Patents
Literature
86 results about "Amicyanin" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Amicyanin is a type I copper protein that plays an integral role in electron transfer. In bacteria such as Paracoccus denitrificans, amicyanin is part of a three-member redox complex, along with methylamine dehydrogenase (MADH) and cytochrome c-551i.
Composition and method for making high-protein and low-carbohydrate food products
InactiveUS20050129823A1Effectively reducing the net carbohydrate total of the traditional productIncrease nutritionDough treatmentBaking mixturesAdditive ingredientProtein isolate
Conventional food compositions for use in making baked goods and extruded food products are improved by reducing the carbohydrate content. This is done by substituting the conventional flour in whole or in part by a combination of starch that is resistant to amylase digestion and / or from about 1-150 baker's percent of a first proteinaceous ingredient comprising at least about 70% by weight protein, and a second proteinaceous ingredient selected from the group consisting of (i) between about 0.5-100 baker's percent of a wheat protein isolate product; (ii), between about 0.5-100 baker's percent of a wheat protein concentrate product; (iii) between about 0.5-100 baker's percent of a devitalized wheat gluten product; (iv) between about 0.5-20 baker's percent of a fractionated wheat protein product; (v) between about 0.5-20 baker's percent of a deamidated wheat gluten product; (vi) between about 0.5-30 baker's percent of a hydrolyzed wheat protein product; and (vii) any combination of ingredients (i) to (vi).
Owner:MGP INGREDIENTS
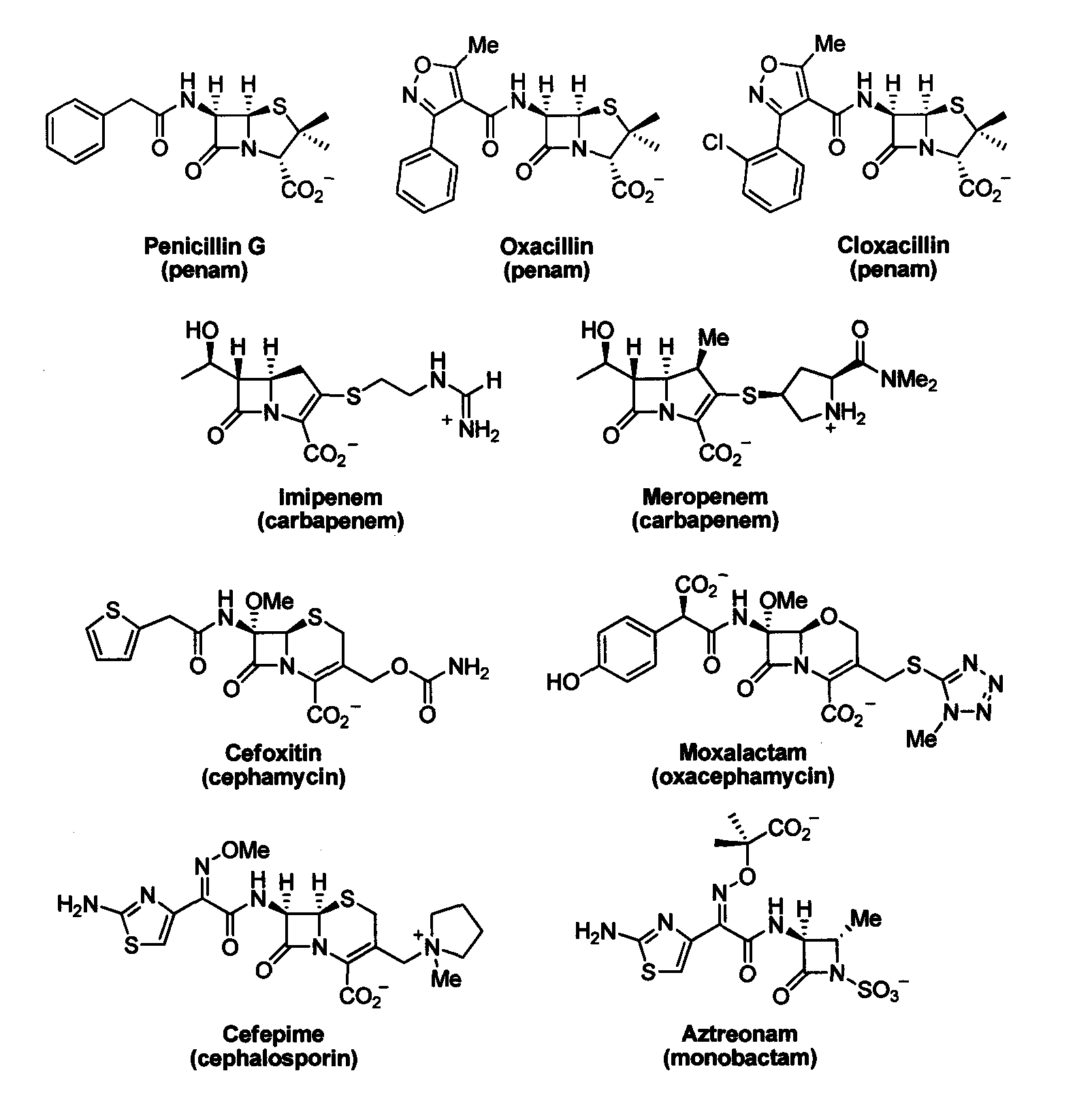
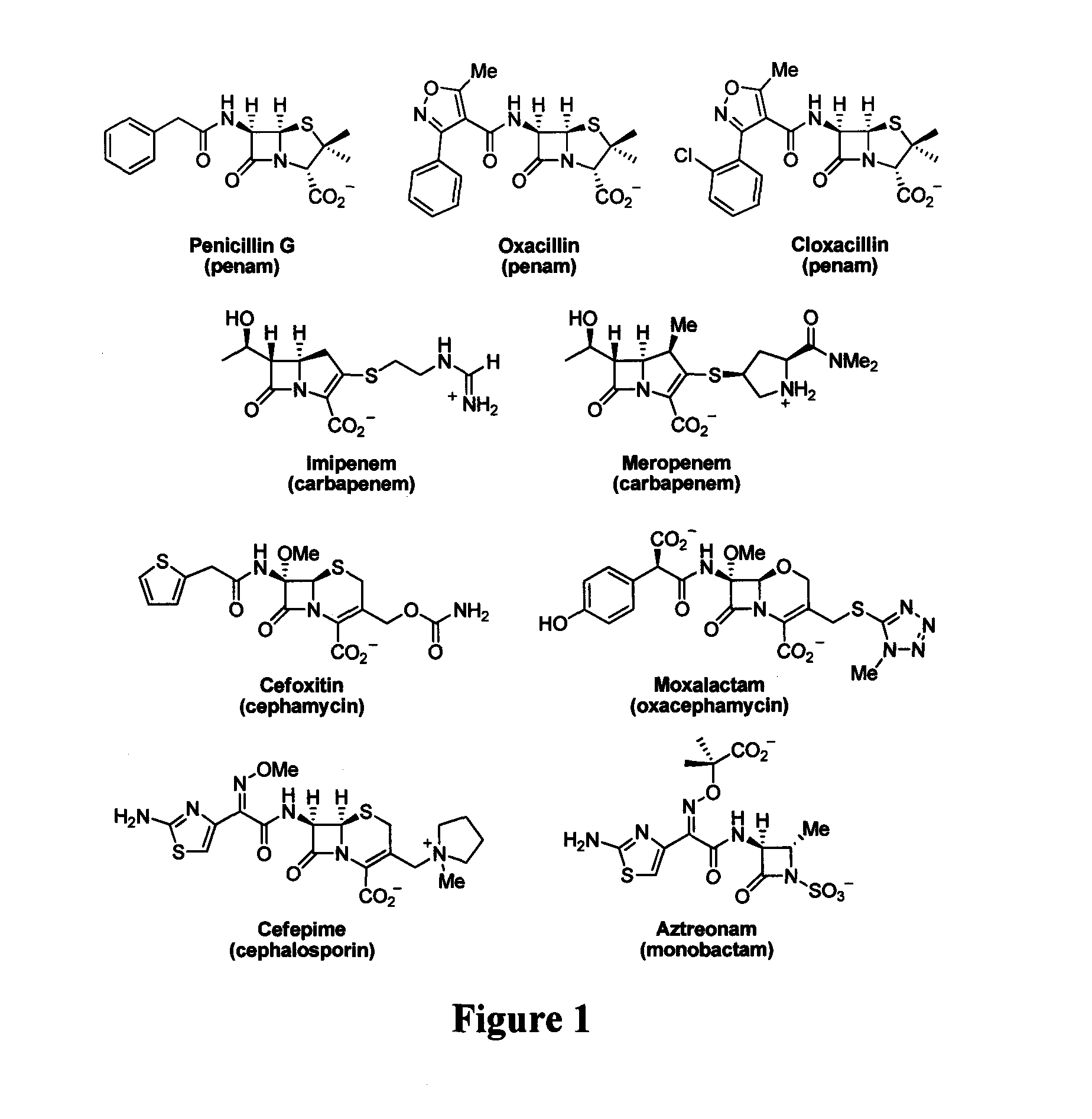
Device and method for detecting antibiotic-inactivating enzymes
ActiveUS20050089947A1Facilitated releaseRobust in vitroMicrobiological testing/measurementBiological material analysisΒ lactamasesMicroorganism
A method for determining whether a microorganism produces an AmpC β-lactamase is disclosed in which a culture of a microorganism suspected of producing a β-lactamase that inactivates a β-lactam-containing antibiotic is admixed with an effective amount of each of i) a β-lactam-containing antibiotic, ii) a β-lactamase inhibitor to which AmpC β-lactamase is resistant, and iii) a permeabilizing agent for the microorganism present in a non-growth-inhibiting microorganism-permeabilizing amount to form an assay culture. That assay culture in maintained under appropriate culture conditions and for a time period sufficient to determine the interaction of the microorganism with the AmpC β-lactamase resistant inhibitor and antibacterial compound, and thereby determine the presence of an AmpC β-lactamase, wherein a positive test indicates the presence of an AmpC β-lactamase.
Owner:CREIGHTON UNIVERSITY
Dietary and other compositions, compounds, and methods for reducing body fat, controlling appetite, and modulating fatty acid metabolism
InactiveUS20050154064A1Inhibit liver fat accumulationLower Level RequirementsBiocidePeptide/protein ingredientsDietary supplementIngested food
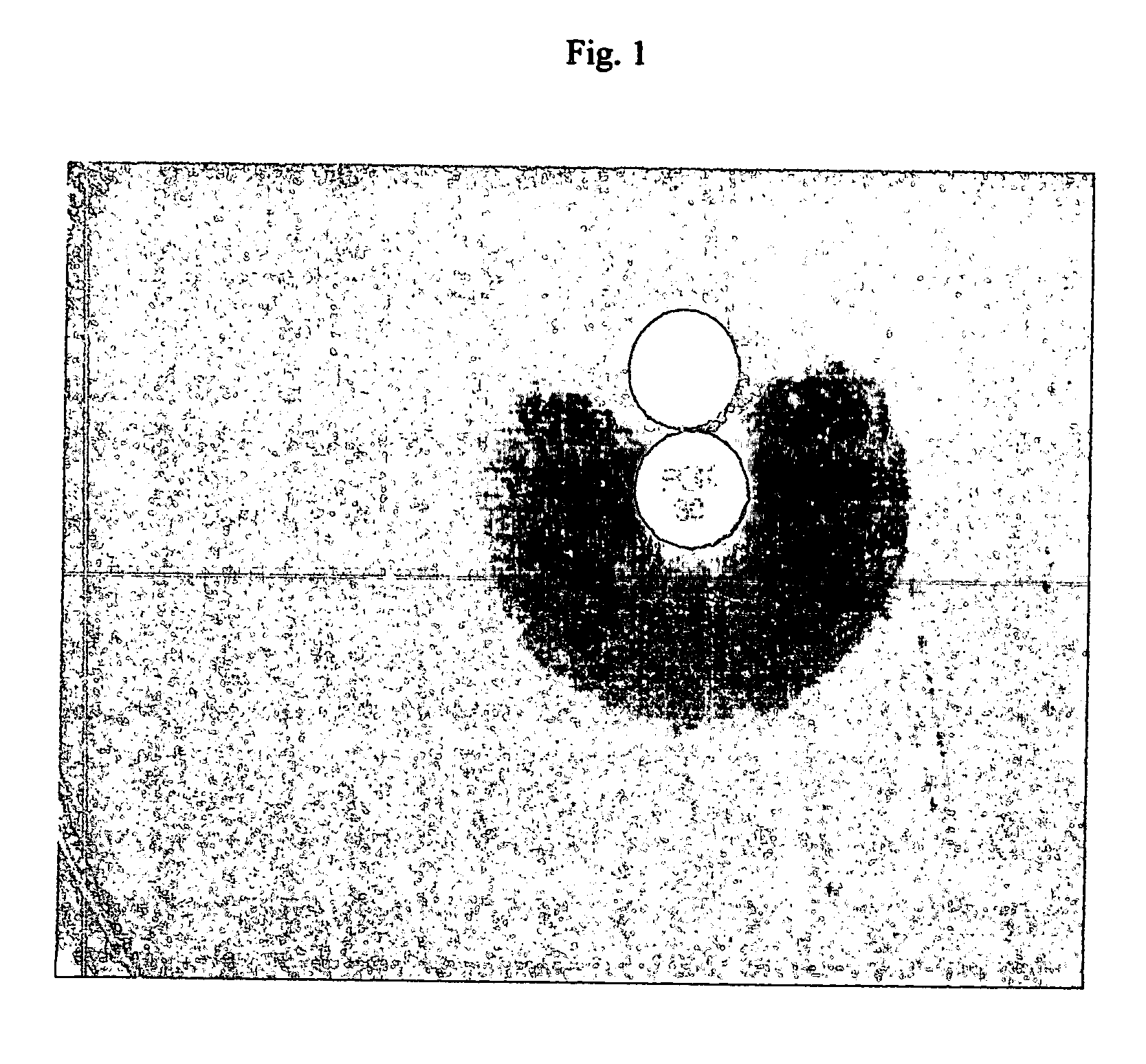
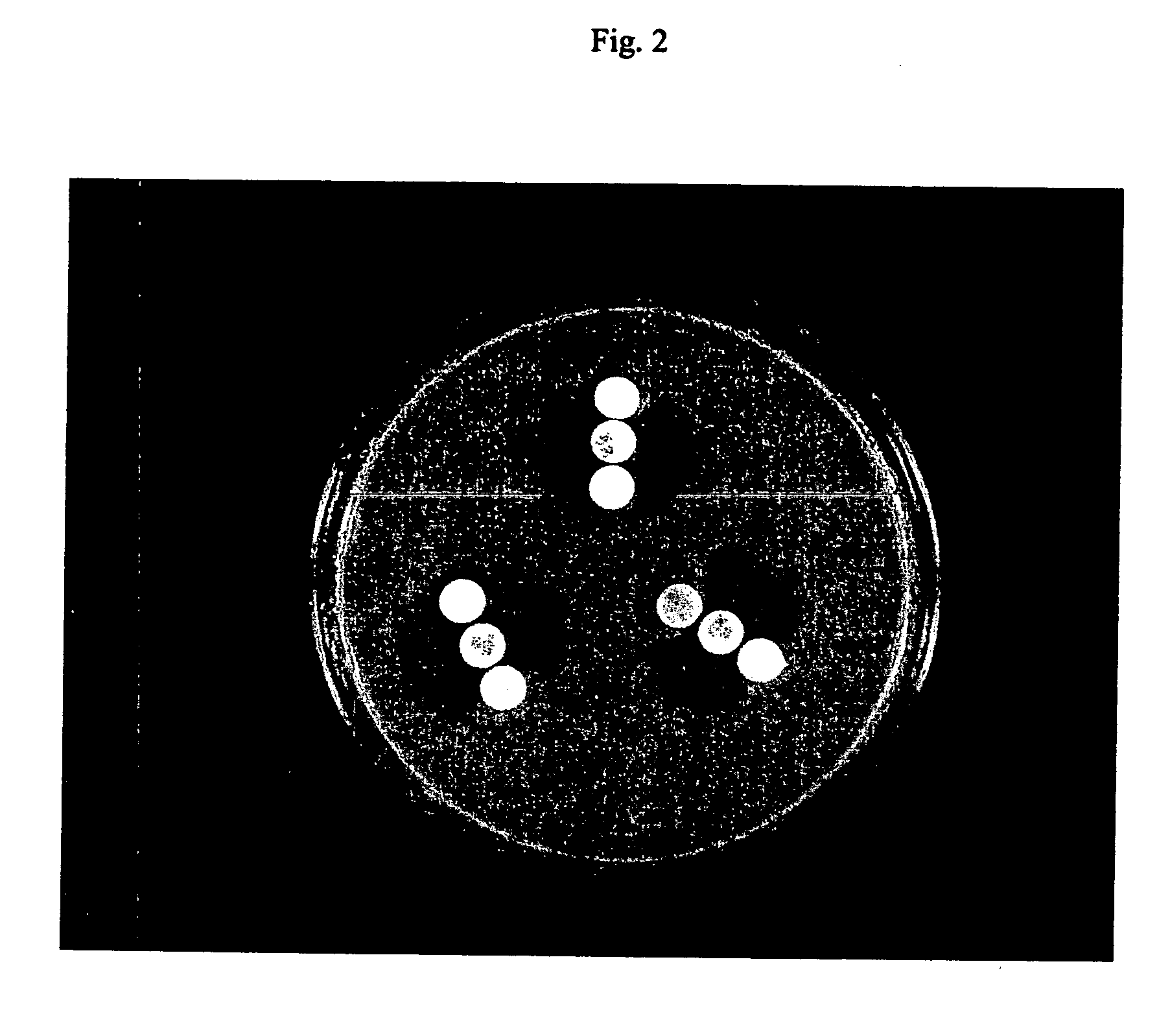
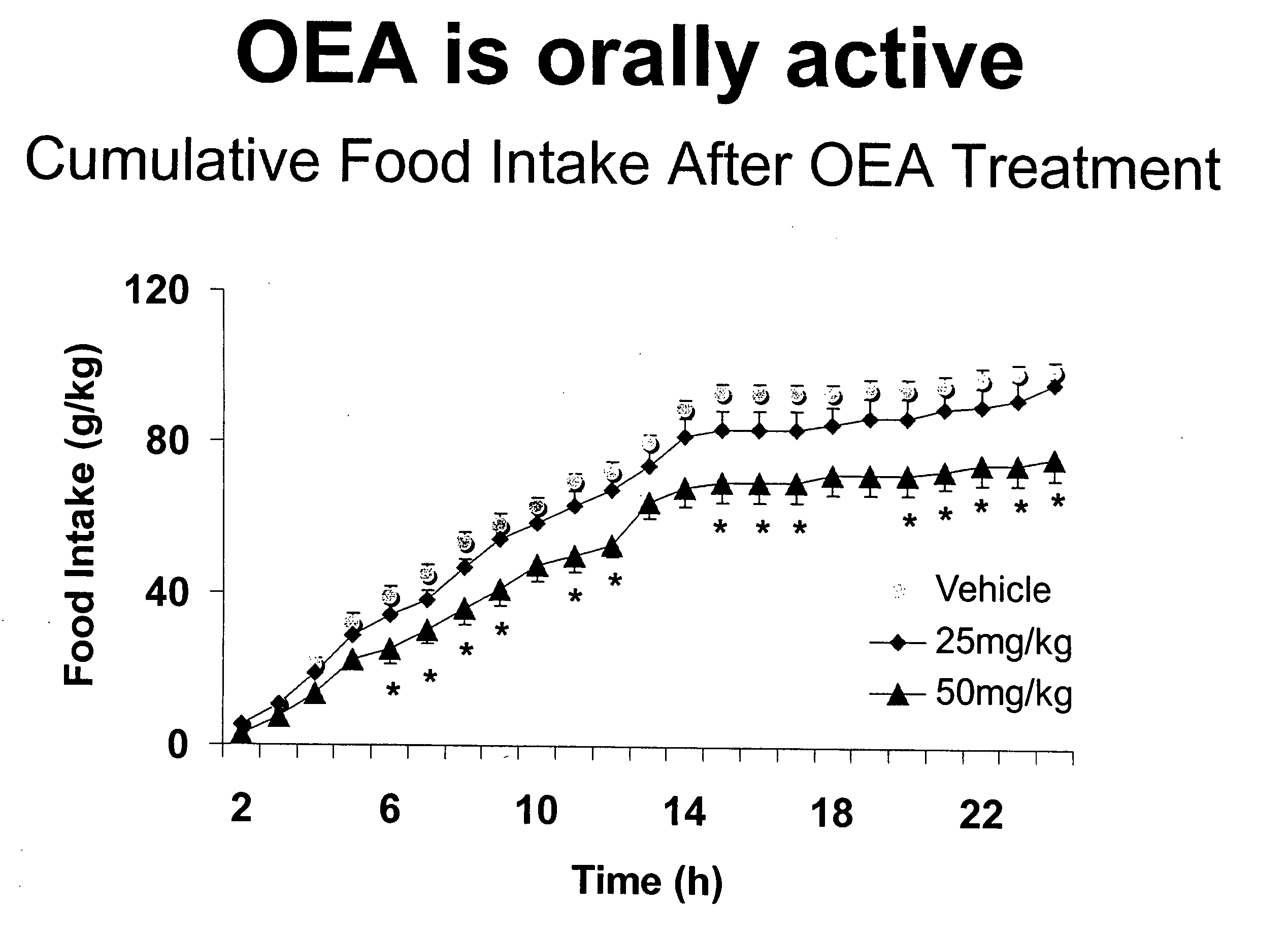
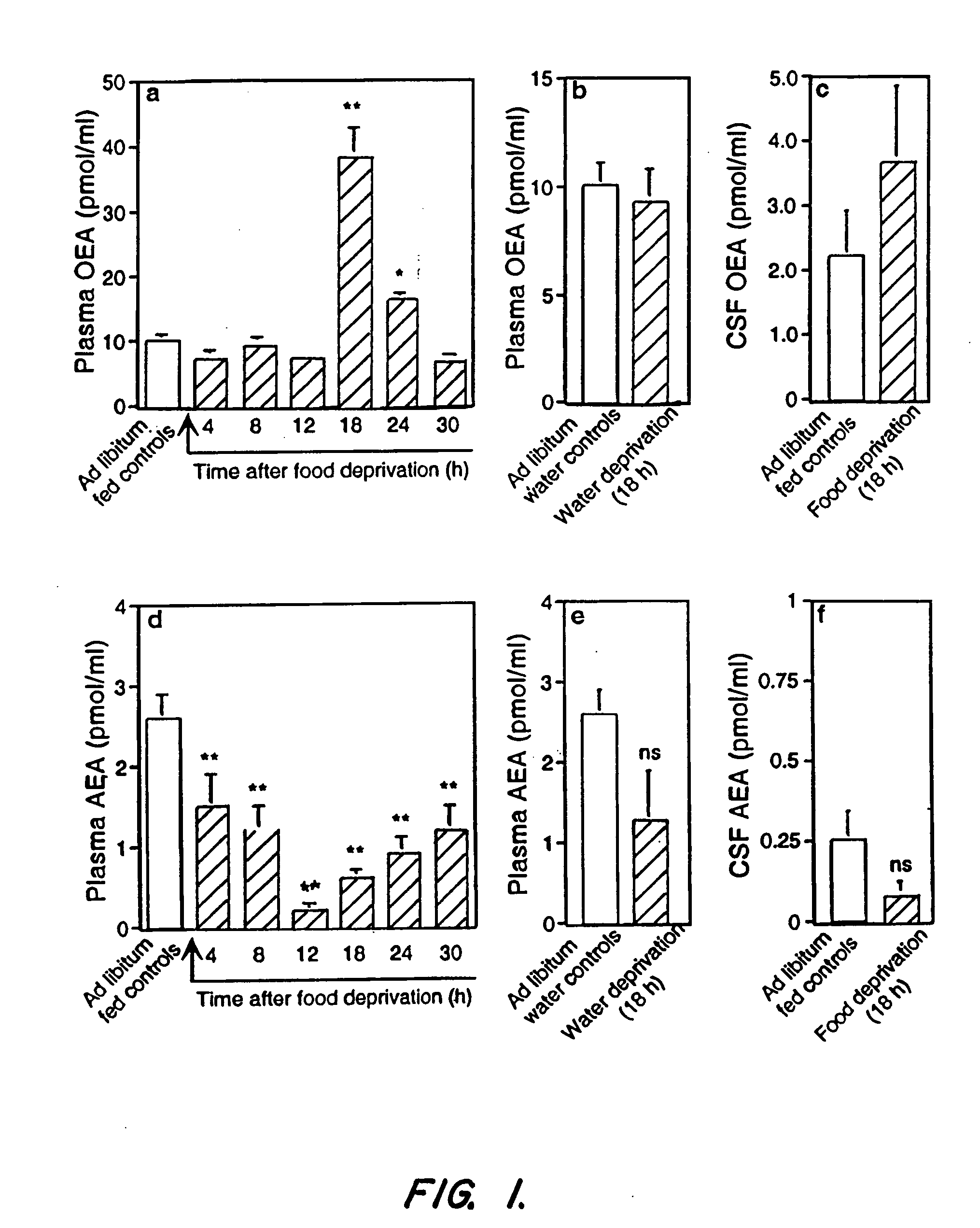
Methods, pharmaceutical, dietary supplement, and nutraceutical compositions, and compounds for reducing body weight, modulating body lipid metabolism, and reducing food intake in mammals are provided. The compounds of the invention include fatty acid ethanolamide compounds, homologues and analogs of which the prototype is the endogenous fatty acid ethanolamide, oleoylethanolamide.
Owner:RGT UNIV OF CALIFORNIA
Method for improving fruit color
A method for promoting the colouring on fruit (apple, pear, peach, grape, etc) features that the 5-aminolevulinic acid (ALA), or levulinic acid (LA)1 or glutamic acid (Glu) is applied to the fruit.
Owner:NANJING AGRICULTURAL UNIVERSITY
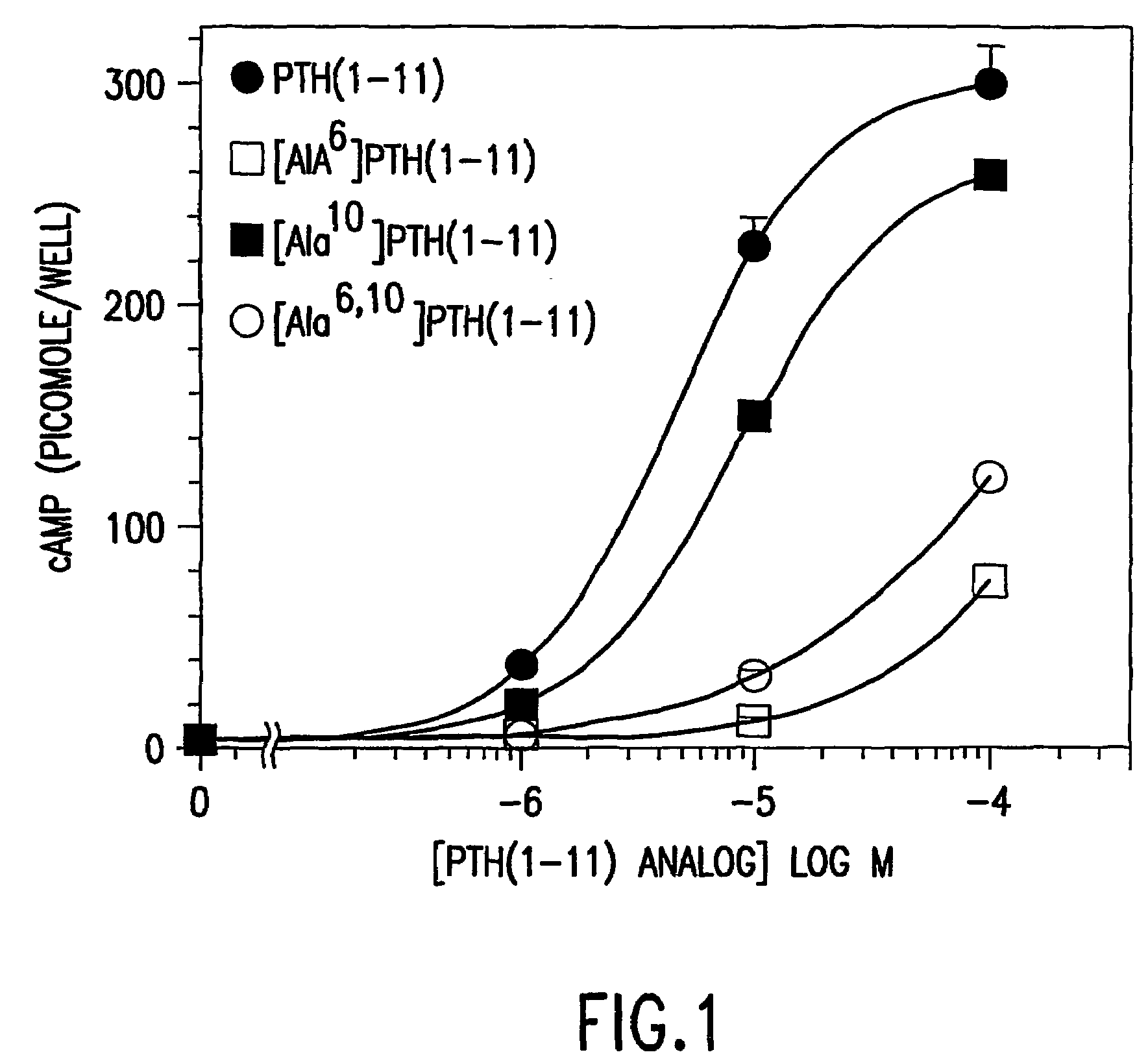
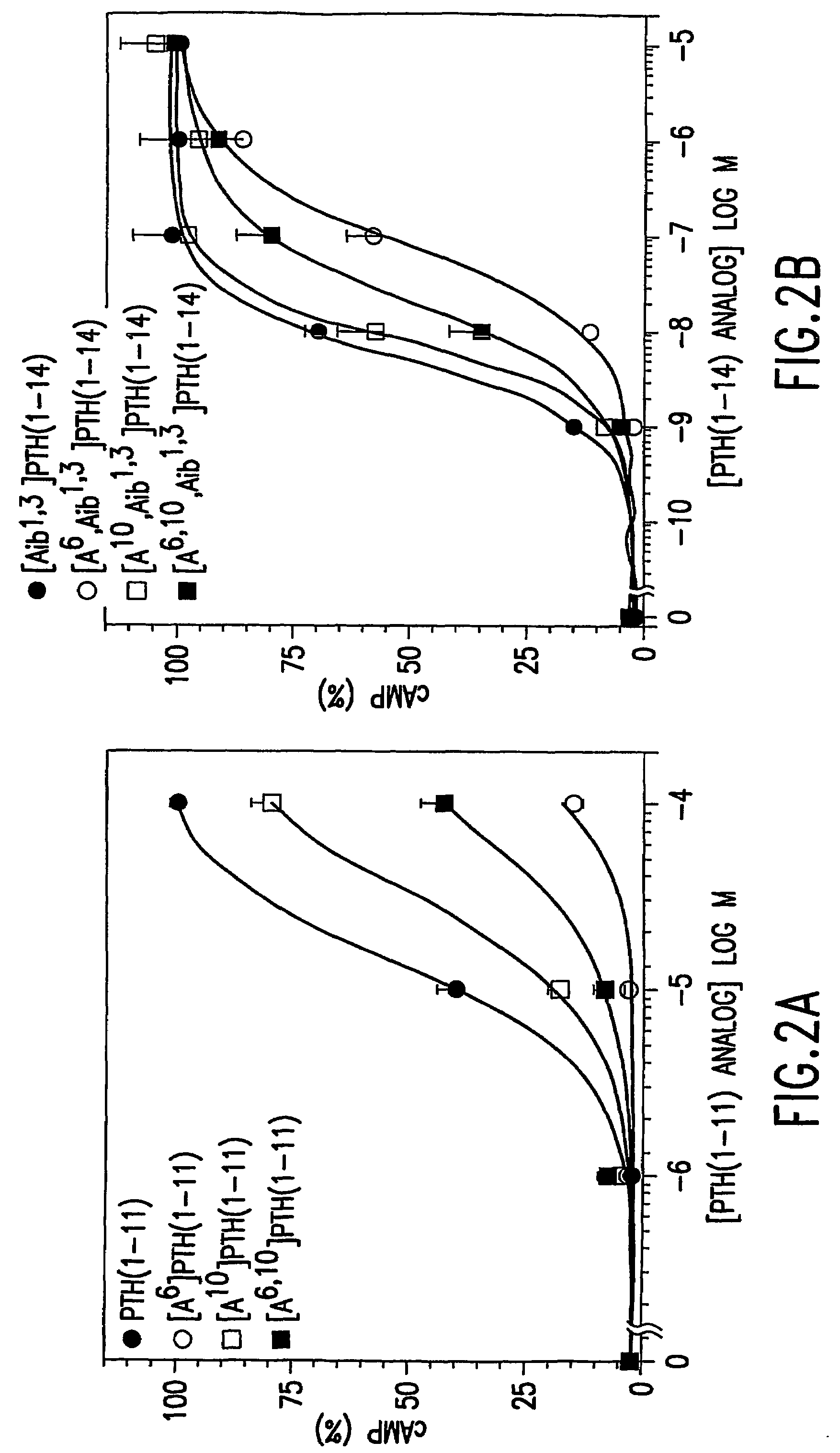
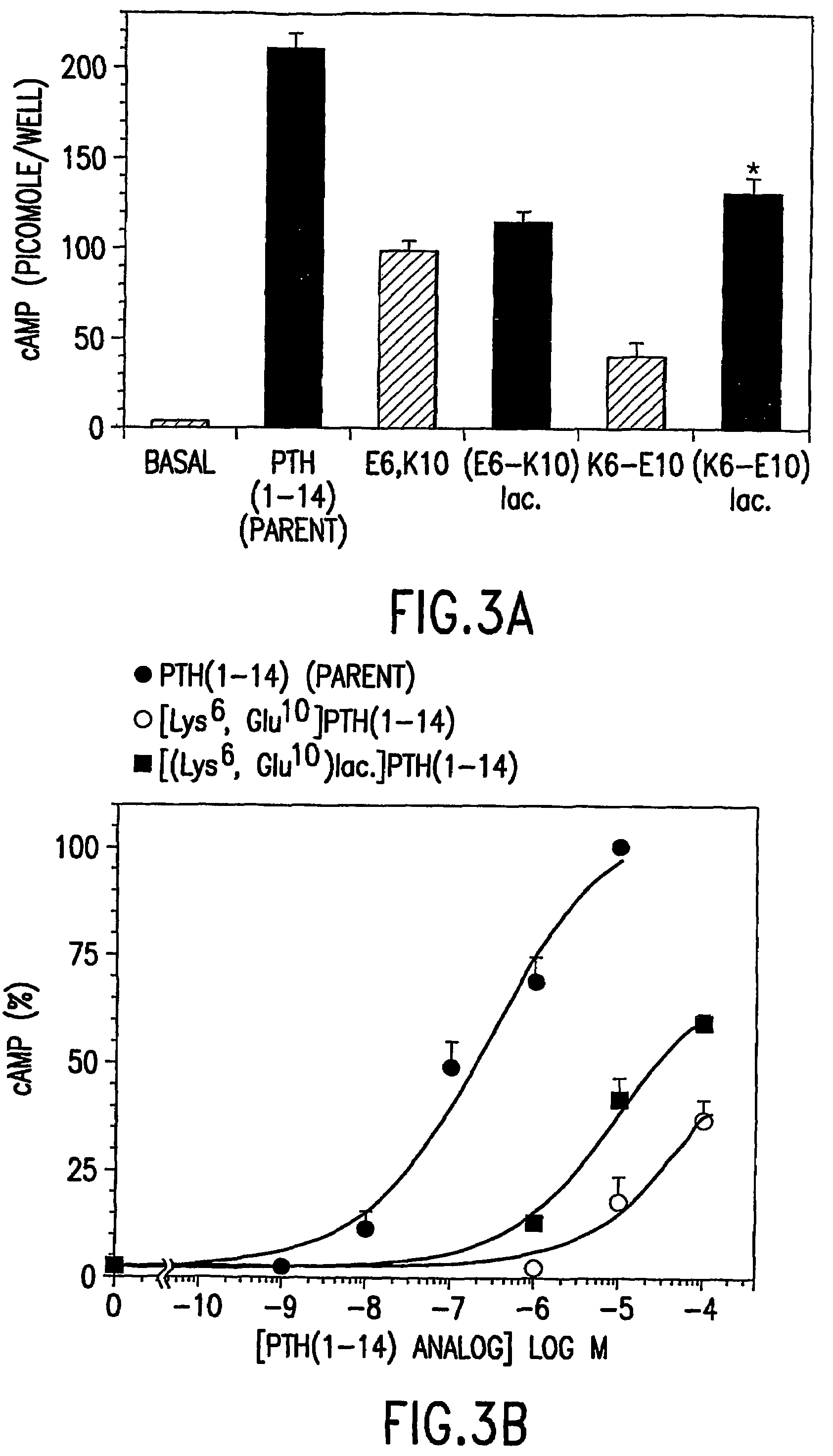
Conformationally constrained parathyroid hormone (PTH) analogs with lactam bridges
Owner:THE GENERAL HOSPITAL CORP
Method of preventing, controlling and ameliorating urinary tract infections and supporting digestive health by using a synergistic cranberry derivative, a d-mannose composition and a proprietary probiotic blend
InactiveUS20110064706A1Preventing, controlling or ameliorating urinary tract infectionsReduced pHBiocideOrganic active ingredientsEscherichia coliDry weight
A method and composition prevents, controls and ameliorates urinary tract infections caused by E. coli by administering a therapeutically effective amount of a human dietary supplement composition comprising a cranberry derivative or proanthocyanidin containing concentrate, D-mannose and a proprietary probiotic blend. The cranberry derivative or proanthocyanidin containing concentrate comprises from about one percent (1.0%) by weight to about 95 percent (95.0%) by weight on a dry weight basis. The composition is formulated for delivering a D-mannose unit dosage between about 0.5 to about 5.0 grams per dose. A proprietary probiotic blend is included to provide 5 billion CFU per dose.
Owner:US NUTRACEUTICALS LLC
Method for obtaining plant hairy roots with high anthocyanin content
ActiveCN103695460AHigh in anthocyaninsVector-based foreign material introductionAngiosperms/flowering plantsPlant tissueBhlh genes
The invention discloses a method for obtaining plant hairy roots with high anthocyanin content, and relates to the technical field of plants. The method comprises the following steps of respectively constructing an MYB gene and a bHLH gene AmDEL for regulating anthocyanin anabolism into a carrier PJAM 1502 and a carrier pK7WG2D to obtain PJAM1502::AmROS1 and pK7WG2D::AmDEL; converting PJAM1502::AmROS1 and pK7WG2D::AmDEL into agrobacterium rhizogenes to obtain the agrobacterium rhizogenes containing plasmids PJAM1502::AmROS1 and pK7WG2D::AmDEL; and inducing out hairy roots with high anthocyanin content after utilizing the converted agrobacterium rhizogenes to infect plant tissue organs for co-culturing and screening-culturing. The method disclosed by the invention simultaneously expresses transcription factor MYB gene and bHLH gene for regulating anthocyanin anabolism to obtain the special plant hairy roots with high anthocyanin content.
Owner:CHINA ACAD OF SCI NORTHWEST HIGHLAND BIOLOGY INST
Preparation method of silk fibre with function of removing formaldehyde
The invention discloses a preparation method of silk fibre with a function of removing formaldehyde, mainly comprising the following steps: (1) dissolving procyanidin with the weight equivalent to 8-12 percent of the silk fibre into water, adding 0.2-0.5 milliliters of glacial acetic acid per each litre of water into the solution and uniformly stirring, adding ferrous sulphate into the solution according to the mass ratio of the ferrous sulphate to the procyanidin of 1 to 1-3 and stirring, adding a sodium bicarbonate solution into the solution to regulate the pH value of the mixing solution to 5-8, increasing the temperature of the mixing solution to 50-60 DEG C, and preserving the heat of the mixing solution for 20-40 minutes; (2) adding 0.5 gram of peregal O and 2-10 grams of salt intoeach litre of the obtained mixing solution and uniformly stirring; (3) adding the silk fibre into the obtained mixing solution and processing for 1-12 hours under the conditions that temperature is 25-90 DEG C and liquor ratio is 1 to 20-40, and then taking the silk fibre out and washing and drying the silk fibre. The obtained silk fibre can enable the removal rate of the formaldehyde contained inindoor air to achieve more than 75 percent.
Owner:ZHEJIANG SCI-TECH UNIV
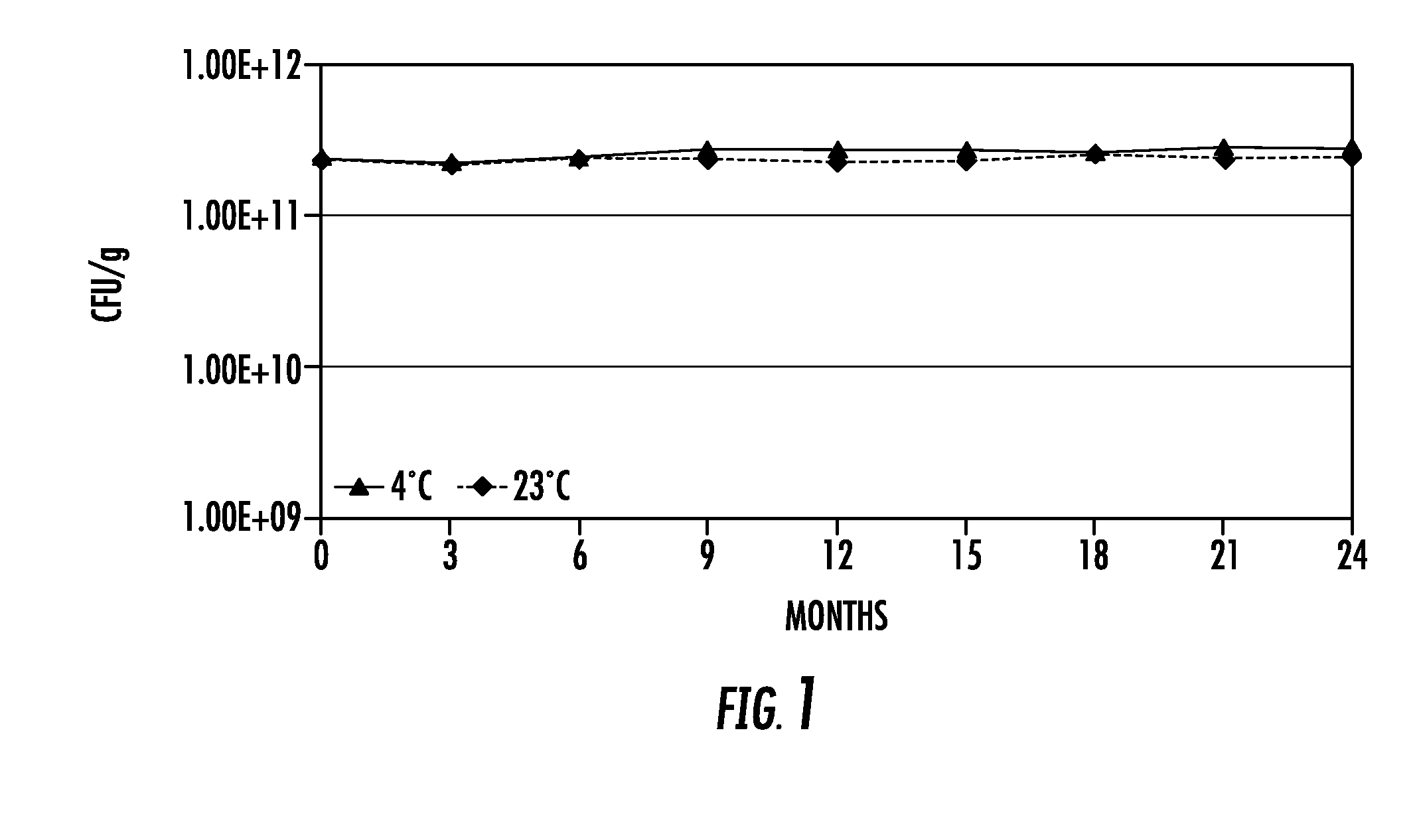
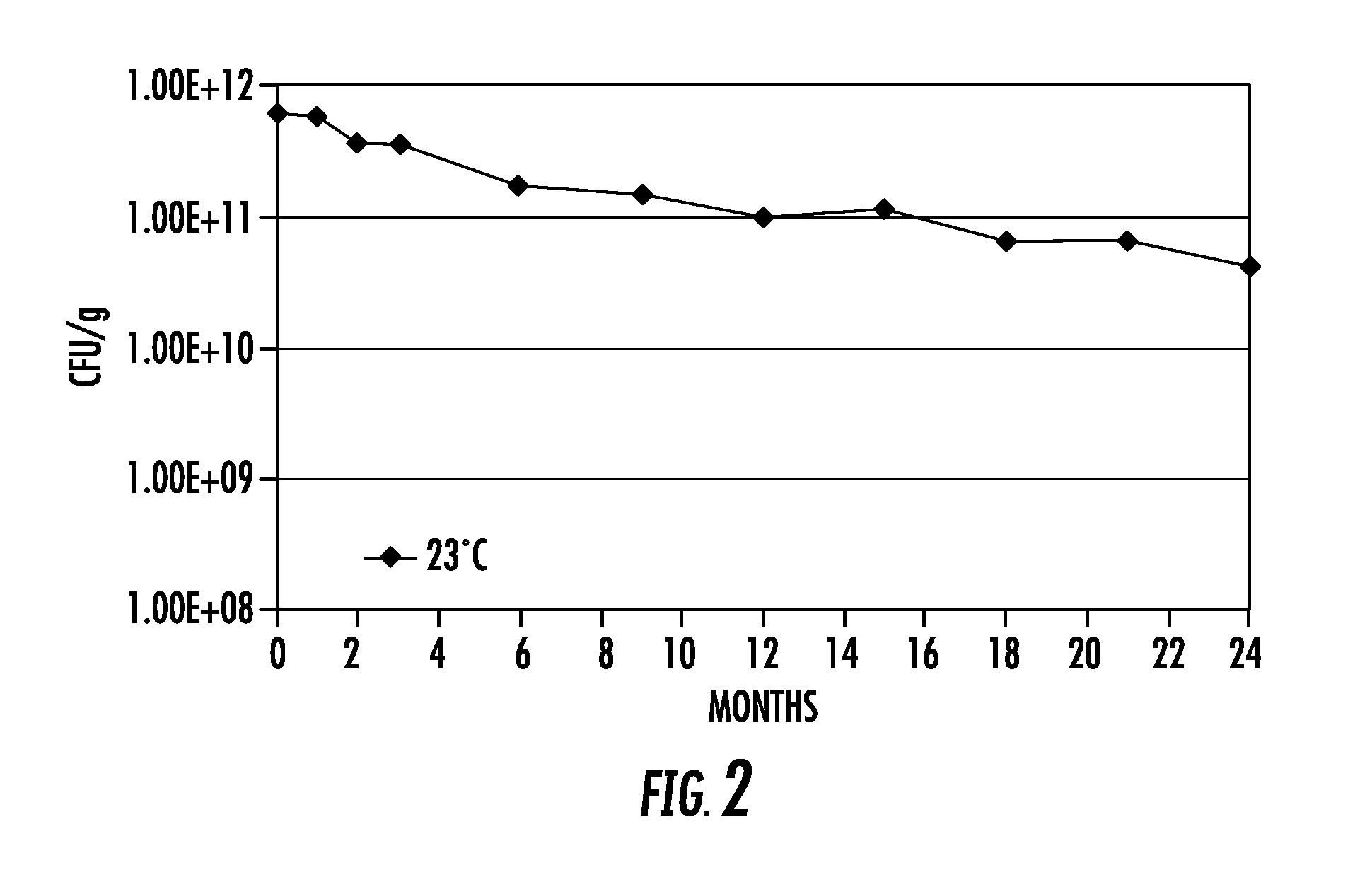
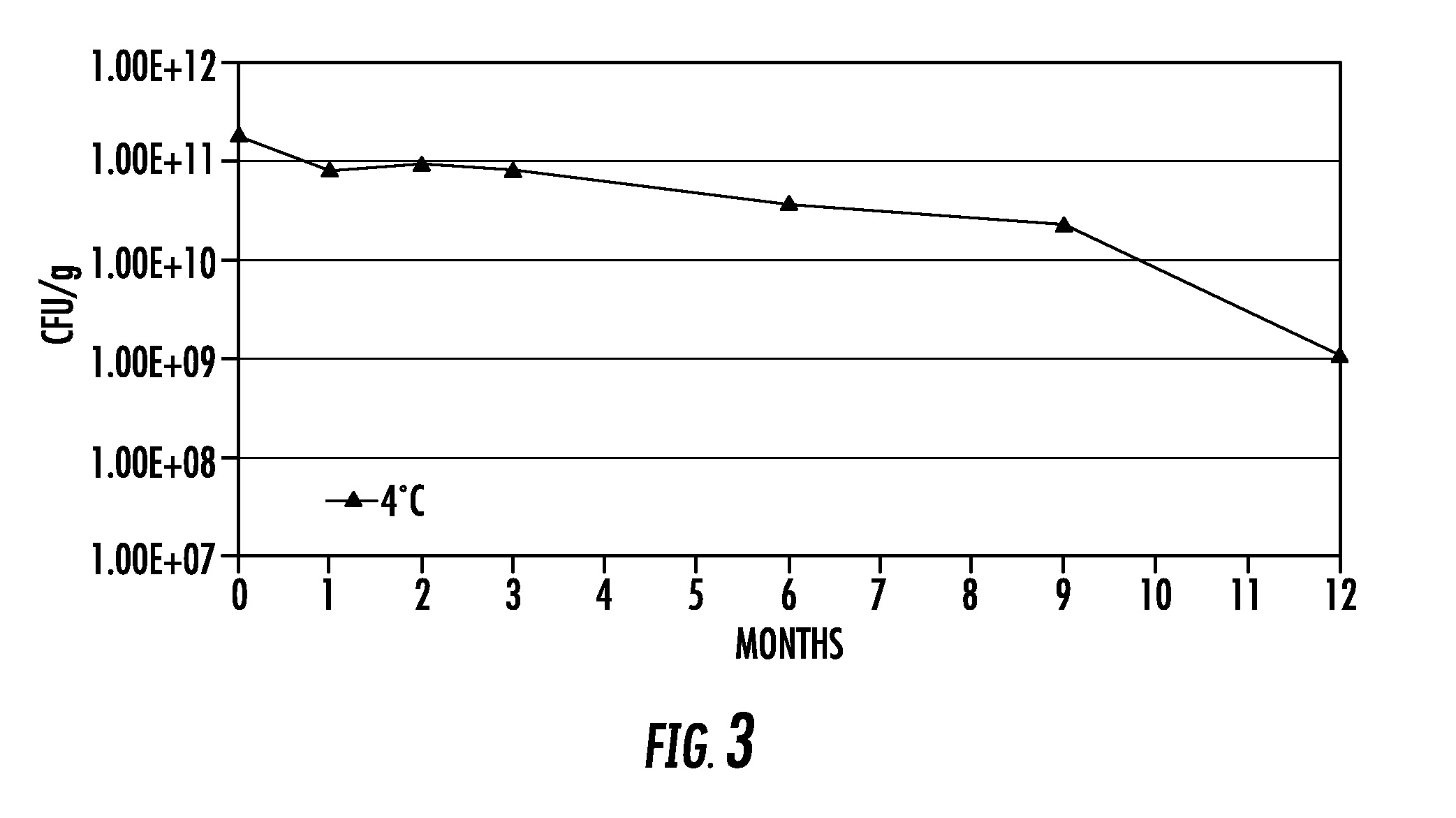
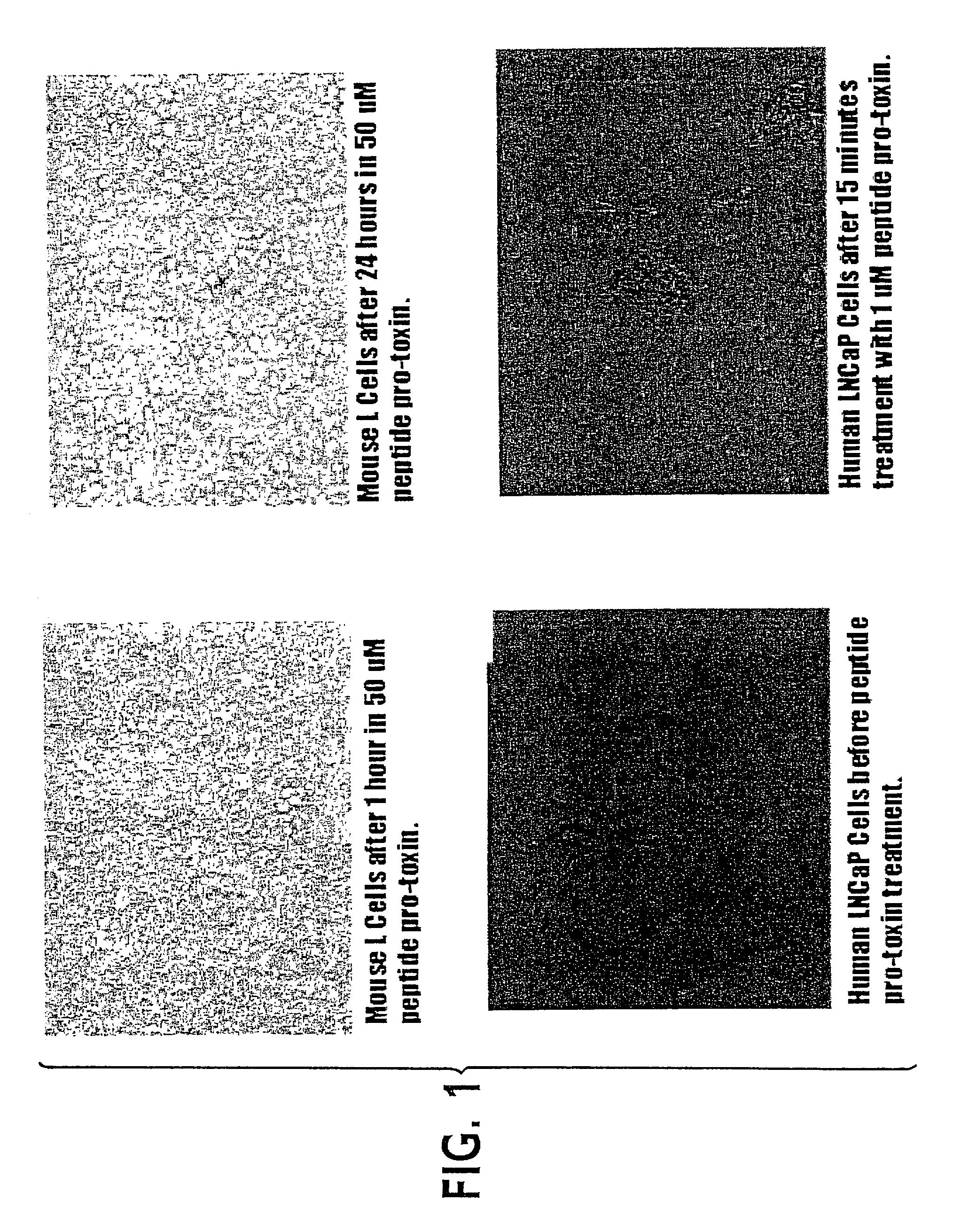
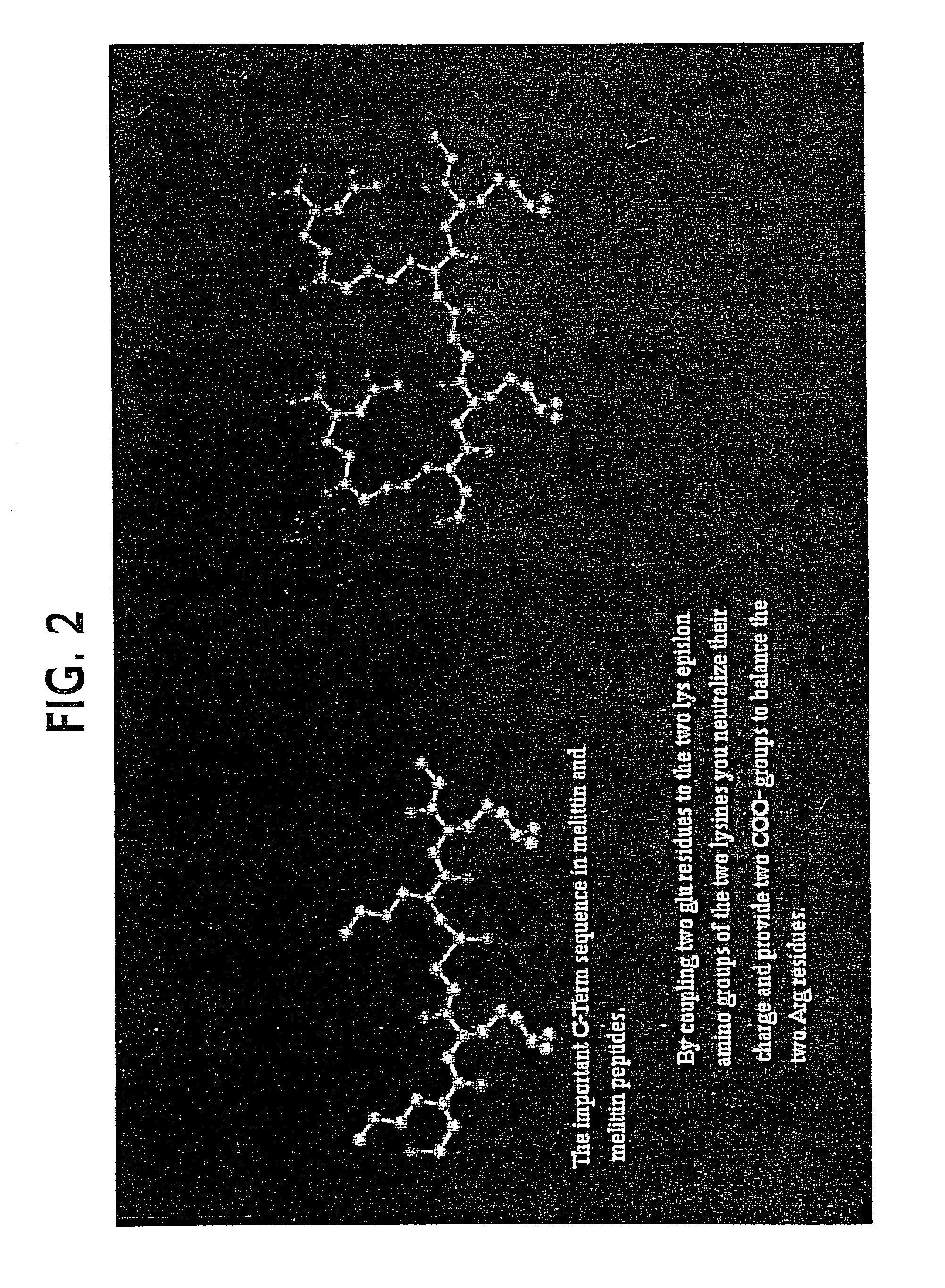
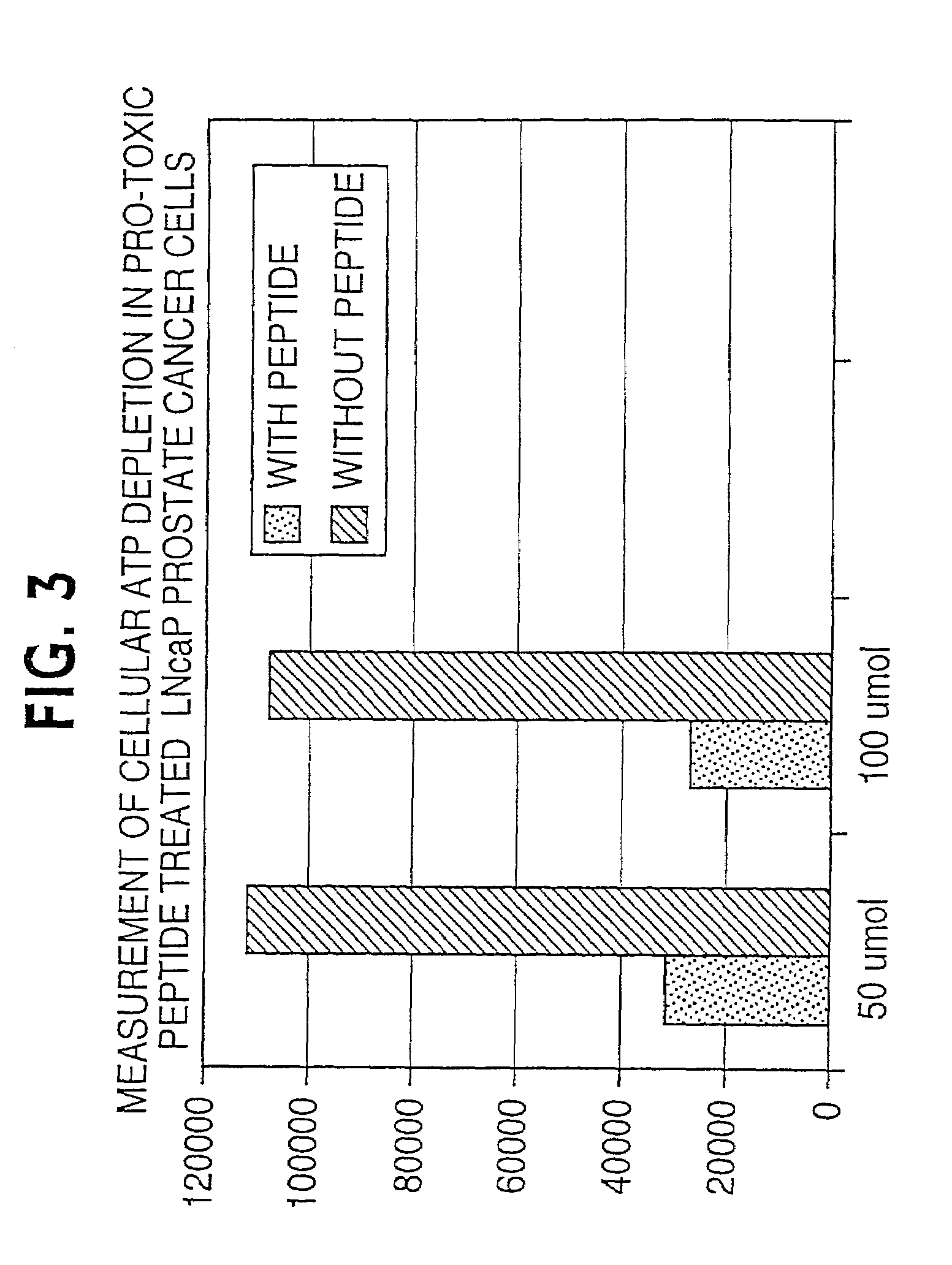
Lytic peptide prodrugs
InactiveUS7456146B2Peptide/protein ingredientsImmunoglobulins against animals/humansLytic peptideProteinase activity
A cytotoxin can be rendered non-toxic by charge neutralizing the amino acids salient to pore assembly and / or sterically inhibiting formation of the peptide's active conformation. In the presence of specific proteases, the inactive peptide or procytotoxin can be activated to assemble into its lytic conformation and selectively destroy a target cell.
Owner:ORBIS HEALTH SOLUTIONS
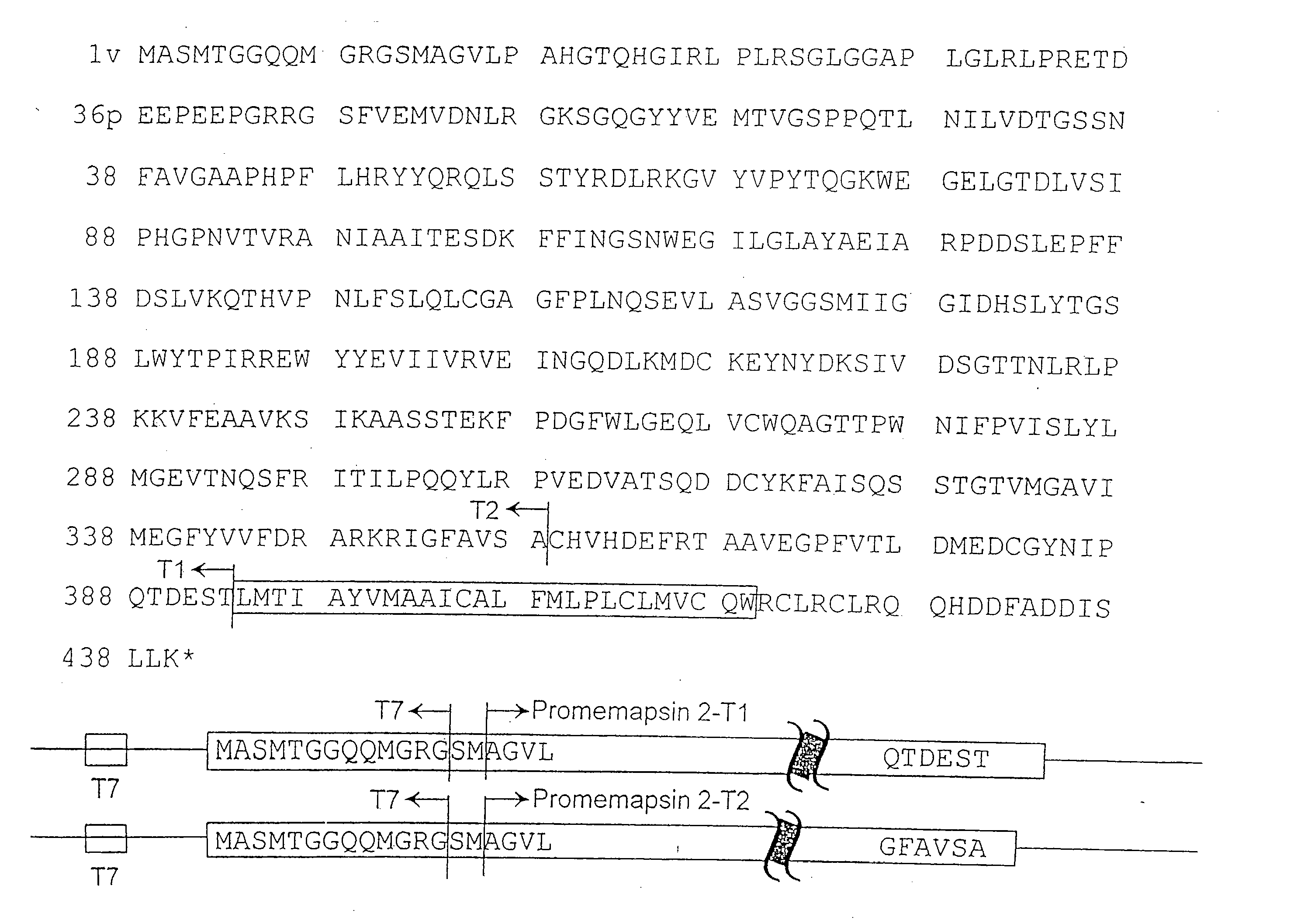
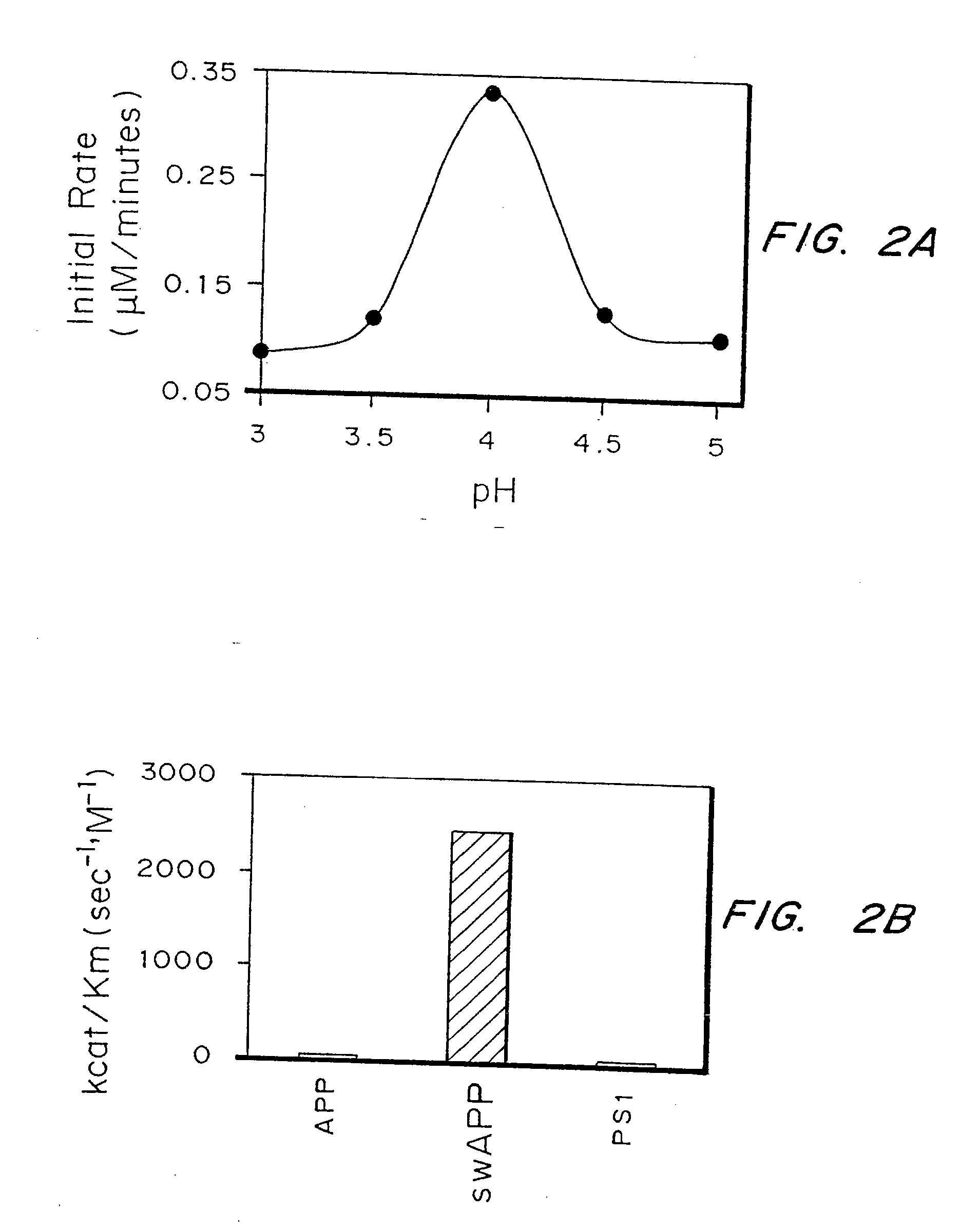
Inhibitors of memapsin 2 and use thereof
InactiveUS20080021196A1Reduce molecular weightGrowth inhibitionNervous disorderHydrolasesDiseaseActive enzyme
Methods for the production of purified, catalytically active, recombinant memapsin 2 have been developed. The substrate and subsite specificity of the catalytically active enzyme have been determined. The substrate and subsite specificity information was used to design substrate analogs of the natural memapsin 2 substrate that can inhibit the function of memapsin 2. The substrate analogs are based on peptide sequences, shown to be related to the natural peptide substrates for memapsin 2. The substrate analogs contain at least one analog of an amide bond which is not capable of being cleaved by memapsin 2. Processes for the synthesis of two substrate analogues including isosteres at the sites of the critical amino acid residues were developed and the substrate analogues, OMR99-1 and OM99-2, were synthesized. OM99-2 is based on an octapeptide Glu-Val-Asn-Leu-Ala-Ala-Glu-Phe (SEQ ID NO:28) with the Leu-Ala peptide bond substituted by a transition-state isostere hydroxyethylene group (FIG. 1). The inhibition constant of OM99-2 is 1.6×10−9 M against recombinant pro-memapsin 2. Crystallography of memapsin 2 bound to this inhibitor was used to determine the three dimensional structure of the protein, as well as the importance of the various residues in binding. This information can be used by those skilled in the art to design new inhibitors, using commercially available software programs and techniques familiar to those in organic chemistry and enzymology, to design new inhibitors to memapsin 2, useful in diagnostics and for the treatment and / or prevention of Alzheimer's disease.
Owner:THE BOARD OF TRUSTEES OF THE UNIV OF ILLINOIS +1
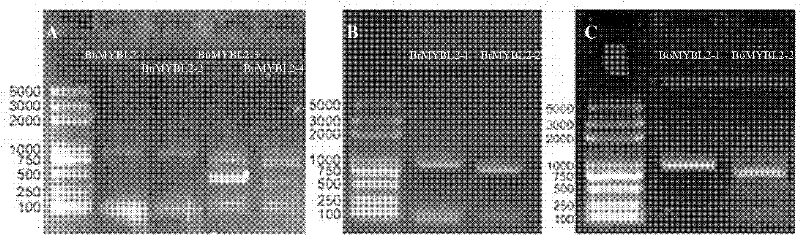
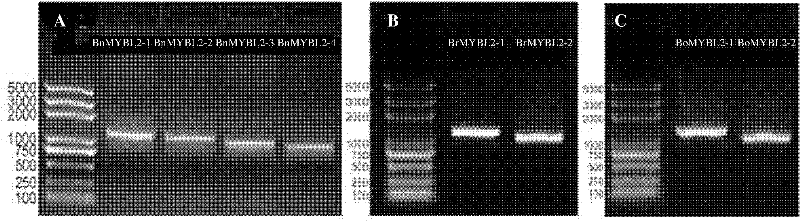
Brassica napus, parent species Chinese cabbage, cabbage MYBL2 (v-myb avian myeloblastosis viral oncogene homolog-like 2) gene family and application thereof
InactiveCN102226193AReduced seed coat pigmentSeed coat thinningFermentationVector-based foreign material introductionBrassicaHigh homology
The invention discloses a Brassica napus, parent species Chinese cabbage, cabbage MYBL2 (v-myb avian myeloblastosis viral oncogene homolog-like 2) gene family. The Chinese cabbage MYBL2 gene family comprises a BrMYBL2-1 gene and a BrMYBL2-2 gene. The cabbage MYBL2 gene family comprises a BoMYBL2-1 gene and a BoMYBL2-2 gene. The Brassica napus MYBL2 gene family comprises a BnMYBL2-1 gene, a BnMYBL2-2 gene, a BnMYBL2-3 gene, a BnMYBL2-4 gene. Eight MYBL2 genes have higher homology. The invention also discloses the application of the above gene families in molecular breeding with characters, such as plant seed coat color, seed coat thickness and the like. After a just overexpression plant carrier is built and double 10 number in the black Brassica napus variety, the Brassica napus is compared with the contrast of the explant of a non-transgenosis, transgenosis plants normally grow and develop, pigments such as Proanthocyanidins in the seed coat are reduced, and meanwhile the seed coat becomes thin to form the transgenosis yellow seed character.
Owner:SOUTHWEST UNIVERSITY
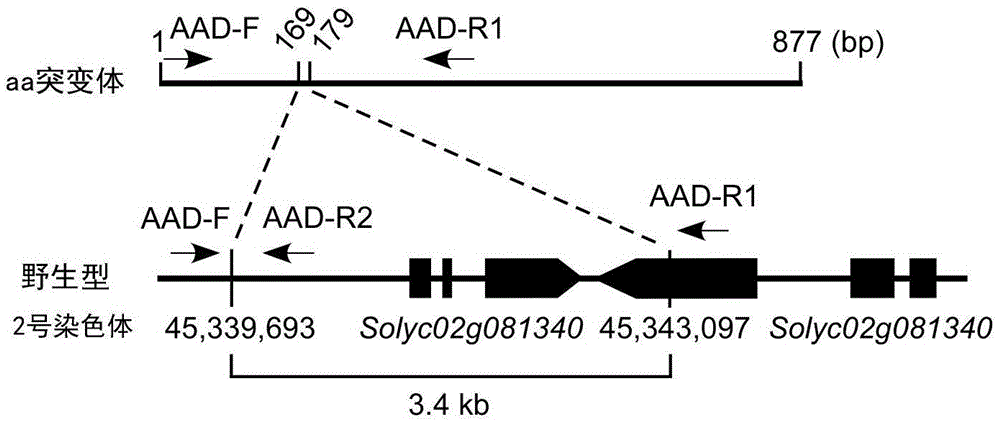
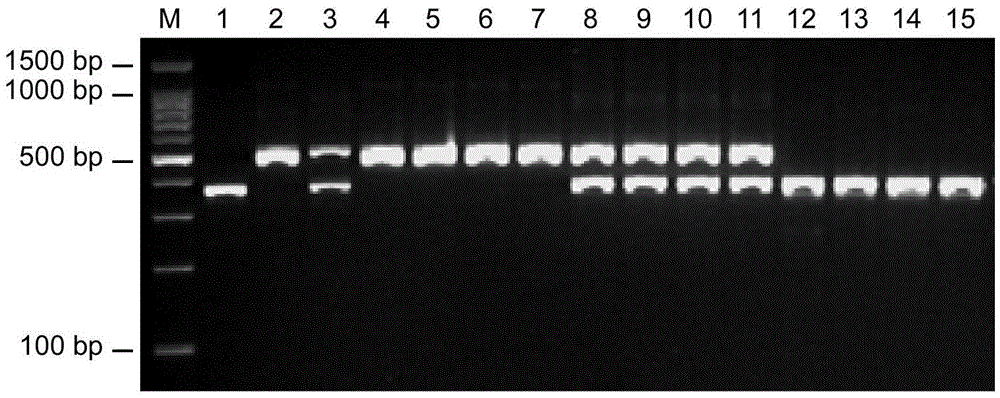
Insertion/deletion (InDel) molecular marker AAD closely-linked to tomato anthocyanin absent character and application thereof
ActiveCN105525016AShorten the breeding cycleImprove breeding efficiencyMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceMolecular breeding
The invention relates to the field of molecular breeding, and in particular to an insertion / deletion (InDel) molecular marker AAD closely-linked to the tomato anthocyanin absent character and application thereof. The physical position of the InDel molecular marker closely-linked to the tomato anthocyanin absent character is No. 45339693 base to No. 45343097 base on chromosome 2 of tomato. The invention can replace a traditional method of selecting a green plant to ensure that a breeding material of tomato contains an anthocyanin absence (AA) mutation site. An AA site hybrid plant in the breeding material with normal phenotype is assist-selected by mean of the molecular marker, the breeding period can be shortened, and the breeding efficiency can be improved.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
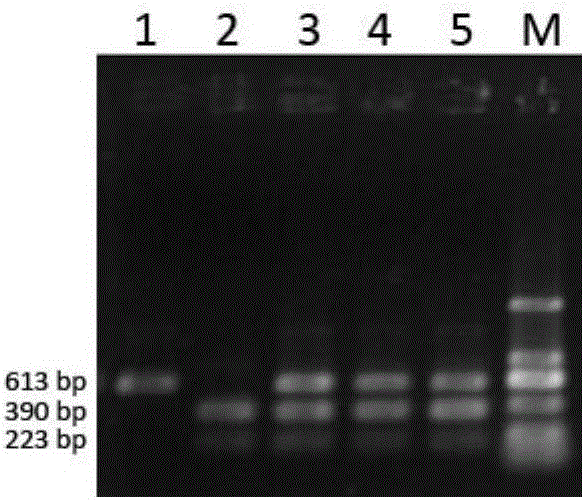
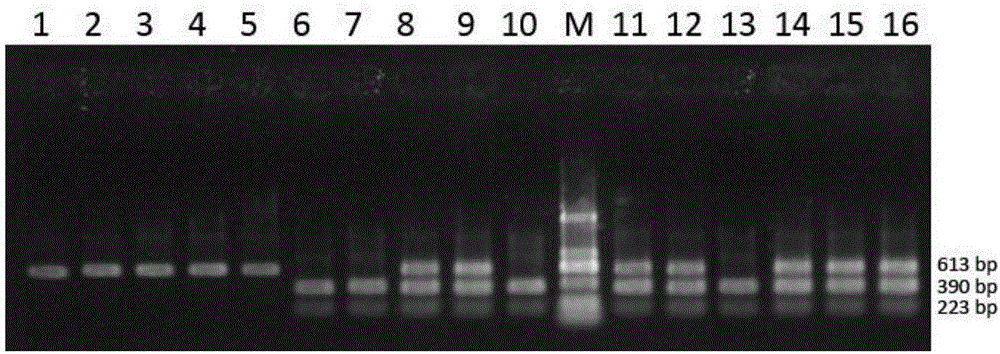
CAPS (cleaved amplified polymorphic sequence) molecular marking method for identifying solanum lycopersicum with purple black striped pericarp and application
ActiveCN106755357AGuaranteed accuracyAvoid the transfer stepMicrobiological testing/measurementGenomic DNAGel electrophoresis
The invention discloses a CAPS (cleaved amplified polymorphic sequence) molecular marking method for identifying solanum lycopersicum with purple black striped pericarp and an application. The sequence of a molecular marker carried by the solanum lycopersicum with purple black striped pericarp is a nucleotide base sequence shown as SEQ ID NO.1, PCR amplification is performed with genomic DNA of the solanum lycopersicum with purple black striped pericarp as a template, nucleotide base sequences shown as SEQ ID NO.3-4 are amplification primers, amplification and cleavage are performed, a specific band different from other solanum lycopersicum pericarp colors can be obtained after cleavage, and the molecular marking method is applied to breed improvement and breeding of the solanum lycopersicum. Compared with the prior art, the CAPS molecular marking method has the advantages that the polymorphism of a cleaved fragment is observed through PCR amplification, cleavage and gel electrophoresis separation, the step of membrane transfer in the traditional RFLP (restricted fragment length polymorphism) analysis is omitted, the operation is simplified, the precision of the RFLP technology is kept, the polymorphism detecting probability is higher, the solanum lycopersicum with purple black striped pericarp is obtained rapidly and applied to breed improvement and assisted breeding, and the method has important theoretical and practical meanings for culturing new varieties of solanum lycopersicum containing rich anthocyanidin.
Owner:JIANGSU ACADEMY OF AGRICULTURAL SCIENCES
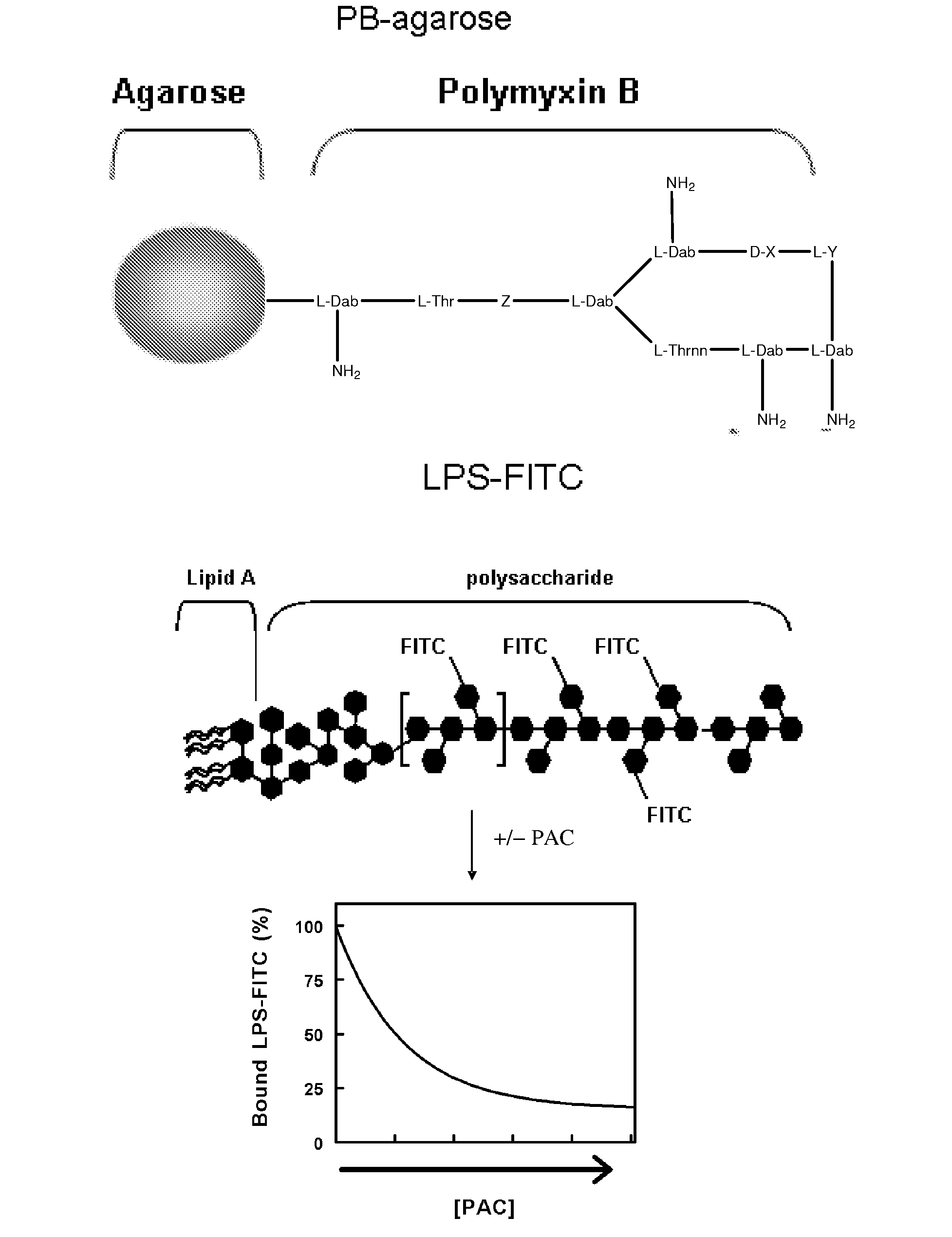
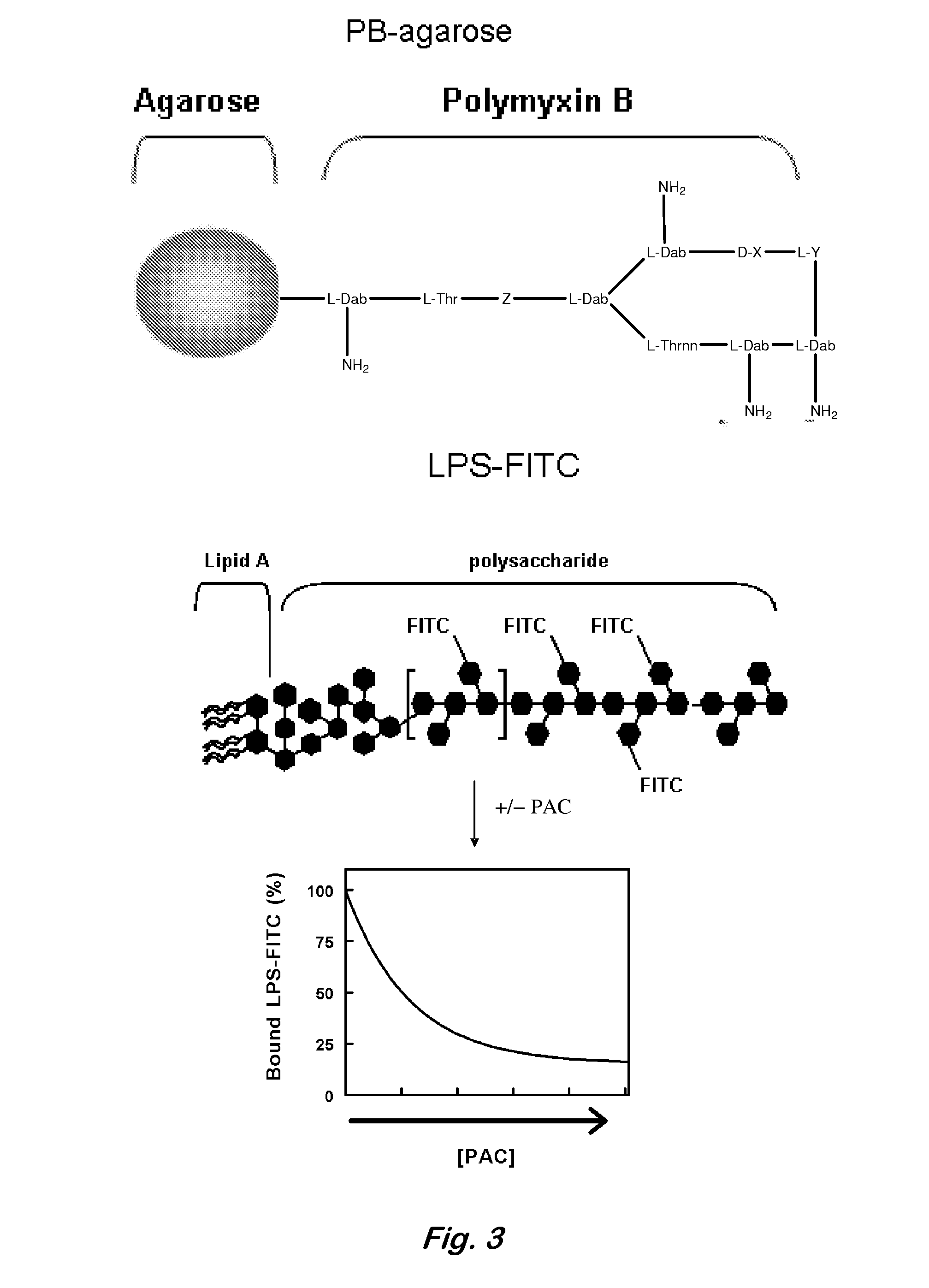
Applications of the binding interaction of proanthocyanidins with bacteria and bacterial components
InactiveUS20080064050A1Antibacterial agentsOrganic active ingredientsBacteroidesBacterial composition
A composition having: a proanthocyanidin; and a macromolecule, an assembly of macromolecules, a semi-solid, or a solid surface to which the proanthocyanidini is immobilized.
Owner:THE UNITED STATES OF AMERICA AS REPRESENTED BY THE SECRETARY OF THE NAVY
Gene PpRd for regulating fruit flesh cyanin synthesis and application thereof
ActiveCN104531724AHigh feasibilityShorten breeding timeMicrobiological testing/measurementPlant peptidesEngineered geneticGene expression level
The invention discloses a gene PpRd for regulating fruit flesh cyanin synthesis and application thereof, relating to plant gene engineering and biotechnology. The gene PpRd nucleic acid sequence is disclosed as SEQ ID NO.1, and the protein sequence coded by the gene PpRd is disclosed as SEQ ID NO.2. The application of the PpRd gene is implemented by (1) designing a molecular marker WPS23 according to the sequence of the PpRd gene, and applying the molecular marker WPS23 in marker auxiliary screening in breeding; and (2) application of expression protein sequence and gene functions: detecting and screening cyanidin content related phenotype according to the expression level of the gene, or enhancing or lowering the gene expression level by genetic transformation to obtain the expected flesh color character. The invention obtains a key gene for cyanin accumulation. When the PpRd gene is used for marker auxiliary selection to instruct hybridization breeding of peaches, the screening can be performed in advance, thereby saving the breeding time and enhancing the breeding efficiency.
Owner:WUHAN BOTANICAL GARDEN CHINESE ACAD OF SCI
Medicinal composition for treating infection with drug-resistant staphylococcus aureus
InactiveUS20060235076A1High antibacterial activityIncrease infectionAntibacterial agentsBiocideDiseaseDisinfectant
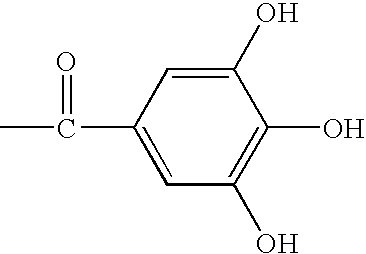
The invention relates to a therapy of an infection with a drug resistant bacterium wherein a characteristic of multivalent phenol derivatives and / or extracts from “Tara” which enhance the activity of β-lactam antibiotics on the drug resistant bacteria is utilized. Specifically, the invention relates to an enhancer of β-lactam antibiotics comprising a multivalent phenol derivative or its pharmaceutically acceptable salt, a pharmaceutical composition comprising a β-lactam antibiotic and a multivalent phenol derivative and / or extract from “Tara” for the therapy of an infection with a drug resistant bacterium, a disinfectant for the same, and a functional food comprising a multivalent phenol derivative and / or extract from “Tara”.
Owner:MICROBIOTECH
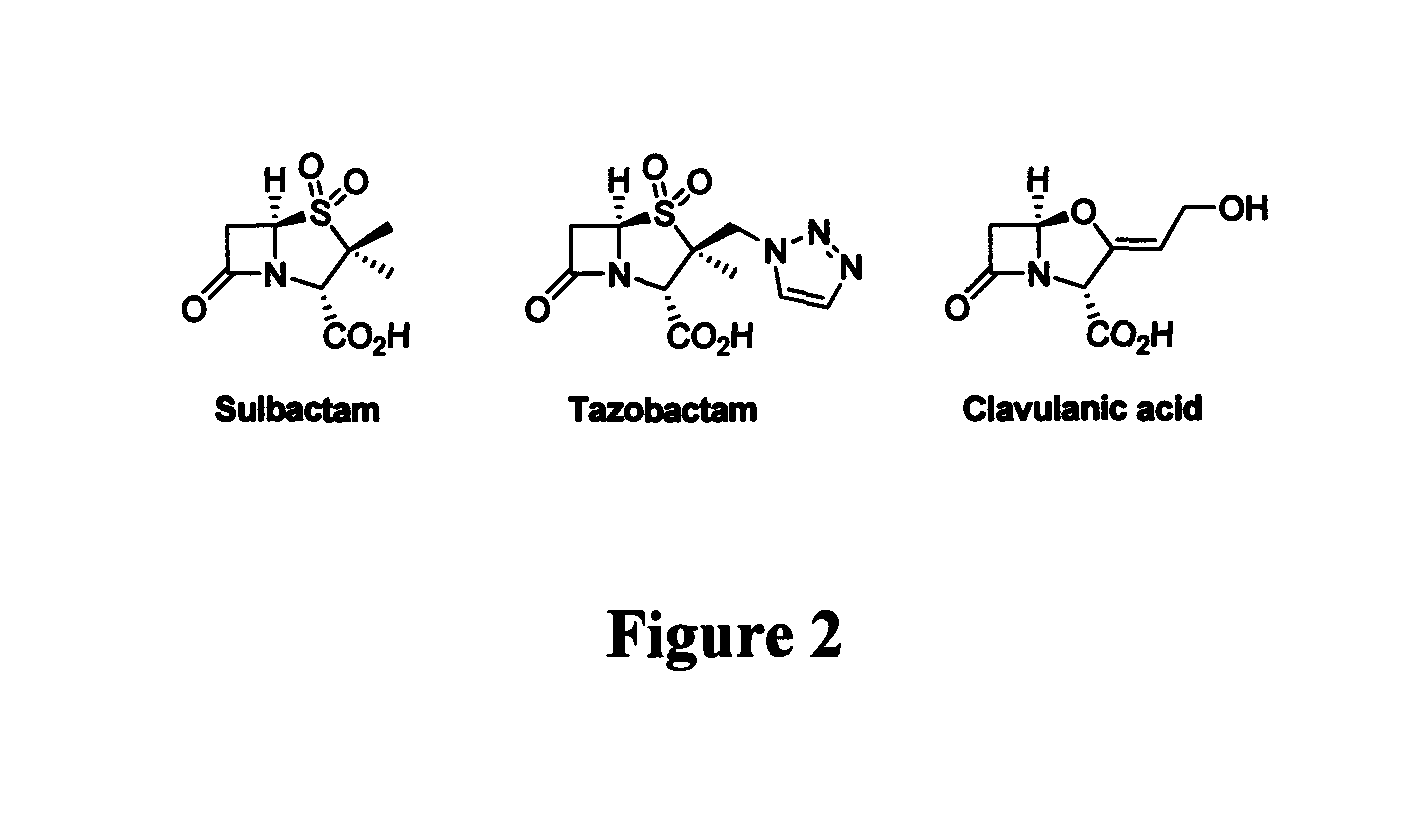
Bate-lactamase inhibitors
The present invention relates to broad spectrum β-lactamase inhibitors. More particularly, the invention relates to inhibitors of Class B metallo (MBL) and Class D (OXA) β-lactamases. A method of treating a bacterial infection is provided, wherein the method comprises administering to a mammalian patient in need of such treatment a compound of formula (I)whereinR1 is selected fromR2 is selected fromwith certain provisos as herein defined;in combination with a pharmaceutically acceptable β-lactam antibiotic in an amount which is effective for treating the bacterial infection.
Owner:DMITRIENKO GARY I +4
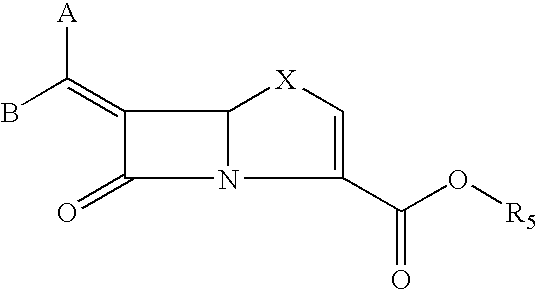
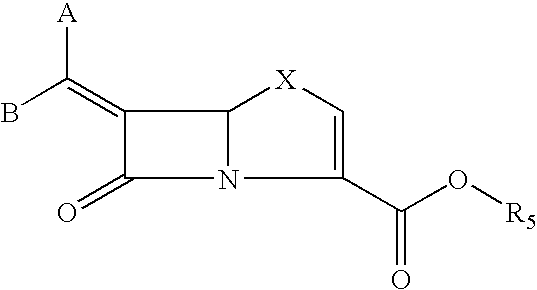
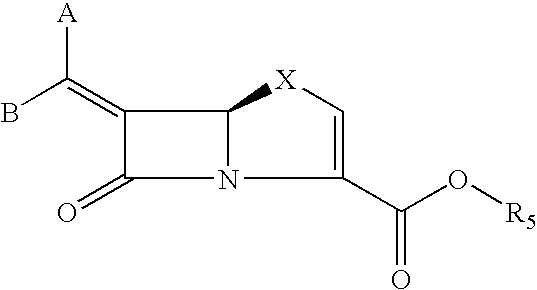
Tricyclic 6-alkylidene-penems as class-D beta-lactamases inhibitors
This invention relates to certain tricyclic 6-alkylidene penems which act as a inhibitor of class-D enzymes. β-Lactamases hydrolyze β-lactam antibiotics, and as such serve as the primary cause of bacterial resistance. The compounds of the present invention when combined with β-lactam antibiotics will provide an effective treatment against life threatening bacterial infections. In accordance with the present invention there are provided compounds of formula I which are useful for treatment of bacterial infections having class-D enzymes associated therewith: wherein: One of A and B denotes hydrogen and the other an optionally substituted fused tricyclic heteroaryl group; and X is S or O.
Owner:WYETH
Composition with anti-oxidation and metabolism-promotion functions
InactiveCN101011424AMature cultivationMature extraction technologyHydroxy compound active ingredientsMetabolism disorderAlcoholProanthocyanidin
The invention relates to a compound with oxidation resistance and metabolism acceleration functions. The inventive compound is formed by Cordyceps sinensis fungus, cyanin and veralk alcohol. The compound uses the Cordyceps sinensis fungus and grape extractive as main material, with low cost and wide sources. The invention has simple production.
Owner:蔡洪亮
Topical compositions and methods for influencing electromagnetic radiation on cutaneous extracellular matrix protein production
Disclosed herein is a composition that is suitable for influencing electromagnetic radiation on cutaneous extracellular matrix protein production. The composition contains (a) a proanthrocyanin, a polyphenol or a proanthrocyanin / polyphenol-containing plant extract, absorbed or adsorbed onto an inert carrier, where component (a) is responsive to UV, IR or visible light; (b) a photo-activatable amino acid or amino acids blend; and (c) a dermatologically-acceptable vehicle, wherein component (a) is present in an amount of from about 15 wt % to about 35 wt % based on the total amount of component (a) and component (b), said component (b) is present in an amount of from about 65 wt % to about 85 wt % based on the total amount of component (a) and component (b), and said component (c) is present in an amount of from about 90 wt % to about 99.95 wt % based on the total weight of the composition.
Owner:MARY ROSE SCI LLC
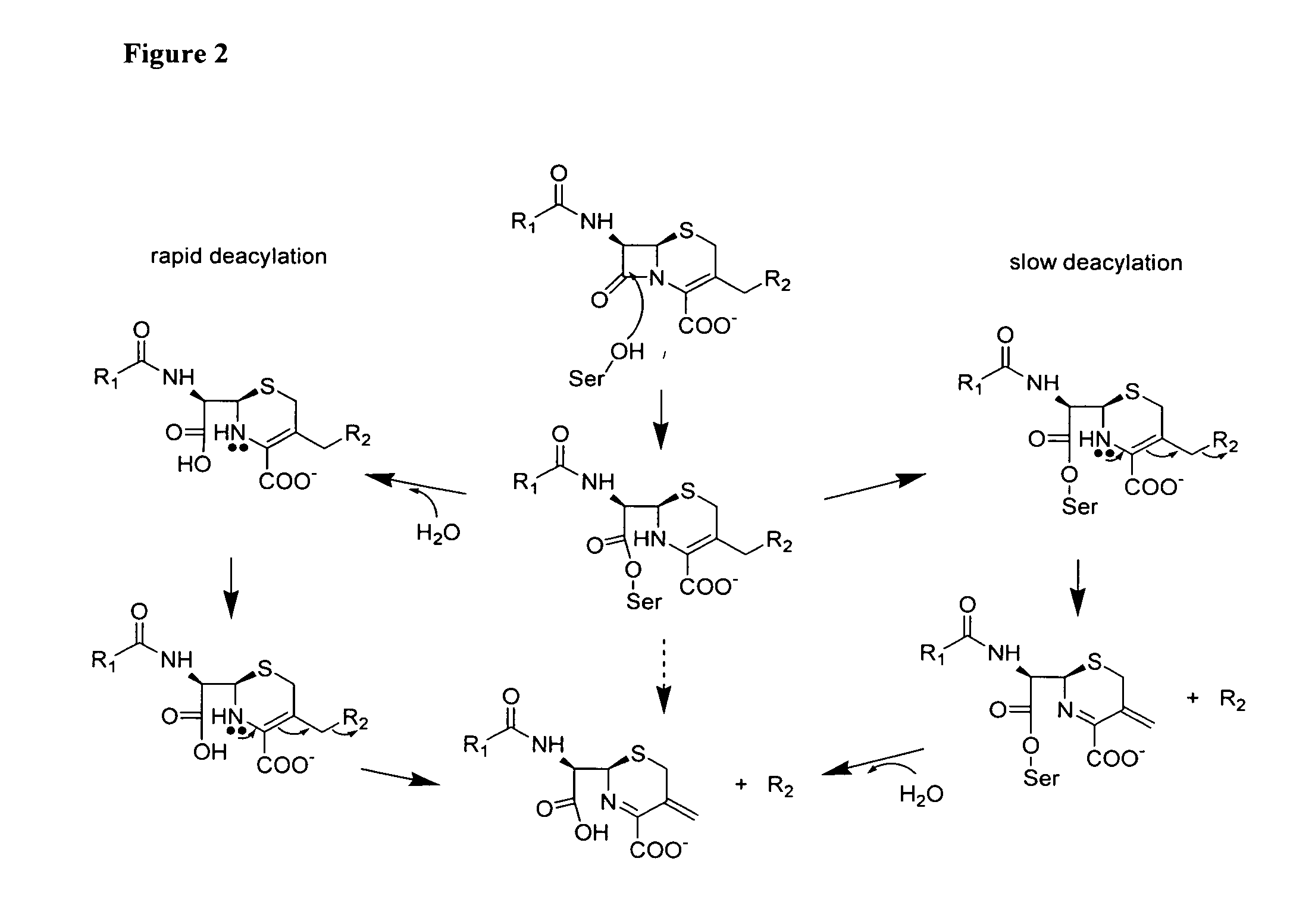
Sterically-awkward beta-lactamase inhibitors
InactiveUS20050245498A1Good effectRemarkable effectOrganic chemistryHeterocyclic compound active ingredientsΒ lactamasesAntibiotic Y
Sterically-awkward 6-β-substituted P-lactam compounds as inhibitors of β-lactamase activity, such compounds as can be used in conjunction with one or more β-lactam antibiotics in a system for treatment of a β-lactam resistant bacterial infection.
Owner:NORTHWESTERN UNIV
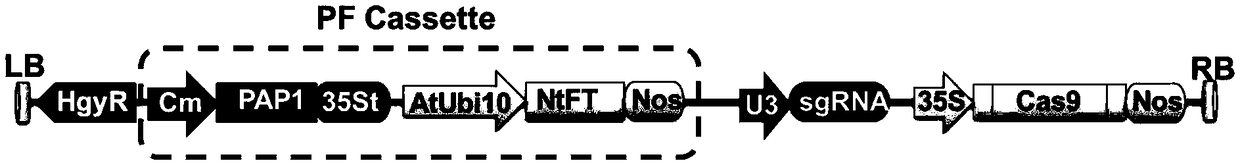
Recombinant vector for rapidly obtaining non-GMO genome editing plants and method for using same
ActiveCN109234310AShorten flowering timeShorten the timeVector-based foreign material introductionAngiosperms/flowering plantsGenome editingNucleotide
Provided are a recombinant vector for rapidly obtaining non-GMO genome editing plants and a method for using the same. The recombinant vector comprises an expression element containing PAP1 and NtFT;The original vector is a vector of CRISPR / Cas9 for plant gene editing containing sgRNA gene, Cas9 gene and screening tag gene; The NtFT expression element produces proteins that promote early flowering and the PAP1 expression element produces proteins that promote anthocyanin biosynthesis in plants. The invention can greatly shorten the time for obtaining the gene editing mutation offspring, and screens the offspring plants by plant color, thus not only retaining the offspring of the target gene mutation, but also ensuring that the mutation offspring does not contain the transgenic fragment, and simultaneously can significantly improve the gene editing efficiency. The invention can be used for preparing the gene editing mutation offspring, and can greatly shorten the time for obtaining thegene editing mutation offspring. The method of the invention is simple, fast, cheap and efficient.
Owner:YUNNAN ACAD OF TOBACCO AGRI SCI
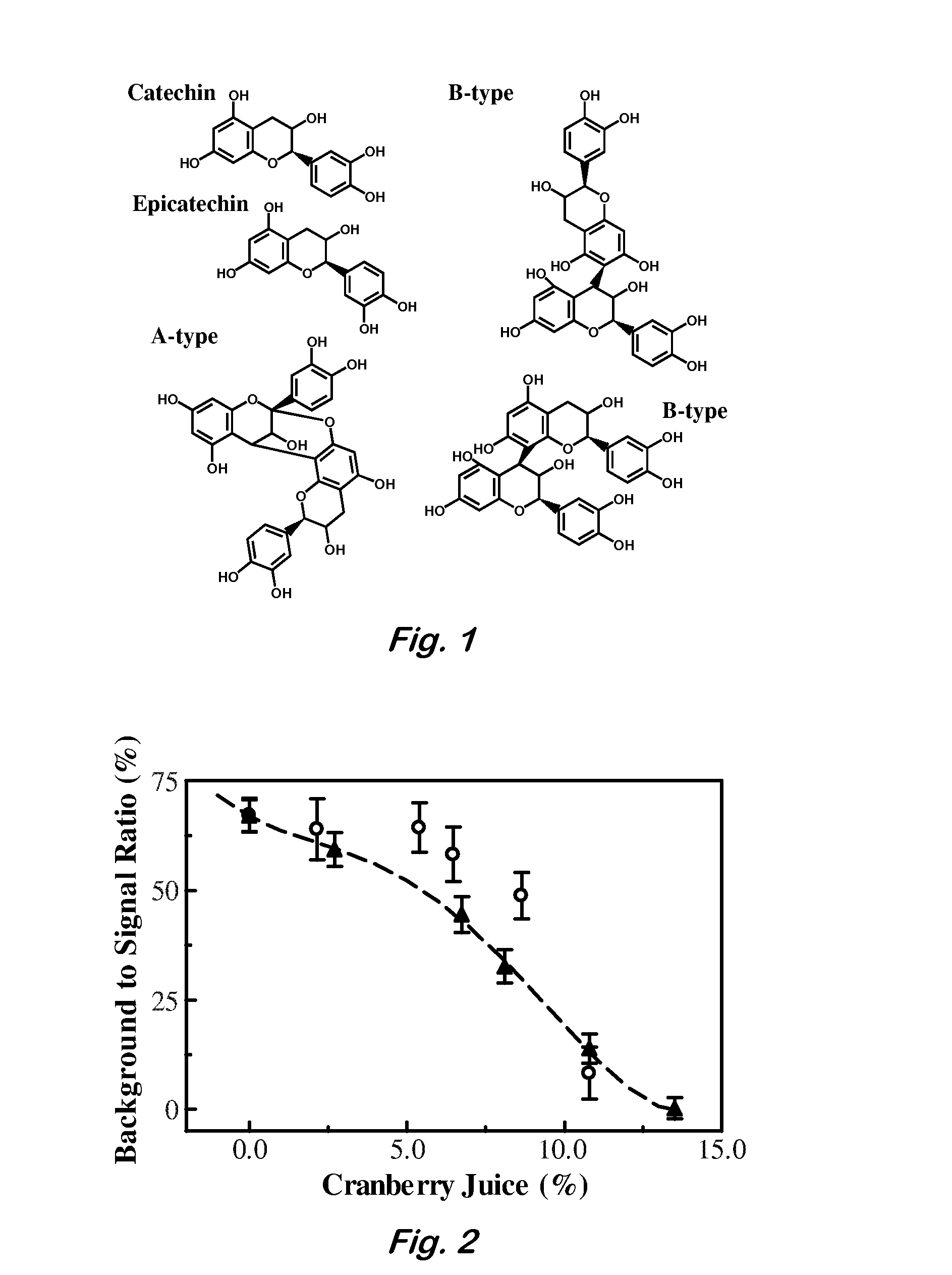
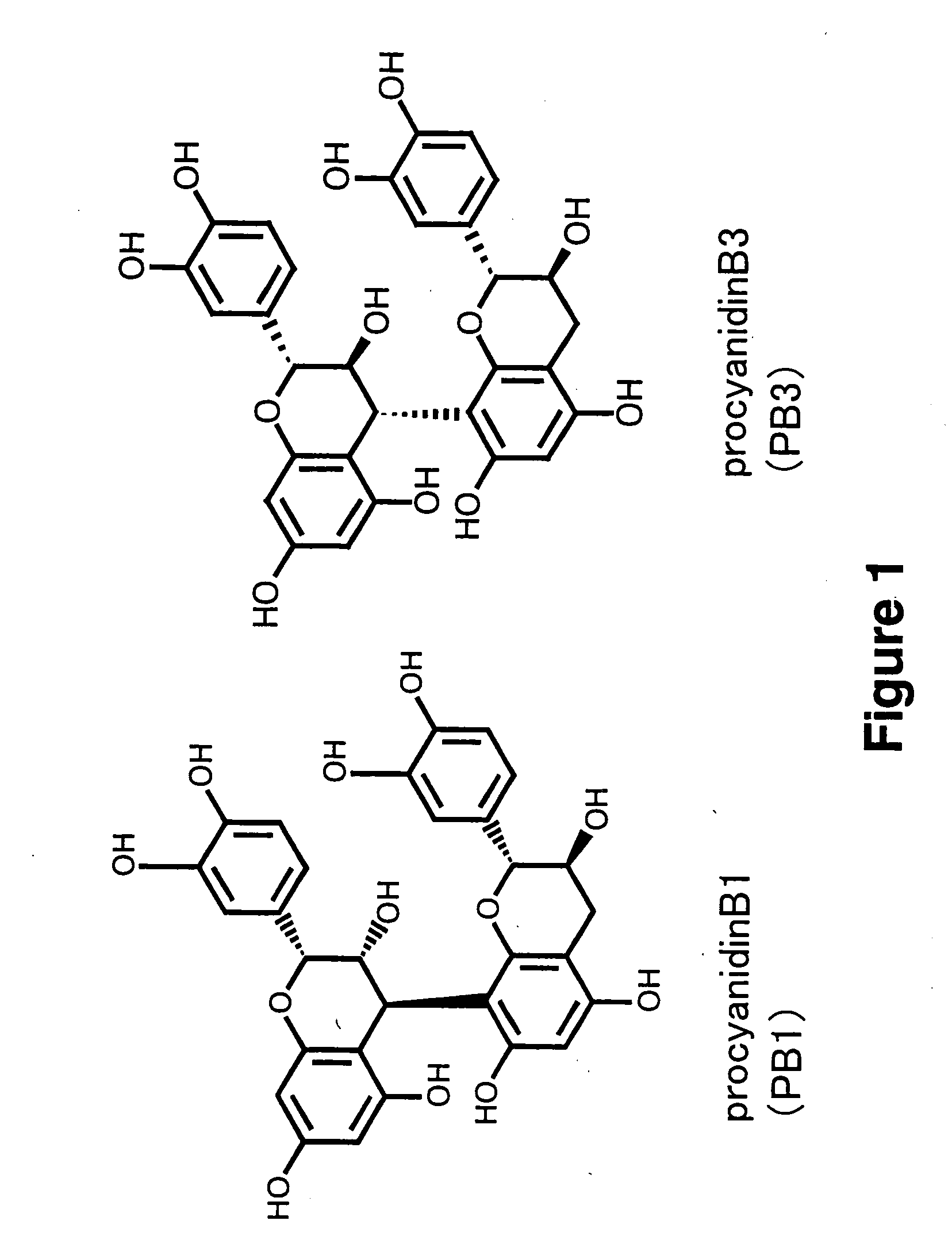
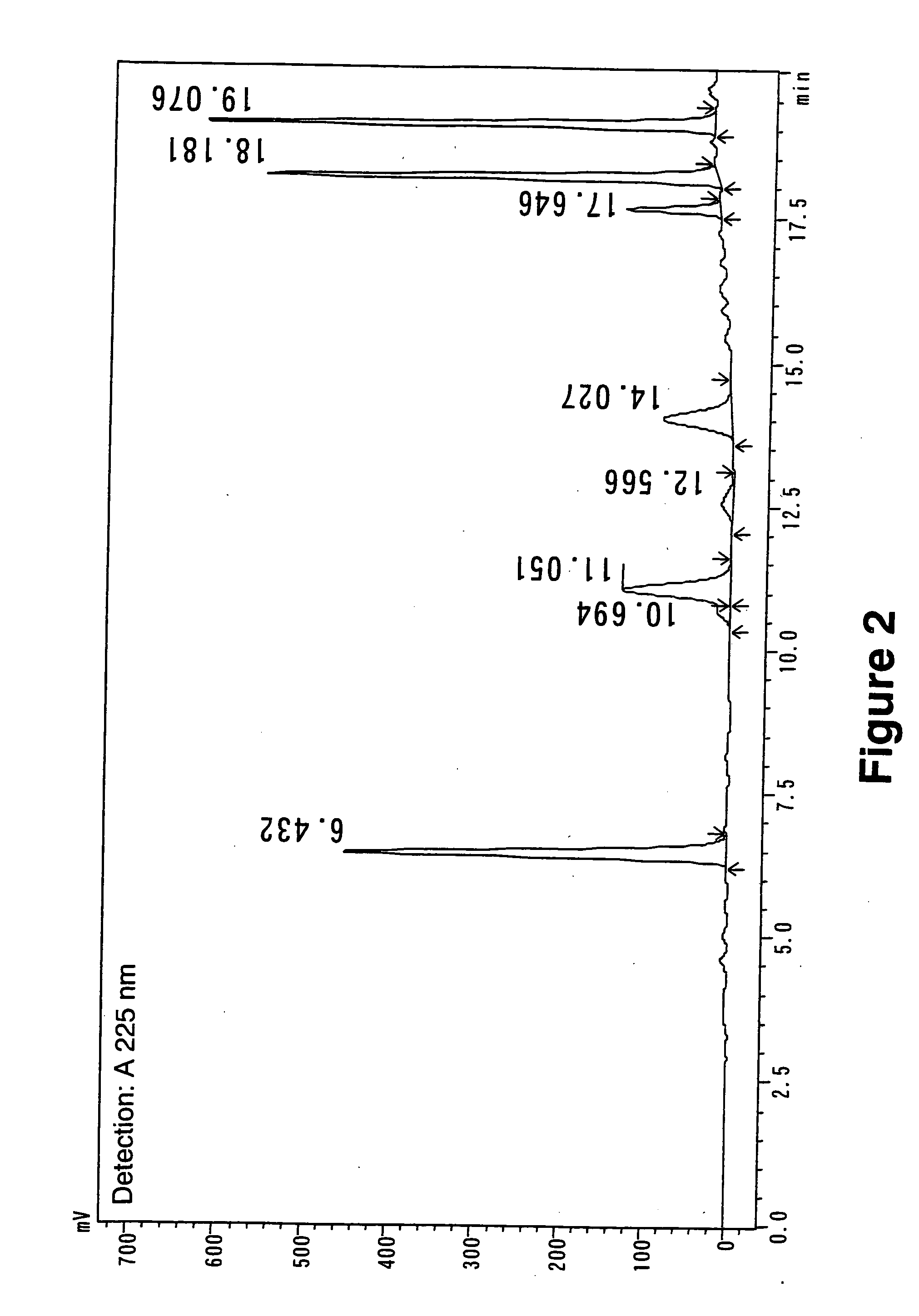
Method for Procyanidin Analysis
InactiveUS20090215186A1Quantitative precisionComponent separationTesting beveragesChromatography columnHplc mass spectrometry
The present invention relates to a novel method for quantifying procyanidin and flavan-3-ols contained in natural products, foods and drinks, pharmaceuticals and / or cosmetics. The present invention provides a method for quantifying procyanidin (a generic name for a mixture of catechin n-mer; n≧1) contained in test samples such as natural products, foods and drinks, pharmaceuticals and / or cosmetics, which comprises a) pretreating a test sample using a column to remove contaminants which affect the analysis of procyanidin and flavan-3-ol; and b) effecting high performance liquid chromatography (HPLC) to separate and quantify procyanidin and flavan-3-ols in the solution treated to remove contaminants during the above pretreatment step a).
Owner:SUNTORY HLDG LTD
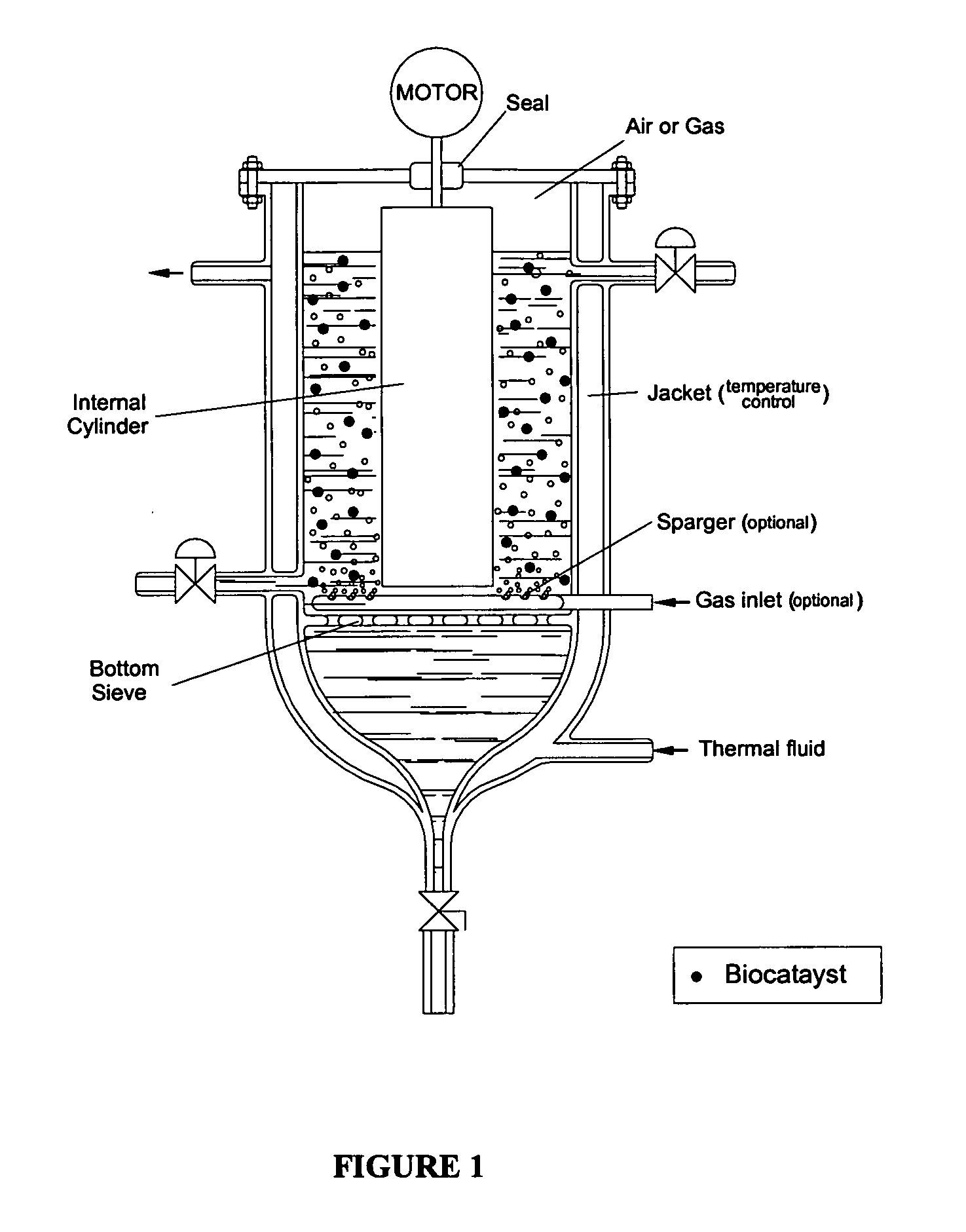
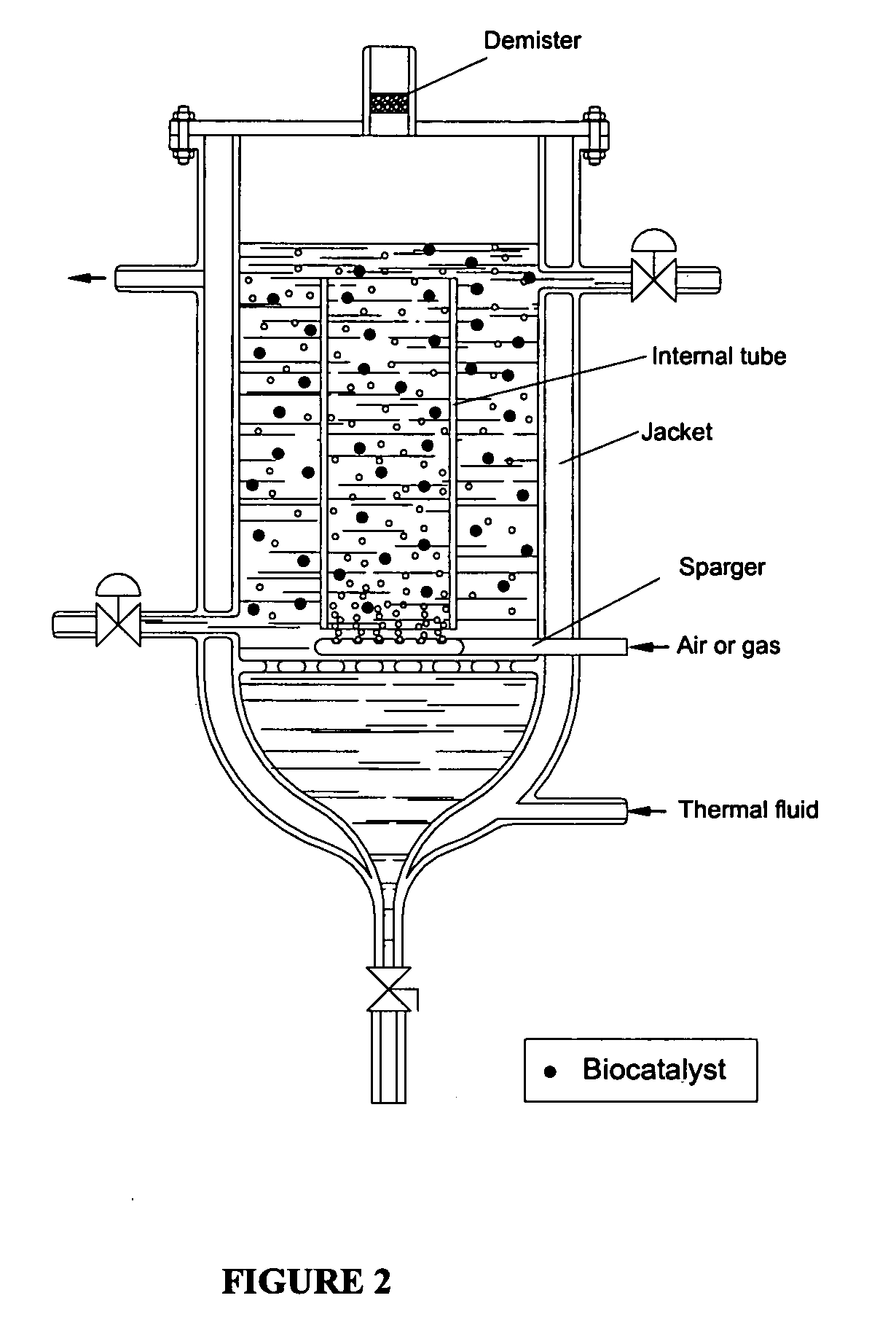
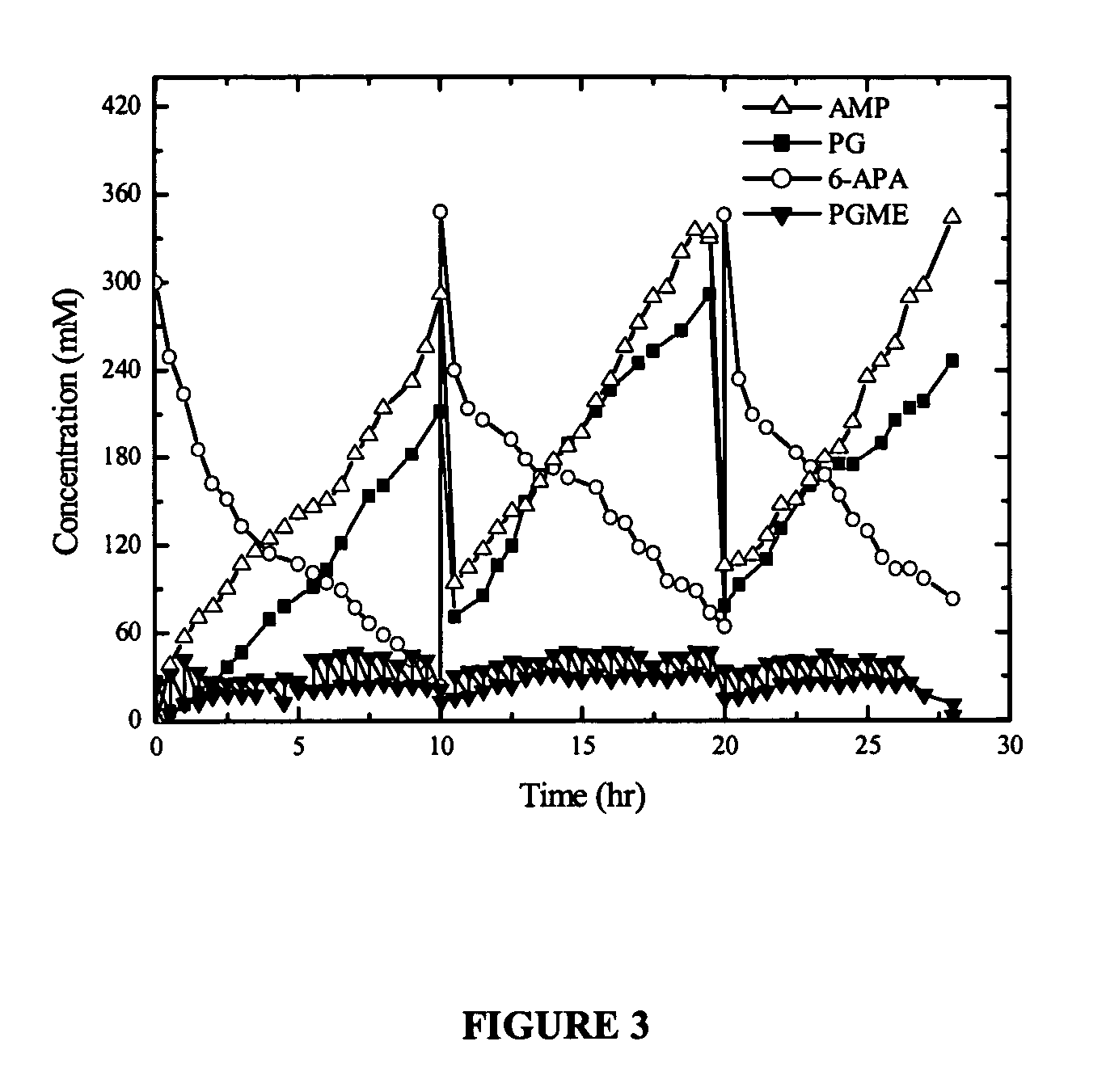
Process for protection of insoluble enzymatic biiocatalysts, biocatalyst obtained thereof and bioreactor with the immobilized biocatalyst
InactiveUS20060127971A1Easy to separateBioreactor/fermenter combinationsBiological substance pretreatmentsEnzymatic synthesisAntibiotic Y
The purpose of the present invention is to provide a process for protection of enzymatic biocatalysts, constituted by a primary support, wrapped by a secondary matrix. More particularly, the invention is embodied as a bioreactor with a configuration that preserves the physical integrity and catalytic activity of the biocatalyst after long runs, and at the same time allows an easy separation of the protected biocatalyst from the solid, precipitated during the reaction course. Still more particularly, the invention refers to the use of the bioreactor herein described for the enzymatic synthesis of β-lactam antibiotics, including the insoluble enzymatic catalysts, which have a primary matrix for immobilization involved by a secondary matrix, which is able to preserve 100% of the catalyst physical integrity and approximately 100% of its catalytic activity.
Owner:UNIVE FEDERAL DE SAO CARLOS +1
Eggplant flavanone 3-hydroxylase SmF3H as well as gene and application thereof
InactiveCN105087508AHigh in anthocyaninsAnthocyanin content noOxidoreductasesFermentationNucleotideArabidopsis thaliana
The invention discloses eggplant flavanone 3-hydroxylase SmF3H as well as a gene and application thereof; nucleotide sequences of cDNA and gDNA of the gene are respectively as shown in SEQ ID NO.1 and SEQ ID NO.2; and the amino sequence coded by the gene is as shown in SEQ ID NO.3. The nucleotide sequence of the promoter sequence of the gene is as shown in SEQ ID NO.4. The gene is expressed in root, stem, leaf, flower, peel and pulp, and the expression amount in the stem is significantly higher than that in other tissues. Under low-temperature stress, the expression amount achieves a maximum value at 12thh, which is 2.34 times as much as that unprocessed. An over-expression vector of the eggplant SmF3H is constructed and is converted into arabidopsis thaliana, discovering that anthocyanin contents in stem and fruit pod are significantly improved. The invention provides theoretical basis for improving plant quality by virtue of gene engineering technology in the future so as to obtain highly anti-oxidative medicines or food; therefore, the eggplant flavanone 3-hydroxylase SmF3H is quite high in application value.
Owner:SHANGHAI JIAO TONG UNIV
Brown cotton fiber procyanidine transfer-related GST protein gene and application thereof
The invention discloses brown cotton fiber procyanidine transfer-related GST protein gene and application thereof. The gene has a nucleotide sequence shown as SEQ ID NO.1 and an amino acid sequence shown as SEQ ID NO.2. According to the invention, the gene is found to be capable of controlling a formation of fiber pigment and improving plant color and luster through gene coded sequence analysis and genetic transformation of Arabidopsis thaliana; thus, reference basis is provided for utilizing a gene engineering technology to genetically improve brown cotton color characters, and a novel path is provided for cultivating a new brown cotton variety with stable fiber pigment.
Owner:ANHUI AGRICULTURAL UNIVERSITY
Analysis method for researching formation of variegation of nelumbo nucifera based on combination of transcriptomics with proteomics TMT
PendingCN110331225ADecor optimizationQuantitative and qualitativeMicrobiological testing/measurementBiological material analysisBiologyGene association
The invention relates to an analysis method for researching formation of variegation of nelumbo nucifera based on combination of transcriptomics with proteomics TMT, and provides a new method for disclosing a molecular mechanism of the formation of the variegation of plants. Samples of "Dasajin" nelumbo nucifera petals are collected, through transcriptome Illumina HiSeq sequencing, expression difference genes on an anthocyan metabolic pathway are screened, besides, proteins in the samples are extracted, the concentration of the proteins is determined, the quality is detected, through TMT labelmarking and 2D-LC-MS analysis, bioinformatics analysis is performed through combining with the proteomics, the proteins for adjusting and controlling expressing level differences on the anthocyan metabolic pathway are screened, the genes having differences in the transcriptome are related into the proteomics, and key proteins and corresponding genes for adjusting and controlling the variegation of "Dasajin" nelumbo nucifera are screened, so that the purpose of understanding a manner of the proteins and the genes for adjusting the formation of the variegation of the "Dasajin" nelumbo nuciferais achieved, and a new thinking and a new method are provided for the formation mechanism of the variegation of other plants.
Owner:CHINA THREE GORGES CORPORATION
Complete sequence of anthocyanidin reductase gene from Rubus ideaus L. and preparation method thereof
InactiveCN107312783AHigh proanthocyanidin contentMicrobiological testing/measurementOxidoreductasesOpen reading frameComplete sequence
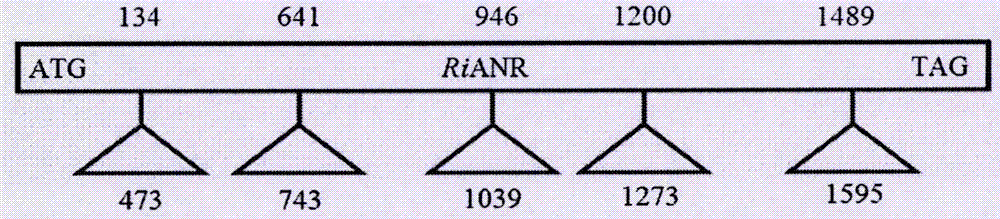
The invention discloses a complete sequence of an anthocyanidin reductase gene from Rubus ideaus L. and a preparation method thereof. According to the invention, Rubus ideaus L. is used as a raw material, a method of combined RT-PCR technology and RACE technology is employed, and the full length cDNA of the anthocyanidin reductase gene (RiANR) from Rubus ideaus L. is cloned; the cDNA comprises an open reading frame with a size of 1020 bp and codes a protein composed of 339 amino acids and having a theoretical isoelectric point of 5.81 and a theoretical molecular weight of 36413.8 D, which are identical to the results of prokaryotic expression SDS-PAGE electrophoresis detection. Cloning and expression analysis are carried out on the anthocyanidin reductase RiANR gene from Rubus ideaus L. in the invention, so a base material is provided for further exploration of the regulation and control mechanism of proanthocyanidin from Rubus ideaus L., and important gene resources are provided for acquiring novel crop varieties with high proanthocyanidin content through genetic improvement.
Owner:YUNCHENG UNIVERISTY
Substituted 3-amino-thieno[2,3-b]pyridine-2-carboxylic acid amide compounds and processes for preparing and their uses
Disclosed are compounds of formula (I): wherein R1 and R2 are defined herein, which are useful as inhibitors of the kinase activity of the IκB kinase (IKK) complex. The compounds are therefore useful in the treatment of IKK mediated diseases including autoimmune diseases inflammatory diseases and cancer. Also disclosed are pharmaceutical compositions comprising these compounds and processes for preparing these compounds.
Owner:BOEHRINGER INGELHEIM PHARMA INC
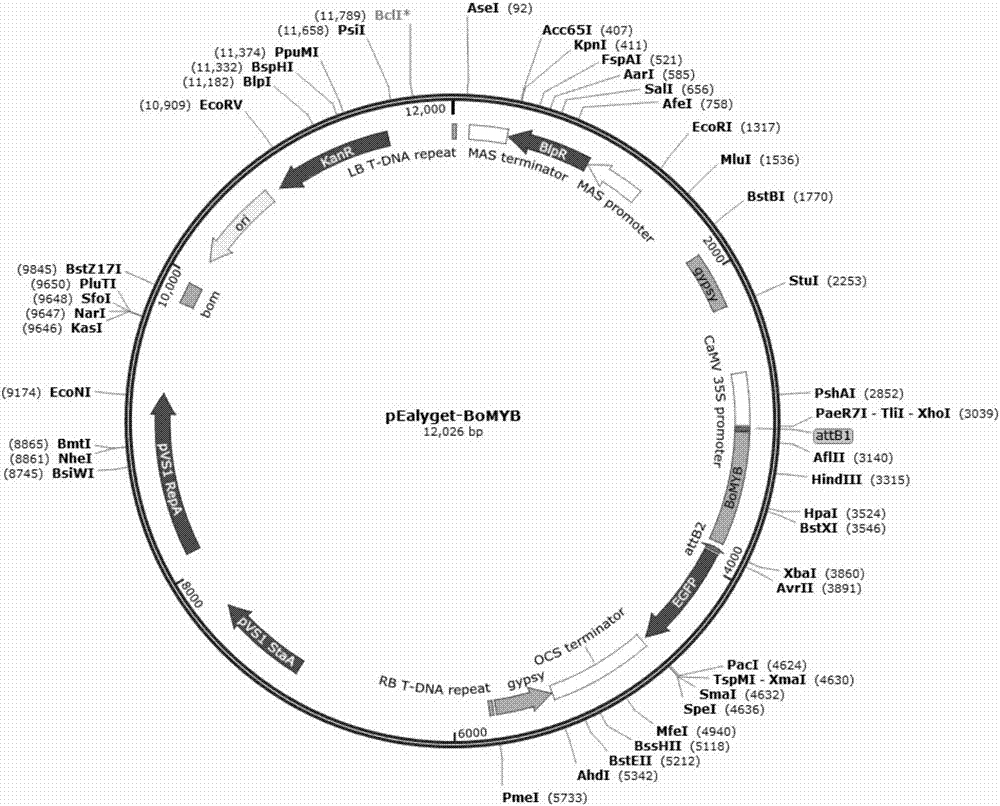
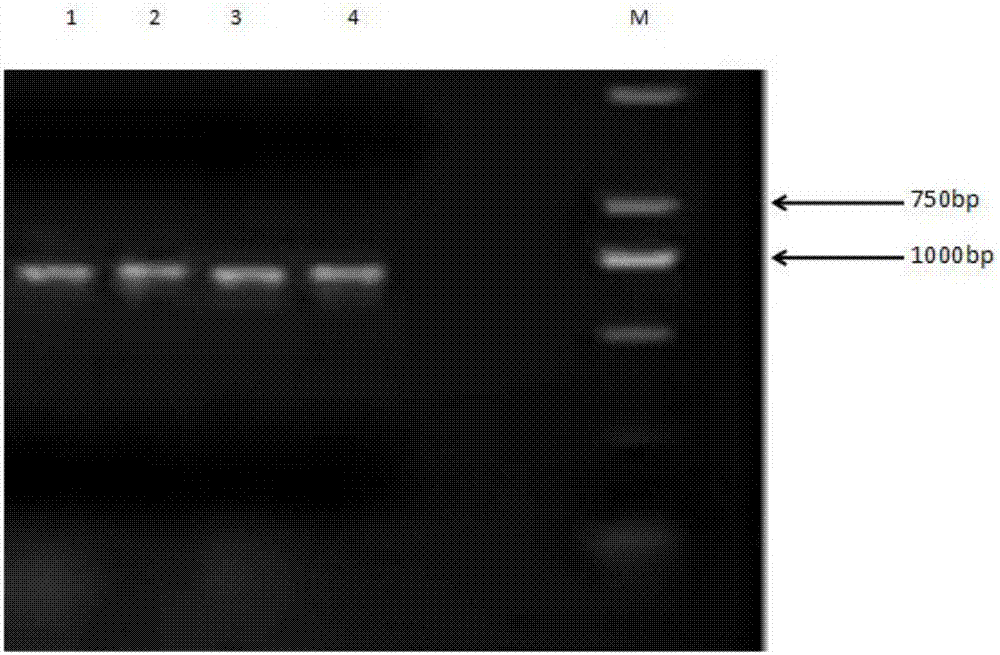
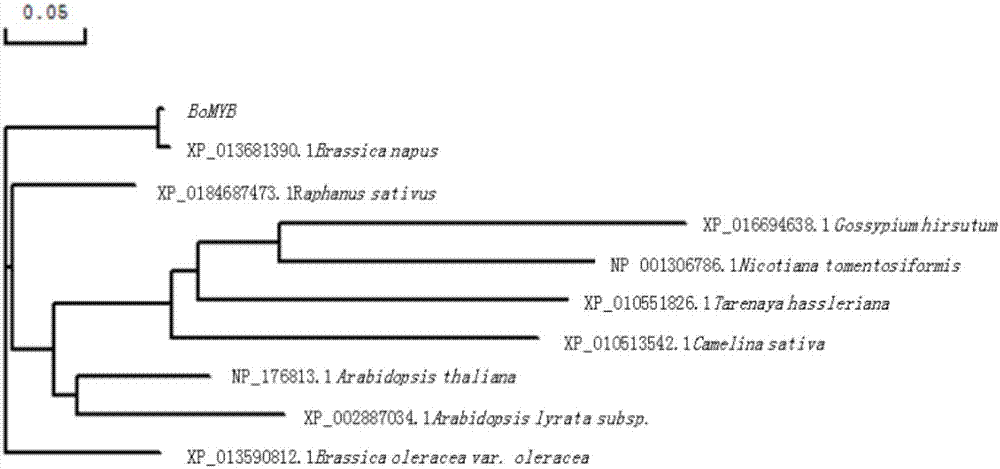
Ornamental collard anthocyanin relevant R2R3-MYB gene BoMYB and expression vector and application thereof
The invention discloses an ornamental collard anthocyanin relevant R2R3-MYB gene BoMYB and an expression vector and application thereof. The cDNA sequence of the BoMYB gene is shown as SEQ ID NO.1. The BoMYB gene can be used for regulating and controlling the biosynthesis of the anthocyanin. The collard anthocyanin purple leaf property regulating and control gene BoMYB is obtained by a homologouscloning method; a real-time fluorescent quantitative PCR method is used for studying the expression mode of the BoMYB gene in different tissues; analysis results show that the tissue specificity is realized in the BoMYB expression; the gene expression quantity is relevant to the anthocyanin content in each tissue. The heterologous expression verification in arabidopsis proves that the accumulationof the anthocyanin in the transgenic arabidopsis plants can be increased through the BoMYB overexpression.
Owner:QIQIHAR UNIVERSITY
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com









![Substituted 3-amino-thieno[2,3-b]pyridine-2-carboxylic acid amide compounds and processes for preparing and their uses Substituted 3-amino-thieno[2,3-b]pyridine-2-carboxylic acid amide compounds and processes for preparing and their uses](https://images-eureka.patsnap.com/patent_img/d3764cbf-0ed1-4030-903e-b63516843403/US20050288285A1-20051229-C00001.png)
![Substituted 3-amino-thieno[2,3-b]pyridine-2-carboxylic acid amide compounds and processes for preparing and their uses Substituted 3-amino-thieno[2,3-b]pyridine-2-carboxylic acid amide compounds and processes for preparing and their uses](https://images-eureka.patsnap.com/patent_img/d3764cbf-0ed1-4030-903e-b63516843403/US20050288285A1-20051229-C00002.png)
![Substituted 3-amino-thieno[2,3-b]pyridine-2-carboxylic acid amide compounds and processes for preparing and their uses Substituted 3-amino-thieno[2,3-b]pyridine-2-carboxylic acid amide compounds and processes for preparing and their uses](https://images-eureka.patsnap.com/patent_img/d3764cbf-0ed1-4030-903e-b63516843403/US20050288285A1-20051229-C00003.png)