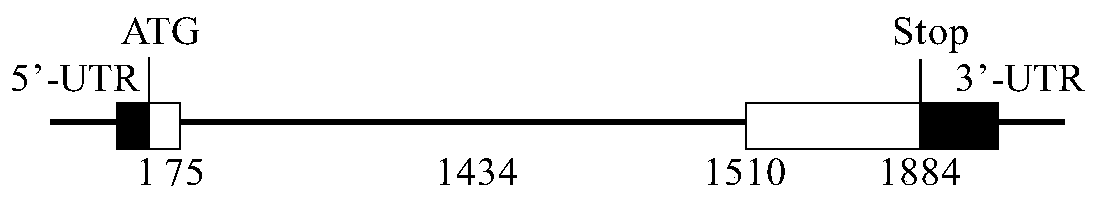
The invention discloses an adenovirus vector and its application in preparing HCV (hepatitis C virus) cells and mouse models. The invention provides a recombinant adenovirus vector which is obtained by inserting nucleotides shown by the sequence 1 in the sequence table in the adenovirus vector HDAd (helper-dependent adenovirus). The experiments prove that the invention provides a third generation adenovirus vector of a recombinant HCV 2a type full-length genome, the prepared adenovirus can be used for preparing HCV whole-genome cell models or mouse models, and the obtained models can be used for drug screening of anti-HCV drugs, so as to be used for HCV vaccine evaluation and researches of the pathogenic mechanism.
Owner:MICROBE EPIDEMIC DISEASE INST OF PLA MILITARY MEDICAL ACAD OF SCI
A barley calmodulin gene hvcam1 and its application in salt tolerance
ActiveCN107653249BImprove salt toleranceGuaranteed normal growthPlant peptidesFermentationBiotechnologyHordeum vulgare
Belonging to the technical field of biological genetic engineering, the invention in particular relates to a hordeum vulgare calmodulin gene HvCAM1 and salt tolerant application thereof. The hordeum vulgare calmodulin gene HvCAM1 has a nucleotide sequence shown as SEQ ID No:1 and a full-length genome sequence 2124 bp. According to the invention, firstly a hordeum vulgare calmodulin coded gene is obtained by cloning and is named as HvCaM1, and the transcript cDNA full-length nucleotide sequence is shown as SEQ ID No:2, and the full-length cDNA sequence is 728 bp. The invention also utilizes RNAi interference method to silence the HvCAM1 gene successfully in hordeum vulgare Golden Promise, also utilizes the material to verify the negative regulation effect of HvCAM1 gene under salt stress can effectively enhance the salt tolerance of hordeum vulgare. At the same time, overexpression of the HvCAM1 gene in hordeum vulgare Golden Promise cannot enhance the salt tolerance of hordeum vulgare.
Owner:ZHEJIANG UNIV
A kind of gii.4 type norovirus genome amplification primer and amplification method
ActiveCN105039330BEfficient amplificationDNA preparationDNA/RNA fragmentationOpen reading frameQuarantine
The invention discloses a GII.4 type norovirus genome amplification primer and an amplification method. In the present invention, six pairs of primers are used as the upstream and downstream primers of the amplification primers, and the RNA of GII.4 type norovirus is used as the template to carry out RT-PCR amplification to obtain amplification products respectively, and then perform nucleic acid sequence sequencing on the amplification products , and then spliced and compared to obtain the full-length sequence of the GII.4 norovirus genome. According to the main popular genotypes of GII.4 Norovirus, the present invention designs a "4+1+1" segmentation amplification strategy according to the three open reading frames contained in the genome, and designs corresponding amplification according to the conserved region Primer; it can effectively amplify the whole genome sequence of GII.4 type norovirus. The invention can be widely used in medical and health care, inspection and quarantine, etc., which have norovirus detection requirements and corresponding scientific research fields.
Owner:GUANGDONG INST OF MICROBIOLOGY GUANGDONG DETECTION CENT OF MICROBIOLOGY +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com